Abstract
Pathogenicity islands (PAIs) are genetic elements whose products are essential to the process of disease development. They have been horizontally (laterally) transferred from other microbes and are important in evolution of pathogenesis. In this study, a comprehensive database and search engines specialized for PAIs were established. The pathogenicity island database (PAIDB) is a comprehensive relational database of all the reported PAIs and potential PAI regions which were predicted by a method that combines feature-based analysis and similarity-based analysis. Also, using the PAI Finder search application, a multi-sequence query can be analyzed onsite for the presence of potential PAIs. As of April 2006, PAIDB contains 112 types of PAIs and 889 GenBank accessions containing either partial or all PAI loci previously reported in the literature, which are present in 497 strains of pathogenic bacteria. The database also offers 310 candidate PAIs predicted from 118 sequenced prokaryotic genomes. With the increasing number of prokaryotic genomes without functional inference and sequenced genetic regions of suspected involvement in diseases, this web-based, user-friendly resource has the potential to be of significant use in pathogenomics. PAIDB is freely accessible at http://www.gem.re.kr/paidb.
INTRODUCTION
Pathogenicity islands (PAIs) are a subset of horizontally-acquired genomic islands (GIs) that are present in various microbial pathogens, and contain virulence-associated genes (1,2). Bacterial pathogenicity/virulence determinants that can be found in PAIs include the type III secretion system (e.g. LEE PAI in pathogenic Escherichia coli and Hrp PAI in Pseudomonas syringae), superantigen (e.g. SaPI1 and SaPI2 in Staphylococcus aureus), colonization factor (e.g. VPI in Vibrio cholerae), iron uptake system (e.g. SHI-2 in Shigella flexneri) and enterotoxin (e.g. espC PAI in E.coli and she PAI in S.flexneri). Widespread presence of PAIs in pathogens is due to their efficient mechanisms of horizontal transfer (3). Although PAIs are loosely defined entities, many of them can be identified by features such as the presence of virulence genes, biased G+C content and codon usage and association with tRNA genes, mobile sequence elements or repeated sequences at their boundaries (4,5).
Acquisition of PAIs by horizontal gene transfer (HGT) is an important mechanism in the development of disease-causing capability and the evolution of bacterial pathogenesis (6). Most of the computational methods for identification of PAIs in microbial genomes are based solely on the detection of putative GIs, which are compositionally different from the rest of the genome in their base composition and codon usage (7–9). Some of these predictions could be wrong, because they often result in GIs that do not contain pathogenicity/virulence genes, rather than PAIs (1). In this regard, a complementary approach involving detection of potential pathogenicity/virulence genes by homology searches is required. A computational method for identifying PAIs in sequenced prokaryotic genomes by combining a homology-based method, and detection of anomalies in genomic composition has been previously developed (10). The method detected 23 out of 27 PAIs in 17 strains which are closely related to the hosts carrying queried PAI loci.
Infectious diseases of animals, plants and humans caused by bacterial pathogens are a major challenge in global public health care. Rapid spread of novel pathogens and highly virulent strains demands a new approach for developing antimicrobial agents (11). This necessity prompted the trend of genome-wide study of microbial pathogenicity, called pathogenomics (6,12,13). A comprehensive database for virulence factors of pathogens would be pivotal in the studies of pathogenomics. Until now, online database servers have been constructed to detect horizontally transferred genes (14), GIs (15,16), insertion sequences (17), or mobile genetic elements (18). Recently, VFDB, a database for bacterial virulence factors, was constructed for bacterial pathogens of medical importance (19). In the current study, PAIDB is reported which is dedicated to provide comprehensive information on all known PAIs and potential PAI regions in prokaryotic genomes. An automatic identification system was also constructed for predicting potential PAI regions in query sequences.
