Abstract
Three independent databases of eukaryotic genome size information have been launched or re-released in updated form since 2005: the Plant DNA C-values Database (www.kew.org/genomesize/homepage.html), the Animal Genome Size Database (www.genomesize.com) and the Fungal Genome Size Database (www.zbi.ee/fungal-genomesize/). In total, these databases provide freely accessible genome size data for >10 000 species of eukaryotes assembled from more than 50 years' worth of literature. Such data are of significant importance to the genomics and broader scientific community as fundamental features of genome structure, for genomics-based comparative biodiversity studies, and as direct estimators of the cost of complete sequencing programs.
INTRODUCTION
Eukaryotic genome size data are becoming increasingly important both as the basis for comparative research into genome evolution and as direct estimators of the cost and difficulty of genome sequencing programs for an expanding sphere of non-model organisms (1–3). Nuclear DNA content data for >10 000 species of plants, animals and fungi are made freely available through three independent databases of eukaryotic genome size that have been either launched or re-released since 2005: the Plant DNA C-values Database (http://www.kew.org/genomesize/homepage.html) (4), the Animal Genome Size Database (www.genomesize.com) (5) and the Fungal Genome Size Database (www.zbi.ee/fungal-genomesize) (6).
Genome sizes are typically given as gametic nuclear DNA contents (‘C-values’) either in units of mass (picograms, where 1 pg = 10−12 g) or in number of base pairs (in eukaryotes, most often in megabases, where 1 Mb = 106 bases). These are directly interconvertible as 1 pg = 978 Mb (or 1 Mb = 1.022 × 10−3 pg) (7). The majority of modern genome size estimates are based on either Feulgen densitometry (more recently using computerized image analysis) or flow cytometry, although DNA reassociation kinetics, bulk fluorometry, static fluorometry, electrophoretic methods, quantitative real-time PCR and complete genome sequencing have also been used. Data from all such measurements are compiled into the databases along with updated taxonomy, analytical details and other relevant information (e.g. chromosome number) where available.
The first genome size estimates were conducted in the late 1940s, and the earliest attempt at a comprehensive list was provided ∼25 years later by Sparrow et al. (8). M.D. Bennett and colleagues carried on this important effort by publishing a series of lists for botanical genome size data beginning in 1976. Unfortunately, zoological and mycological counterparts were not forthcoming for another 30 years, aside from a few taxon-specific compilations based on a small number of sources [e.g. (9)] or online lists of limited scope [e.g. Database of Genome Sizes (http://www.cbs.dtu.dk/databases/DOGS/) and DBA Mammalian Genome Size Database (http://www.unipv.it/webbio/dbagsdb.htm)]. The databases described below, therefore, provide the first truly comprehensive catalogues of eukaryotic genome size data and represent a much-needed resource for members of the genomics community.
PLANT DNA C-VALUES DATABASE
Early development
By 1995, four major lists of angiosperm genome sizes had been published, which together contained data for 2802 species (10–13). Although the lists were well used (collectively, they have been cited >1500 times as on August 2006), it became increasingly cumbersome to determine whether a particular species was listed. It was therefore decided to pool the values into a single database and release them on the internet. The resulting Angiosperm DNA C-values Database was compiled by M.D. Bennett and I.J. Leitch and was coded by the Information Services Department at the Royal Botanic Gardens, Kew; it went live in April 1997. Between 1997 and 2001, two updates of the Angiosperm DNA C-values Database were released and the Pteridophyte DNA C-values Database was added.
Based on the evident utility and high usage of these two databases, efforts were initiated to construct counterparts for other plant taxa as data became available. Ultimately, this led to the assembly of the overarching Plant DNA C-values Database, which was made available through Java-based queries of a SyBase database and was released in September 2001. Initially, it contained C-values for 3864 species from the four land plant groups [angiosperms, gymnosperms, pteridophytes (comprising monilophytes and lycophytes) and bryophytes], with available genome size estimates for three algal groups (Rhodophyta, Chlorophyta and Phaeophyta) added to Release 3.0 in December 2004.
Coverage and features of the Plant DNA C-values Database, Release 4.0
Release 4.0 of the Plant DNA C-values Database, launched in October 2005, contains genome sizes for 5150 species including 4427 angiosperms, 207 gymnosperms, 87 pteridophytes, 176 bryophytes, and 253 algae compiled from over 550 publications or personal communications (4). Tables 1 and 2 provide a breakdown of absolute and relative coverage of the major plant groups. Table 1 also gives the minimum, maximum and mean C-values for each of the major groups and shows that genome sizes in plants range from 0.01 pg in some unicellular algae (e.g. Cyanidium caldarium) to 127.4 pg in the tetraploid angiosperm Fritillaria assyriaca. Most of these data have been acquired through either Feulgen densitometry of stained root tip squashes (63%) or flow cytometry of freshly chopped leaf material (31%) using a range of plant calibration standards. These data are accessible through a variety of search options that allow users to either analyze C-value data across different groups of plants (by clicking on the Plant DNA C-values Database icon), or by searching within taxonomically specific subsections of the database (by clicking on the appropriate plant group icon).
