Abstract
Protein classification by machine learning algorithms is now widely used in structural and functional annotation of proteins. The Protein Classification Benchmark collection (http://hydra.icgeb.trieste.it/benchmark) was created in order to provide standard datasets on which the performance of machine learning methods can be compared. It is primarily meant for method developers and users interested in comparing methods under standardized conditions. The collection contains datasets of sequences and structures, and each set is subdivided into positive/negative, training/test sets in several ways. There is a total of 6405 classification tasks, 3297 on protein sequences, 3095 on protein structures and 10 on protein coding regions in DNA. Typical tasks include the classification of structural domains in the SCOP and CATH databases based on their sequences or structures, as well as various functional and taxonomic classification problems. In the case of hierarchical classification schemes, the classification tasks can be defined at various levels of the hierarchy (such as classes, folds, superfamilies, etc.). For each dataset there are distance matrices available that contain all vs. all comparison of the data, based on various sequence or structure comparison methods, as well as a set of classification performance measures computed with various classifier algorithms.
INTRODUCTION
Classification of proteins is a fundamental technique in computational genomics which is carried out, to a large extent, by automated machine learning methods (1). Application of machine learning techniques to proteins is a delicate task since the known protein groups—such as those of domain-types and protein families—are highly variable in most of their characteristics (e.g. average sequence length, number of known members, within-group similarity, etc.). A further problem is the complexity of the calculations, since a system capable of testing and comparing machine learning algorithms should include (i) datasets and classification tasks; (ii) sequence/structure comparison methods; (iii) classification algorithms; and (iv) a validation protocol.
Even though the application of machine learning algorithms to protein classification is a frequent topic in the literature, it is often quite difficult to compare the performance of a new classification method with the figures published on other methods. In our opinion this is mainly because (i) the published results are often based on different and sometimes by then obsolete databases and program versions, (ii) the fine-tuning of the program parameters is sometimes not described in sufficient detail and finally, (iii) the classification performance is characterized by various, often ad hoc chosen performance measures and validation protocols.
In order to get a reliable estimate of the performance, an algorithm needs to be tested on not only one, but many protein groups selected from a well-curated database. For instance, an algorithm may be efficient in classifying protein superfamilies into families, but less efficient in classifying folds into superfamilies. In other words, one can choose to conduct a test at different levels of a classification hierarchy, and within each of these levels one can define many different classification tasks. The choice of the test/train groups is also critical. It is well known that once a group of proteins has been identified, it is relatively easy to recognize new members of the group. On the other hand, each new genome may contain new subtypes of the already known groups (say new families within a known superfamily), which are often not recognized by the classification algorithms trained on the old examples. In other words, it is important to know how a given algorithm generalizes to novel subtypes. This ability can be estimated by a method that we term ‘knowledge based cross-validation’ by which we determine how the a priori known subtypes (e.g. protein families within a superfamily) can be recognized, based on other known subtypes (2–4).
In view of the above difficulties and the number of new genomes sequenced, it is critically important to define benchmark datasets for assessing the accuracy of classification algorithms. The goal of the Protein Classification Benchmark collection is to provide a standardized set of protein data and procedures that makes it easier to compare new methods with the established ones. The collection is based on two general ideas: (i) since protein groups are highly variable, the performance of an algorithm has to be tested on a wide range of classification tasks, such as the recognition of all the protein families in a given database; (ii) the utility of a classifier is determined by its ability to recognize novel subtypes of the existing proteins. The collection is primarily meant for those interested in developing sequence or structure comparison algorithms and/or machine learning methods for protein classification.