METHODS
Definition of terms
In this study, a ‘PAI-like region’ is a predicted genomic region that is homologous to known PAI(s) and contains at least one homolog of the pathogenicity/virulence genes on the PAI loci. If a PAI-like region overlaps GI(s), it is considered to be a ‘candidate PAI (cPAI)’ (10). Many of the PAIs, such as Hrp PAI and LIPI-1, have DNA compositions similar to the core genomes, because they are believed to be introduced to the host genome long ago or transferred from a phylogenetically close strain (20). In the current detection scheme, elements of such characteristics can be included in the category of PAI-like regions which do not overlap GI(s), and are designated as ‘non-probable PAIs (nPAIs)’ to distinguish them from cPAIs.
Data collection
Sequence files of complete prokaryotic genomes were downloaded from the NCBI FTP server (ftp://ftp.ncbi.nih.gov). To collect GenBank accessions of the PAI locus, the GenBank database and literature (4,21) were searched for the words ‘PAI’ or ‘genomic island’ in their description or text. We also added PAIs that were reported in genome sequencing papers in a GenBank-like flat file format. They were extracted from the original genome files. Exhaustive literature surveys were carried out to identify pathogenicity/virulence genes contained in each of the PAI loci (10). Virulence factors denoted in VFDB (19) were also reviewed. A PAI was considered a genetic element incorporated into the chromosome by HGT and encoding more than one virulence factor (5). In this regard, resistance islands that did not contain virulence genes such as SCCmec of S.aureus (22) and plasmid- or phage-encoded virulence gene clusters such as CTX prophage of V.cholerae (23) were excluded.
Identification of genomic regions homologous to PAIs
Methods for detecting GIs, PAI-like regions and cPAIs have been described in earlier work (10). In each of the genome sequences, homologs of each open reading frame(s) [ORF(s)], RNA gene(s) and repeat region(s) of all the PAI loci were searched at the nucleotide level and then at the amino acid level using BLAT (24) and BLASTP (25), respectively. Genomic strips corresponding to each PAI locus were obtained by identifying regions containing four or more homologs of the genes from the same PAI accession, and by merging the neighboring regions. Overlapping or adjacent genomic strips corresponding to the same or different kind of PAI loci were fused into a large region. Among these regions, PAI-like regions were identified by checking the presence of at least one gene homologous to a virulence gene on the PAI loci. Likewise, genomic regions containing four or more potentially foreign genes in a 10-gene window were identified, and subsequently merged into a GI. A gene was considered as a foreign gene if its G+C content (>1.5 σ) and codon usage (P-value < 0.05) were both aberrant (10). The method was used to predict GIs in the genome of Hahella chejuensis (26). Finally, a cPAI was considered only if the PAI-like region partly or entirely spanned GI(s), and nPAI was a PAI-like region that did not span GI(s).
RESULTS
PAIDB is implemented in a MySQL relational database and is freely accessible at http://www.gem.re.kr/paidb/. The basic functionalities of the PAIDB are to ‘Browse’ the stored data and to ‘Search’ the database with a user-chosen input. Another feature is an application program for predicting PAI-like regions with the sequence of a user's interest.
DB contents
PAIDB contains 112 kinds of PAIs and associated 889 GenBank accessions including 87 PAIs from sequenced genomes (Table 1). They are either part or all of the reported PAI loci from 497 strains of pathogenic bacteria. The 293 complete prokaryotic genomes available at GenBank as of January 2006 were searched using the above algorithm, producing 546 PAI-like regions. Among them, 310 cPAIs were detected in 81 pathogenic and 37 non-pathogenic bacterial strains (Tables 2 and 3).
Table 1.