Table 1.
Minimum, maximum and mean 1C DNA amounts for each of the major plant groups in the Plant DNA C-values Database (Release 4.0, October 2005), together with the current level of species representation of C-value data
| Min. (pg) | Max. (pg) | Mean (pg) | Number of species in the Plant DNA C-values Database | Approximate number of species recognizeda | % Representation in the Plant DNA C-values Database | |
|---|---|---|---|---|---|---|
| Algae | ||||||
| Chlorophyta | 0.01 | 19.60 | 1.68 | 91 | 6 500 | 1.4 |
| Rhodophyta | 0.01 | 1.40 | 0.41 | 118 | 6 000 | 1.9 |
| Phaeophyta | 0.10 | 0.90 | 0.44 | 44 | 1 500 | 2.9 |
| Bryophytes | 0.17 | 2.05 | 0.51 | 176 | 18 000 | ∼1.0 |
| Pteridophytes | ||||||
| Lycophytes | 0.17 | 11.97 | 1.95 | 9 | 900 | ∼1.0 |
| Monilophytes | 0.44 | 72.68 | 11.86 | 78 | 11 000 | ∼0.7 |
| Gymnosperms | 2.30 | 36.00 | 18.50 | 207 | 730 | ∼28.4 |
| Angiospermsb | 0.06 | 127.40 | 6.51 | 4427 | 250 000 | ∼1.8 |
Table 2.
Representation of gymnosperms and angiosperms at different taxonomic levels in the Plant DNA C-values Database (Release 4.0, October 2005)
| Group | No. in database | Approximate number of species recognized | % Representation |
|---|---|---|---|
| Angiosperms | |||
| Order | 43 | 45 | 96 |
| Family | 219 | 450 | 49 |
| Genera | 1126 | 13 000 | 9 |
| Species | 4427 | 250 000 | ∼1.8 |
| Gymnosperms | |||
| Order | 4 | 4 | 100 |
| Family | 17 | 17 | 100 |
| Genera | 55 | 83 | 66 |
| Species | 207 | 730 | ∼28.4 |
The database contains information where available for the following fields:
Plant group (e.g. angiosperm, gymnosperm and pteridophyte).
Family.
Genus.
Species.
Taxonomic authority.
Genome size, for which users have the choice of outputting data in pg or Mb and choosing among 1C, 2C and 4C DNA content. (In plants, the measurement of genome size by Feulgen microdensitometry involves determining the amount of staining in mitotic or meiotic dividing cells, typically prepared from actively dividing root tips. The most suitable stage for measurement was considered to be prophase, as chromatin in metaphase or telophase is too condensed. As prophase cells contain a fully replicated genome with a 4C DNA amount it is these values that were reported in publications. Today, 2C values are usually given.)
Ploidy level.
Chromosome number.
Method used to estimate the genome size.
Information on taxonomic vouchers that may exist for the species analyzed.
The full bibliographic reference from which the original data were taken.
At the end of each output, the number of records returned and summary statistics (minimum, maximum, mean and standard deviations) of these records are given.
Additional search options further enhance the flexibility of the database:
All versus prime estimates. Where multiple genome size estimates exist for a given species, users have the choice of outputting all estimates or only the ‘prime’ estimate. The availability of additional, non-prime estimates for a species provides the user with an indication of the range of values that have been reported. In some cases the differences point to genuine intraspecific variation (e.g. Zea mays) but in others they highlight discrepancies attributable to either taxonomic or methodological errors in genome size estimation (14,15). Recent reviews covering potential problems in genome size estimation include those by Greilhuber (15) for Feulgen densitometry and Dolezel and Bartos (16) for flow cytometry.
Wild card searches. An asterisk (*) can be used to indicate wild cards in searches that include only partial names.
- From/to searches. To restrict searches based on numeric data (i.e. chromosome number, ploidy level, DNA amount), users can set criteria in the ‘from’ and ‘to’ boxes of the query page. As examples, user may use this feature:
- To limit the results of a query to taxa with diploid chromosome numbers between 18 and 36 (inclusive) by entering 18 in the ‘from’ box and 36 in the ‘to’ box for chromosome numbers.