CLASSIFICATION TASKS AND BENCHMARK TESTS
A classification task is the subdivision of a dataset into +train, +test, −train and −test groups. Given such a subdivision, one can train a classifier and evaluate its performance. A benchmark test is a collection of several classification tasks defined on a given database. At present the collection contains 34 benchmark tests consisting of 10–490 classification tasks. There is a total of 6405 classification tasks, 3297 on protein sequences, 3095 on protein structures and 10 on protein coding regions in DNA. A typical test refers to the prediction of novel subtypes within protein superfamilies, folds or taxonomic groups, etc. As a comparison we have included benchmark tests that are based on random subdivision of the datasets according to a 5-fold cross-validation scheme. The benchmark tests were selected so as to represent various degrees of difficulty. For instance, the sequences in orthologous groups of the COG database (5) are closely related to each other within the group, while there are relatively weak similarities between the groups. On the other hand, protein families of SCOP (6) or homology groups of CATH (7) are less closely related to each other in terms of sequence similarity and the similarities between groups are also weak. Finally, sequences of the same protein in different organisms that can be divided into taxonomic groups represent a case where both the within-group and between-group similarities are high.
From the computational point of view, a classification task is described as a ‘cast-vector’ that assigns a membership code (+test, +train, −test, −train) to each entry in a given database. A benchmark test is an ensemble of such cast-vectors which is represented in the form of a ‘cast-matrix’ or membership table. In a cast-matrix each column vector represents a classification task. For each benchmark test a cast-matrix is deposited as a tab-delimited ASCII file, using a format described by Liao and Noble (2).
PROTEIN DATA
The collection contains datasets of protein sequences, 3D structures and in a few cases, reading frame DNA sequences of the same molecules. The sequences are deposited in concatenated FASTA format (http://www.ncbi.nlm.nih.gov/blast/fasta.shtml), the structures are in PDB format (http://www.rcsb.org/static.do?p=file_formats/pdb/index.html or http://www.pdb.org/).
PROTEIN COMPARISON DATA
Dataset versus dataset comparison data are deposited in the form of symmetrical distance matrices stored in the form of tab-delimited ASCII files. The methods include sequence comparisons such as BLAST (8), Smith–Waterman (9), Needleman–Wunsch (10), compression-based distances (11) and the local alignment kernel (12). The structure comparison algorithm included is PRIDE2 (13). These data can then be used directly in nearest neighbor classification schemes as well as for the training of kernel methods.
MACHINE LEARNING ALGORITHMS
Results are deposited for nearest neighbor (1NN), support vector machines (SVM) (14), artificial neural networks (ANN) (15), random forest (RF) (16) and logistic regression (LogReg) (17) learning algorithms. In general, the input of these algorithms is a feature vector whose parameters are comparison scores calculated between a protein of interest and the members of the training set.
PERFORMANCE MEASURES AND VALIDATION PROTOCOL
The primary evaluation protocol used in this database is standard receiver operator characteristic (ROC) analysis (18). This method is especially useful for protein classification as it includes both sensitivity and specificity, and it is based on a ranking of the objects to be classified (19). The ranking variable is a number, such as a BLAST score, or an output variable produced by a machine learning algorithm. For nearest neighbor classification, the ranking variable is the similarity/distance between a test example and the nearest member of the positive training set, which corresponds to one-class classification with outlier detection. For SVM, the distance from the separating hyperplane can be used as a ranking variable. The analysis is then carried out by plotting sensitivity versus 1−specificity at various threshold levels, and the resulting curve is integrated to give an ‘area under curve’ or AUC value. For perfect ranking, AUC = 1.0 and for random ranking AUC = 0.5 (18).
As a benchmark test contains several ROC experiments, one can draw a cumulative distribution curve of the AUC values. The integral of this cumulative curve, divided by the number of the classification experiments is in [0,1], the higher values represent the better classifier performances (2). Alternatively, the average AUC can be used as summary characteristics for a database, and this value is given for each benchmark test within the database.
BENCHMARK RESULTS AND PROGRAMS
Nearest neighbor performance data are deposited for all benchmark tests and all comparison methods. The program used for the calculation of the results was written in R (20) and its code is deposited at the database site. This program takes a cast-matrix and a distance matrix as the input, and carries out either 1NN classification. The program is downloadable from the site and is written in such a way that it can easily be modified for testing other classification algorithms. In addition, SVM, ANN, RF and LogReg results are deposited for a few other datasets. The results were produced with open source software written in JAVA (21) or in R.