Statistics of PAI loci and related genes in PAIDB (as of April 2006)
| Pathogen (number of strains)a | Number | |||
|---|---|---|---|---|
| Kinds of PAIs | GenBank accessions | Virulence genes | ORFs | |
| Bacteroides fragilis (8) | 2 | 22 | 2 | 30 |
| Bartonella tribocorum | 1 | 1 | 0 | 35 |
| Citrobacter rodentium | 1 | 1 | 25 | 42 |
| Clostridium difficile | 1 | 1 | 5 | 12 |
| Dichelobacter nodosus | 2 | 4 | 31 | 57 |
| Enterococcus (6) | 2 | 9 | 16 | 270 |
| Erwinia amylovora | 1 | 8 | 30 | 93 |
| Escherichia coli (48) | 26 | 76 | 338 | 1507 |
| Francisella tularensis (4) | 1 | 5 | 4 | 79 |
| Helicobacter (284) | 2 | 478 | 868 | 1063 |
| Listeria (5) | 3 | 24 | 37 | 151 |
| Neisseria (10) | 5 | 14 | 9 | 184 |
| Photorhabdus luminescens | 5 | 5 | 34 | 191 |
| Porphyromonas gingivalis | 1 | 1 | 0 | 5 |
| Pseudomonas (25) | 10 | 38 | 131 | 813 |
| Salmonella (32) | 17 | 70 | 505 | 1194 |
| Shigella (11) | 6 | 16 | 57 | 327 |
| Staphylococcus (14) | 16 | 39 | 188 | 954 |
| Streptococcus pneumoniae | 1 | 1 | 3 | 35 |
| Streptomyces turgidiscabies | 1 | 5 | 11 | 34 |
| Vibrio cholerae (20) | 4 | 38 | 105 | 233 |
| Xanthomonas (9) | 1 | 11 | 207 | 252 |
| Yersinia (12) | 3 | 22 | 75 | 281 |
| Total (497) | 112 | 889 | 2681 | 7842 |
aNumber of strains (>1) that belong to the genus.
Table 2.
Statistics of pathogenic bacteria containing at least one PAI-like region (as of April 2006)
| Strain (number of strains)a | Number | ||
|---|---|---|---|
| PAI-like regionb | cPAIc | nPAId | |
| Acinetobacter sp. ADP1 | 4 | 3 | 1 |
| Agrobacterium tumefaciens (2) | 17 | 0 | 17 |
| Bacillus (7) | 10 | 3 | 7 |
| Bacteroides fragilis | 1 | 1 | 0 |
| Bartonella (2) | 4 | 0 | 4 |
| Bdellovibrio bacteriovorus | 4 | 1 | 3 |
| Bordetella (3) | 19 | 6 | 13 |
| Borrelia (2) | 2 | 0 | 2 |
| Brucella (4) | 10 | 1 | 9 |
| Burkholderia (4) | 41 | 13 | 28 |
| Campylobacter jejuni (2) | 3 | 1 | 2 |
| Chromobacterium violaceum | 4 | 2 | 2 |
| Clostridium perfringens | 2 | 0 | 2 |
| Corynebacterium (2) | 3 | 0 | 3 |
| Enterococcus faecalis | 3 | 1 | 2 |
| Erwinia carotovora | 16 | 4 | 12 |
| Escherichia coli (3) | 60 | 23 | 37 |
| Francisella tularensis | 3 | 2 | 1 |
| Fusobacterium nucleatum | 1 | 0 | 1 |
| Haemophilus (3) | 6 | 2 | 4 |
| Helicobacter (3) | 3 | 3 | 0 |
| Legionella pneumophila (3) | 5 | 0 | 5 |
| Leifsonia xyli | 1 | 0 | 1 |
| Leptospira interrogans (2) | 4 | 0 | 4 |
| Listeria monocytogenes (2) | 10 | 1 | 9 |
| Mycobacterium (5) | 6 | 1 | 5 |
| Neisseria meningitidis (2) | 4 | 3 | 1 |
| Nocardia farcinica | 2 | 0 | 2 |
| Pasteurella multocida | 6 | 2 | 4 |
| Photorhabdus luminescens | 16 | 7 | 9 |
| Propionibacterium acnes | 1 | 0 | 1 |
| Pseudomonas (4) | 46 | 7 | 39 |
| Ralstonia solanacearum | 5 | 2 | 3 |
| Salmonella (5) | 95 | 59 | 36 |
| Shigella (5) | 89 | 28 | 61 |
| Staphylococcus (12) | 71 | 35 | 36 |
| Streptococcus (5) | 14 | 3 | 11 |
| Treponema (2) | 5 | 3 | 2 |
| Tropheryma whipplei (2) | 2 | 0 | 2 |
| Vibrio (4) | 28 | 6 | 22 |
| Xanthomonas (5) | 27 | 6 | 21 |
| Xylella fastidiosa | 2 | 0 | 2 |
| Yersinia (4) | 88 | 28 | 60 |
| Total (115) | 743 | 257 | 486 |
aNumber of strains (>1) that belong to the genus.
bGenomic region that is homologous to known PAI(s) and contains at least one homolog of the pathogenicity/virulence gene on the PAI loci.
cCandidate PAI that is a PAI-like region overlapping genomic island(s).
dNon-probable PAI that is a PAI-like region not overlapping a genomic island.
Table 3.
List of non-pathogenic prokaryotes or those with unconfirmed pathogenicity containing at least one PAI-like region (as of April 2006)
| Strain (number of strains) | Number | ||
|---|---|---|---|
| PAI-like region | cPAI | nPAI | |
| Anabaena variabilis | 1 | 0 | 1 |
| Azoarcus sp. EbN1 | 7 | 1 | 6 |
| Bacillus (5) | 29 | 5 | 24 |
| Bradyrhizobium japonicuma | 4 | 1 | 3 |
| Burkholderia thailandensis | 13 | 3 | 10 |
| Carboxydothermus hydrogenoformans | 1 | 1 | 0 |
| Caulobacter crescentus | 3 | 0 | 3 |
| Clostridium acetobutylicum | 2 | 0 | 2 |
| Colwellia psychrerythraea | 5 | 2 | 3 |
| Corynebacterium (3) | 3 | 0 | 3 |
| Dechloromonas aromatica | 3 | 0 | 3 |
| Deinococcus radiodurans | 2 | 0 | 2 |
| Desulfotalea psychrophila | 1 | 0 | 1 |
| Desulfovibrio (2) | 4 | 3 | 1 |
| Escherichia coli | 15 | 2 | 13 |
| Geobacillus kaustophilus | 5 | 2 | 3 |
| Geobacter (2) | 5 | 1 | 4 |
| Gloeobacter violaceus | 2 | 0 | 2 |
| Gluconobacter oxydans | 1 | 1 | 0 |
| Hahella chejuensisa | 7 | 2 | 5 |
| Halobacterium salinarum | 1 | 0 | 1 |
| Idiomarina loihiensis | 5 | 1 | 4 |
| Lactobacillus (3) | 6 | 2 | 4 |
| Lactococcus lactis | 1 | 1 | 0 |
| Listeria innocua | 3 | 1 | 2 |
| Magnetospirillum magneticum | 3 | 1 | 2 |
| Mannheimia succiniciproducens | 1 | 0 | 1 |
| Mesorhizobium lotia | 4 | 1 | 3 |
| Methanosarcina (2) | 2 | 0 | 2 |
| Methylococcus capsulatus | 1 | 0 | 1 |
| Moorella thermoacetica | 1 | 0 | 1 |
| Nitrobacter winogradskyi | 1 | 0 | 1 |
| Nitrosococcus oceani | 4 | 3 | 1 |
| Nitrosomonas europaea | 4 | 1 | 3 |
| Nitrosospira multiformis | 2 | 0 | 2 |
| Nostoc sp. | 2 | 1 | 1 |
| Oceanobacillus iheyensis | 6 | 1 | 5 |
| Pelagibacter ubique | 1 | 0 | 1 |
| Pelobacter carbinolicus | 2 | 1 | 1 |
| Pelodictyon luteolum | 1 | 0 | 1 |
| Photobacterium profundum | 8 | 0 | 8 |
| Pseudoalteromonas haloplanktisa | 3 | 0 | 3 |