DATABASE STRUCTURE
The database consists of records. Each record contains a benchmark test, which consists of several (10–490) classification tasks defined on a given database. Each record contains at least one distance matrix (an all versus all comparison of the dataset) as well as performance measures (typically ROC analysis results) for all the classification tasks for at least one classification algorithm. The bibliographic references and the details of the calculations are included in Table 1.
Table 1.
Examples of records (benchmark tests) included in the collection
| Benchmark testsa | Data | Classification tasks | Comparison methodsb |
|---|---|---|---|
| Classification of protein domains in SCOP [PCB0001, PCB00003, PDB0005] | 11 944 Protein sequences/or protein structures from SCOP95 (6) | Superfamilies subdivided into families………246 | BLAST, Smith–Waterman, Needleman–Wunsch, LA–kernel, PRIDE2 |
| Folds subdivided into superfamilies………191 | |||
| Classes subdivided into folds………377 | |||
| Classification of protein domains in CATH [PCB00007, PCB00009, PCB00011, PCB00013] | 11 373 Protein sequences/or protein structures from CATH (7) | (H) groups subdivided into S groups………165 | BLAST, Smith–Waterman, Needleman–Wunsch, LA–kernel, PRIDE2 |
| T groups subdivided into H groups………199 | |||
| A groups subdivided into T groups………297 | |||
| Classes subdivided into A groups………33 | |||
| CLassification of phyla based on 3 phospho-glycerate kinase (3PGK) sequences. [PCB00031, PCB00032] | 131 3PGK Protein and DNA sequences (11,29) | Groups of kingdoms (Archaea, Bacteria, Eucarya) subdivided into phyla……10 | BLAST, Smith–Waterman, Needleman–Wunsch, LA–kernel, LZW, PPMZ |
| Functional annotation of unicellular eukaryotic sequences based on prokaryotic orthologs. [PCB00031] | 17 973 Sequences of prokaryotes and unicellular eukaryotes from the COG databases (5) | Orthologous groups subdivided into prokaryotes and eukaryotes………119 | BLAST, Smith–Waterman, Needleman–Wunsch, LA–kernel, LZW, PPMZ |
aThe collection contains a total of 6405 benchmark tests including a total of 3297 protein sequence classification tests, 3095 3D classification tests and 10 DNA (coding region) classification tests. The accession numbers of the records are given in square brackets.
bSee text for the references.
AVAILABILITY
The database and a collection of documents and help files can be accessed at http://hydra.icgeb.trieste.it/benchmark/.
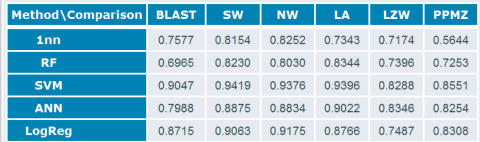
The records can be accessed directly from the homepage (Figure 1). Each record contains statistical data and a detailed description of the methodology used to produce the data and the analysis results. The results are shown as tables of AUC values obtained by ROC analysis (Figure 2) and several detailed table-views can be generated on-line in various formats.
Figure 1.
Details of a record in the database.
Figure 2.
Cumulative results of a benchmark test PCB00033. The underlying dataset is a small subset of SCOP comprising of 55 classification tasks (corresponding to 8 all-α, 15 all-β, 30 α/β and 2 other classes). The numbers represent average AUC values [0,1] obtained by receiver operator curve (ROC) analysis (18). This value is high for good classifiers and is close to 0.5 for random classification. The classification methods include 1NN—Nearest neighbor (30), RF—Random forest (16), SVM—Support Vector Machines (14), ANN—Artificial neural networks (15) and LogReg—Logistic regression (17). The comparison methods include BLAST (8), SW—Smith–Waterman (9), NW—Needleman–Wunsch (10), LZW—Lempel–Ziv compression distance and PPMZ—partial match compression distance (11). The Smith–Waterman algorithm performs better than the other comparison algorithms, especially when used in conjunction with SVM.