| Pseudomonas (3) | 25 | 8 | 17 |
| Psychrobacter arcticus | 1 | 0 | 1 |
| Ralstonia eutropha | 7 | 2 | 5 |
| Rhodobacter sphaeroides | 5 | 0 | 5 |
| Rhodopirellula baltica | 1 | 0 | 1 |
| Rhodopseudomonas palustris | 3 | 0 | 3 |
| Rhodospirillum rubrum | 3 | 0 | 3 |
| Salinibacter ruber | 1 | 0 | 1 |
| Shewanella oneidensis | 4 | 0 | 4 |
| Sinorhizobium melilotia | 8 | 2 | 6 |
| Streptomyces (2) | 3 | 0 | 3 |
| Sulfolobus tokodaii | 1 | 0 | 1 |
| Symbiobacterium thermophiluma | 3 | 1 | 2 |
| Synechococcus (2) | 3 | 1 | 2 |
| Synechocystis sp. PCC6803 | 2 | 0 | 2 |
| Thermoanaerobacter tengcongensis | 1 | 0 | 1 |
| Thermobifida fusca | 2 | 0 | 2 |
| Thiobacillus denitrificans | 1 | 0 | 1 |
| Thiomicrospira crunogena | 1 | 0 | 1 |
| Vibrio fischeria | 9 | 1 | 8 |
| Total (77) | 259 | 53 | 206 |
Descriptions of the titles are the same as Table2.
aStrains that interact with eukaryotic organisms.
Browse
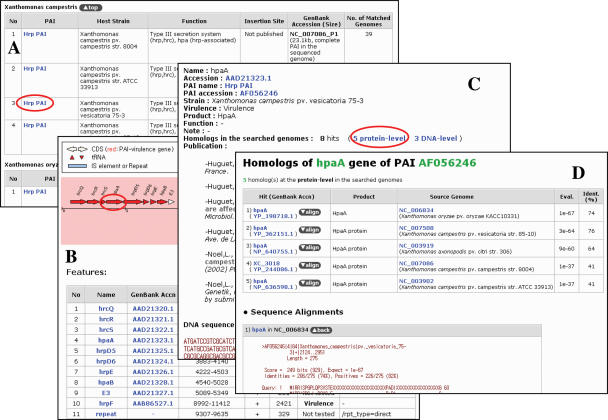
Web pages in the PAIs and Genomes are organized to offer a user-friendly graphic interface with clear visualization of PAIs and computationally-predicted PAI-like regions. The PAIs menu provides a general description on deposited PAIs such as name, host strain, function, insertion site, associated GenBank accessions and number of matched genomes in tabular formats ordered by their host strains (Figure 1). Each PAI name is hyperlinked to the page that shows information on each of the associated accessions and its linear map. Genes and indications of GIs such as tRNA, IS element and repeat region can be clicked to show a description such as existence of virulence, function, references and DNA/protein sequence. If a user wants to know the homologs of a PAI gene at the DNA and protein levels, BLAST scores and sequence alignments of all the homologs found in the searched genomes can be reported.
Figure 1.
A screenshot of PAIs menu. (A) Main page showing a list of PAIs. (B) PAI page for detailed information on each PAI. (C) PAI Gene Information page. (D) Aligned BLAST search of the selected gene against PAI genes. Items clicked on each page for next page were marked in red circles.
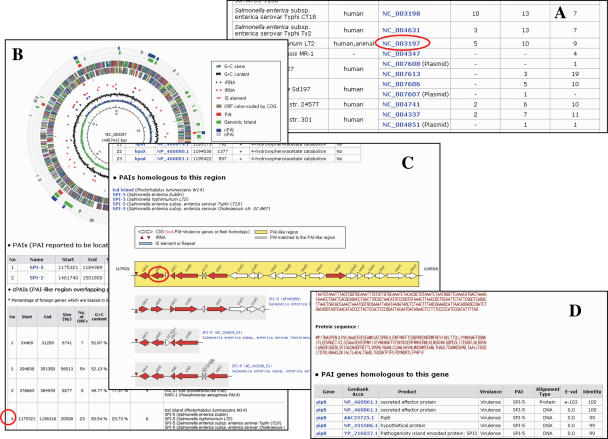
The Genomes menu begins with a list of genomes and a summary of PAIs and PAI-like regions (Figure 2). The Genome Information page for a genome accession shows a circular genome map in which distribution of PAIs, PAI-like regions and GIs. Following is an in-depth description of PAI-like regions such as location, G+C content, percentage of foreign genes, number of homologs of PAI-encoded virulence genes and PAIs homologous to the region. Each of the PAI-like regions is linked to the PAI-like Region page that shows information on basic features located in the region such as name, GenBank accession, position, product, and putative virulence. Putative virulence of a gene is classified according to the results of a BLAST search against PAI-encoded virulence genes. The PAI-like Region page also gives graphical representation of the region and fragments of PAIs matching the region. Clicking the gene name leads to the Gene Information page, which shows details on which of the PAI genes are homologous to the gene.
Figure 2.
A screenshot of Genomes menu. (A) Main page showing a list of genomes containing at least one PAI-like region. (B) Genome Information page for the circular map, PAIs and PAI-like regions of the selected genome. (C) PAI-like Region page shows a PAI-like region (yellow strip) homologous to PAIs (gray strips). Red arrows denote PAI-encoded virulence genes or their homologs. (D) Gene information page of a gene in the PAI-like region. PAI genes homologous to the selected gene are shown or illustrated. Items clicked on each page for next page were marked in red circles.
Search
Search tools provide information on PAI data deposited in PAIDB through text or BLAST searches. Retrieved genes are displayed in a table with name, product, function, host strain and the associated PAI, and users can browse details. Following the table is the option for carrying out a multiple sequence alignment of selected genes by ClustalW (27) and construction of their phylogenetic tree.
PAI Finder
An on-the-fly analysis tool was implemented as a Perl script to predict potential PAI regions in query sequences. Basically, the detection algorithm was set at the gene level (10), and thus, the program takes a multi-sequence query in a FASTA format containing a series of DNA sequences in their original order. It should be noted that the results are genomic regions homologous to PAIs rather than cPAIs (PAI-like regions overlapping GIs), because prediction of GIs requires average G+C content and codon usage at the genome-scale.
With a sequence query, the program finds which PAIs are homologous and which region is likely to be a PAI. The resulting page shows a summary of the potential PAI region that has information on location, number of homologs of PAI-encoded virulence genes and PAIs matched to the region. PAI Finder reports all the regions homologous to the PAIs in our database irrespective of their putative virulence. Thus, users are advised to check the existence of a homolog of PAI-encoded virulence genes to know whether the query sequence(s) contain PAI-like region(s) from the Features table. Each of the regions is linked to the PAI-like Region page showing the putative virulence of each ORF and the PAIs matching the region as described in Browse under RESULTs section. Likewise, a series of tracks of a query gene or a PAI gene leads to detailed information, and a user can infer the function(s) and origin(s) of potential PAIs in their own sequences.
DISCUSSION
Pathogenomics, a genome-scale study of pathogenicity triggered by genomics and bioinformatics, requires comprehensive information on various virulence factors and genes. Identification of PAIs in microbial genomes is essential for understanding the nature of infectious diseases and the evolution of pathogenesis (6). PAIDB provides comprehensive information on PAIs, which are reservoirs of virulence genes, along with potential PAIs in sequenced prokaryotic genomes. PAI Finder can give insights into the functions and origins of potential PAIs in the sequences of user interest, which could be a good starting point for the study of pathogenesis. Further, PAIDB will be useful in developing new antibiotics and vaccines and in designing clinical biosensors that can be used for pathogen detection and disease diagnostics.