SUGGESTIONS FOR USE
The purpose of this collection is to provide benchmark datasets for the development of new protein classification algorithms. In order to benchmark a new comparison algorithm for sequences or structures, the user can download a dataset and calculate a distance matrix. This matrix can then be used by the R programs deposited with the collection, to calculate a performance measure based on one of the available benchmark tests (defined by one of a cast-matrices deposited for the chosen dataset) and the result will be directly comparable with those deposited in the collection.
If the goal is the benchmarking of a new machine learning method, the tests can be performed on an existing distance matrix and a cast-matrix. For example, the new method to be tested can be included as a procedure called by the R scripts downloadable from the site. As the calculations are repeated many times during program development, we have included two mini-datasets (PCB0033, PCB0034), designed for the use of program developers.
SUBMISSION OF NEW DATA
It is our intention to include new data found in the literature and submitted by authors. The new data can include sequence/structure collections subdivided into +train, +test, −train and −test sets, distance matrices and new evaluation results. In order to comply with the data formats, authors intending to submit new data are encouraged to contact the development team at benchmark@icgeb.org.
CONCLUSIONS AND FUTURE DEVELOPMENTS
The bioinformatics literature contains relatively few benchmark datasets (22–28). The distinctive feature of the current collection is the explicit subdivision of the data into +test, +train, −test and −train sets in order to facilitate the comparison of machine learning algorithms. Another important characteristic of the collection is the availability of evaluation results and the detailed documentation of the methodologies. At present, evaluation results are deposited mainly for the smaller datasets. We plan to continuously add evaluation results for the larger datasets and include additional methodologies including Hidden Markov models. At the same time we will augment and improve the tools and interfaces.
Acknowledgments
A. Kocsor was supported by the János Bolyai fellowship of the Hungarian Academy of Sciences. Work at ICGEB was supported in part by grants from the Ministero dell. Università e della Ricerca (D.D. 2187, FIRB 2003 (art. 8), “Laboratorio Internazionale di Bioinformatica”). Funding to pay the Open Access publication charges for this article was provided by ICGEB.
Conflict of interest statement. None declared.
REFERENCES
- 1.Baldi P., Brunak S. Bioinformatics: The Machine Learning Approach, 2nd edn. (Adaptive Computation and Machine Learning) Cambridge, MA: MIT Press; 2001. [Google Scholar]
- 2.Liao L., Noble W.S. Combining pairwise sequence similarity and support vector machines for detecting remote protein evolutionary and structural relationships. J. Comput. Biol. 2003;10:857–868. doi: 10.1089/106652703322756113. [DOI] [PubMed] [Google Scholar]
- 3.Jaakkola T., Diekhans M., Haussler D. Using the Fisher kernel method to detect remote protein homologies. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999:149–158. [PubMed] [Google Scholar]
- 4.Jaakkola T., Diekhans M., Haussler D. A discriminative framework for detecting remote protein homologies. J. Comput. Biol. 2000;7:95–114. doi: 10.1089/10665270050081405. [DOI] [PubMed] [Google Scholar]
- 5.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andreeva A., Howorth D., Brenner S.E., Hubbard T.J., Chothia C., Murzin A.G. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearl F., Todd A., Sillitoe I., Dibley M., Redfern O., Lewis T., Bennett C., Marsden R., Grant A., Lee D., et al. The CATH Domain Structure Database and related resources Gene3D and DHS provide comprehensive domain family information for genome analysis. Nucleic Acids Res. 2005;33:D247–D251. doi: 10.1093/nar/gki024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 9.Smith T.F., Waterman M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 10.Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 11.Kocsor A., Kertesz-Farkas A., Kajan L., Pongor S. Application of compression-based distance measures to protein sequence classification: a methodological study. Bioinformatics. 2006;22:407–412. doi: 10.1093/bioinformatics/bti806. [DOI] [PubMed] [Google Scholar]
- 12.Saigo H., Vert J.P., Ueda N., Akutsu T. Protein homology detection using string alignment kernels. Bioinformatics. 2004;20:1682–1689. doi: 10.1093/bioinformatics/bth141. [DOI] [PubMed] [Google Scholar]
- 13.Gaspari Z., Vlahovicek K., Pongor S. Efficient recognition of folds in protein 3D structures by the improved PRIDE algorithm. Bioinformatics. 2005;21:3322–3323. doi: 10.1093/bioinformatics/bti513. [DOI] [PubMed] [Google Scholar]
- 14.Vapnik V.N. Statistical Learning Theory. New York: John Wiley & Sons; 1998. [Google Scholar]
- 15.Bishop C.M. Neural Networks for Pattern Recognition. Oxford: Clarendon Press; 1995. [Google Scholar]
- 16.Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- 17.Rice J.C. Logistic regression: An introduction. In: Thompson B., editor. Advances in social science methodology. Vol. 3. Greenwich, CT: JAI Press; 1994. pp. 191–245. [Google Scholar]
- 18.Egan J.P. Signal Detection theory and ROC Analysis. New York: 1975. [Google Scholar]
- 19.Gribskov M., Robinson N.L. Use of Receiver Operating Characteristic (ROC) analysis to evaluate sequence matching. Comput. chem. 1996;20:25–33. doi: 10.1016/s0097-8485(96)80004-0. [DOI] [PubMed] [Google Scholar]
- 20.Ihaka R., Gentleman R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996;5:299–314. [Google Scholar]
- 21.Witten I.H., Frank E. Data Mining: Practical Machine Learning Tools and Techniques, 2nd edn. San Francisco, CA: Morgan Kaufmann; 2005. [Google Scholar]
- 22.Antonov A.V., Mewes H.W. BIOREL: the benchmark resource to estimate the relevance of the gene networks. FEBS Lett. 2006;580:844–848. doi: 10.1016/j.febslet.2005.12.101. [DOI] [PubMed] [Google Scholar]
- 23.Chen R., Mintseris J., Janin J., Weng Z. A protein-protein docking benchmark. Proteins. 2003;52:88–91. doi: 10.1002/prot.10390. [DOI] [PubMed] [Google Scholar]
- 24.Mintseris J., Wiehe K., Pierce B., Anderson R., Chen R., Janin J., Weng Z. Protein–Protein Docking Benchmark 2.0: an update. Proteins. 2005;60:214–216. doi: 10.1002/prot.20560. [DOI] [PubMed] [Google Scholar]
- 25.Patton S.J., Wallace A.J., Elles R. Benchmark for evaluating the quality of DNA sequencing: proposal from an international external quality assessment scheme. Clin. Chem. 2006;52:728–736. doi: 10.1373/clinchem.2005.061887. [DOI] [PubMed] [Google Scholar]
- 26.Skolnick J., Kihara D., Zhang Y. Development and large scale benchmark testing of the PROSPECTOR_3 threading algorithm. Proteins. 2004;56:502–518. doi: 10.1002/prot.20106. [DOI] [PubMed] [Google Scholar]
- 27.Thompson J.D., Koehl P., Ripp R., Poch O. BAliBASE 3.0: latest developments of the multiple sequence alignment benchmark. Proteins. 2005;61:127–136. doi: 10.1002/prot.20527. [DOI] [PubMed] [Google Scholar]
- 28.Van Walle I., Lasters I., Wyns L. SABmark—a benchmark for sequence alignment that covers the entire known fold space. Bioinformatics. 2005;21:1267–1268. doi: 10.1093/bioinformatics/bth493. [DOI] [PubMed] [Google Scholar]
- 29.Pollack J.D., Li Q., Pearl D.K. Taxonomic utility of a phylogenetic analysis of phosphoglycerate kinase proteins of Archaea, Bacteria, and Eukaryota: insights by Bayesian analyses. Mol. Phylogenet. Evol. 2005;35:420–430. doi: 10.1016/j.ympev.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Duda R.O., Hart P.E., Stork D.G. Pattern Classification. 2nd edn. New York: John Wiley & Sons; 2000. [Google Scholar]