Among the PAI-like regions, recently transferred PAIs may be preferentially included in the list of cPAIs. An anciently transferred PAI may have lost compositional abnormalities, because the nucleotide composition of the transferred genes is ameliorated to the host genome with a function of time (20). Thus, PAIs, which were introduced long ago or transferred from close relatives, often fall in the category of nPAIs. There is increasingly more information available indicating that genes considered as virulence-associated in certain strains can also be found in non-pathogenic relatives (28). Consequently, the definition of virulence genes is difficult as their function may depend on growth conditions and host niches. Genes that may contribute to competitiveness and fitness (e.g. adhesins, iron uptake systems, proteases) may, under certain conditions, also contribute to virulence. As shown in Table 3, many PAI-like regions found in non-pathogenic strains including symbionts are located in the seemingly core genomes. Genes carried by most of them are functionally related to iron uptake and flagellar biosynthesis (10). Therefore, the unexpected locations of PAI-like regions in non-pathogens should be interpreted as clusters of homologs of virulence genes. It should be noted that confirmation of the involvement of cPAIs in virulence requires biological experiments.
Due to the difficulty in assigning virulence features to a gene, a gene was considered a virulence gene only if it was experimentally validated or reported by literature. Efforts to find more virulence genes such as Virulence Searcher (29) could prove helpful in increasing the inventory of virulence genes in the database. The prediction power of the current method is highly dependent on the query data set of the known PAIs. The next version plans to include resistance islands and plasmid- or phage-encoded virulence gene clusters. As prokaryotic complete genomes and PAI data are rapidly accumulating, PAIDB will continue to be updated on a regular basis. The process of improving the algorithm to detect potential PAIs and PAI Finder has been initiated as well, and the results will be reflected in the later versions of the database. Corrections and comments are welcome and can be sent to jfk@kribb.re.kr.
Acknowledgments
The authors thank Ensoltek Co., Ltd (www.ensoltek.co.kr) for help in the development of the genome map viewer. The authors also thank Seung-Hwan Park, Haeyoung Jeong and Choong-Min Ryu for comments on the work and critical reading of the manuscript. This work was financially supported by the 21C Frontier Microbial Genomics and Applications Center Program, Ministry of Science and Technology, Republic of Korea. Funding to pay the Open Access publication charges for this article was provided by the same program.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schmidt H., Hensel M. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 2004;17:14–56. doi: 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakamura Y., Itoh T., Matsuda H., Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nature Genet. 2004;36:760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- 3.Dobrindt U., Hochhut B., Hentschel U., Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nature Rev. Microbiol. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 4.Hacker J., Kaper J.B. Pathogenicity Islands and The Evolution of Pathogenic Microbes. Berlin: Springer-Verlag; 2002. [Google Scholar]
- 5.Hacker J., Blum-Oehler G., Muhldorfer I., Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 6.Hacker J., Hochhut B., Middendorf B., Schneider G., Buchrieser C., Gottschalk G., Dobrindt U. Pathogenomics of mobile genetic elements of toxigenic bacteria. Int. J. Med. Microbiol. 2004;293:453–461. doi: 10.1078/1438-4221-00290. [DOI] [PubMed] [Google Scholar]
- 7.Karlin S. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 2001;9:335–343. doi: 10.1016/s0966-842x(01)02079-0. [DOI] [PubMed] [Google Scholar]
- 8.Lio P., Vannucci M. Finding pathogenicity islands and gene transfer events in genome data. Bioinformatics. 2000;16:932–940. doi: 10.1093/bioinformatics/16.10.932. [DOI] [PubMed] [Google Scholar]
- 9.Tu Q., Ding D. Detecting pathogenicity islands and anomalous gene clusters by iterative discriminant analysis. FEMS Microbiol. Lett. 2003;221:269–275. doi: 10.1016/S0378-1097(03)00204-0. [DOI] [PubMed] [Google Scholar]
- 10.Yoon S.H., Hur C.G., Kang H.Y., Kim Y.H., Oh T.K., Kim J.F. A computational approach for identifying pathogenicity islands in prokaryotic genomes. BMC Bioinformatics. 2005;6:184. doi: 10.1186/1471-2105-6-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser C.M., Rappuoli R. Application of microbial genomic science to advanced therapeutics. Annu. Rev. Med. 2005;56:459–474. doi: 10.1146/annurev.med.56.062904.144853. [DOI] [PubMed] [Google Scholar]
- 12.Pompe S., Simon J., Wiedemann P.M., Tannert C. Future trends and challenges in pathogenomics. A Foresight study. EMBO Rep. 2005;6:600–605. doi: 10.1038/sj.embor.7400472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crossman L., Cerdeno-Tarraga A., Bentley S., Parkhill J. Pathogenomics. Nature Rev. Microbiol. 2003;1:176–177. doi: 10.1038/nrmicro778. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Vallve S., Guzman E., Montero M.A., Romeu A. HGT-DB: a database of putative horizontally transferred genes in prokaryotic complete genomes. Nucleic Acids Res. 2003;31:187–189. doi: 10.1093/nar/gkg004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mantri Y., Williams K.P. Islander: a database of integrative islands in prokaryotic genomes, the associated integrases and their DNA site specificities. Nucleic Acids Res. 2004;32:D55–D58. doi: 10.1093/nar/gkh059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsiao W., Wan I., Jones S.J., Brinkman F.S. IslandPath: aiding detection of genomic islands in prokaryotes. Bioinformatics. 2003;19:418–420. doi: 10.1093/bioinformatics/btg004. [DOI] [PubMed] [Google Scholar]
- 17.Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leplae R., Hebrant A., Wodak S.J., Toussaint A. ACLAME: a CLAssification of Mobile genetic Elements. Nucleic Acids Res. 2004;32:D45–D49. doi: 10.1093/nar/gkh084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L., Yang J., Yu J., Yao Z., Sun L., Shen Y., Jin Q. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence J.G., Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 21.Kaper J.B., Hacker J. Pathogenicity Islands and Other Mobile Virulence Elements. Washington, DC: American Society for Microbiology Press; 1999. [Google Scholar]
- 22.Hiramatsu K., Cui L., Kuroda M., Ito T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2001;9:486–493. doi: 10.1016/s0966-842x(01)02175-8. [DOI] [PubMed] [Google Scholar]
- 23.Boyd E.F., Moyer K.E., Shi L., Waldor M.K. Infectious CTXφ and the vibrio pathogenicity island prophage in Vibrio mimicus: evidence for recent horizontal transfer between V.mimicus and V.cholerae. Infect. Immun. 2000;68:1507–1513. doi: 10.1128/iai.68.3.1507-1513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kent W.J. BLAT-the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeong H., Yim J.H., Lee C., Choi S.H., Park Y.K., Yoon S.H., Hur C.G., Kang H.Y., Kim D., Lee H.H., et al. Genomic blueprint of Hahella chejuensis, a marine microbe producing an algicidal agent. Nucleic Acids Res. 2005;33:7066–7073. doi: 10.1093/nar/gki1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holden M., Crossman L., Cerdeno-Tarraga A., Parkhill J. Pathogenomics of non-pathogens. Nat. Rev. Microbiol. 2004;2:91. doi: 10.1038/nrmicro825. [DOI] [PubMed] [Google Scholar]
- 29.Underwood A.P., Mulder A., Gharbia S., Green J. Virulence Searcher: a tool for searching raw genome sequences from bacterial genomes for putative virulence factors. Clin. Microbiol. Infect. 2005;11:770–772. doi: 10.1111/j.1469-0691.2005.01210.x. [DOI] [PubMed] [Google Scholar]