- To limit the search to taxa with only 18 chromosomes, by placing this number in both the ‘from’ and ‘to’ boxes.
- To select all records having a diploid number of 18 or greater, by entering 18 in the ‘from’ box and leaving the ‘to’ box empty.
Sorting results. The results of searches are automatically sorted by increasing 1C DNA amounts in picograms. To sort the results by family, genus, species, taxonomic authority, chromosome number or ploidy level, users can select their appropriate choice from the drop-down box under the option ‘Sort by’ at the bottom of the Query form.
In addition to searching the entire database in this way, users can choose to search subsections of the database by selecting the specific plant group of interest (i.e. angiosperms, gymnosperms, pteridophytes, bryophytes or algae) from the homepage. In doing so, the user is provided with additional options for querying and/or outputting that are of unique relevance to each taxon:
- Angiosperms:
- Angiosperm group (i.e. monocots, eudicots or basal angiosperms).
- Life cycle type (i.e. annual, biennial and perennial).
- Family. In particular, users have the choice of displaying either the family name given in the original source of the genome size data or the assigned family following the Angiosperm Phylogeny Group (APG) circumscription (17).
- Gymnosperms:
- Gymnosperm group [i.e. Cycadales, Ginkgoales, Gnetales, Pinaceae or Coniferales II (all conifer families excluding Pinaceae)].
- Sperm flagella number (i.e. multiflagellate or none).
- Pteridophytes:
- Pteridophyte group.
- Spore type (i.e. homosporous or heterosporous).
- Sporangium type (i.e. eusporangiate or leptosporangiate).
- Sperm flagella number (i.e. biflagellate or multiflagellate).
- Bryophytes:
- Bryophyte group (i.e. hornwort, liverwort or moss).
- Algae:
- Algal group (i.e. Chlorophyta, Phaeophyta or Rhodophyta).
Users are required to provide an email address to query the database, which aids in the tracking of usage and in the protection of intellectual property, but otherwise there are no restrictions whatsoever on access.
Besides genome size data, the database includes a summary of the development and release history of the database, instructions on how to search the database, author contact information, links to other databases containing genome size data, and the meeting reports from the international Plant Genome Size meetings, two of which have been held to date (in 1997 and 2003) at the Royal Botanic Gardens, Kew.
USAGE OF THE DATABASE
The Plant DNA C-values Database has been widely used, with >110 000 hits from over 55 countries since its (re-)launch in 2001. On average, the database receives 2000–3000 hits per month with a mean of >60 queries per day, with each query downloading on average 110 genome size estimates. As on August 2006, the database has been cited in ∼130 publications since its initial launch as the Angiosperm DNA C-values Database in 1997.
ANIMAL GENOME SIZE DATABASE
Early development
The first large-scale compilation of animal genome size data was created for an analysis of the correlation between genome size and erythrocyte size in mammals (18), which was later expanded for a similar study in birds (19). Recognizing the severe limitations on the study of animal genome size variation posed by the lack of access to such data, these unpublished datasets were expanded to include data from both vertebrates and invertebrates and were posted online as the Animal Genome Size Database on January 10, 2001. This initial release consisted only of flat text tables and included ∼2900 animal species. As data continued to be added over the ensuing 5 years, the flat table format became increasingly cumbersome in terms of both updates and for the growing number of users.
Coverage and features of the Animal Genome Size Database, Release 2.0
A completely redesigned Release 2.0 of the Animal Genome Size Database was launched on December 24, 2005, meant to coincide approximately with the 5-year anniversary of the database (5). Rather than flat tables, the database has been converted to a MySQL database accessed through a user-friendly website coded in XHTML, CSS, Javascript and PHP. Its search tools also employ some AJAX (Asynchronous Javascript and XML) features, and some Flash charts are used in information display. At the time of this writing, the database contains 5677 records from 601 sources, covering 2953 species of vertebrates and 1323 invertebrates. Reported animal genome sizes range >4000-fold, from ∼0.03 pg in the root-knot nematode Meloidogyne graminicola to ∼133 pg in the marbled lungfish Protopterus aethiopicus. Table 3 provides more detailed breakdowns of the available data, including the ranges, means, and absolute and relative coverage of the major animal groups.
Table 3.
Summary of the content of the Animal Genome Size Database as on August 2006, showing the number of records (i.e. including multiple entries for the same species), species coverage in absolute numbers and percentage of described diversity (in parentheses; note that for many invertebrate taxa only a minority of species have been described), ranges in reported genome sizes, and mean of available genome size data
| Taxon | Records | No. of species (%) | Genome size range (pg) | Mean genome size (pg) |
|---|---|---|---|---|
| Vertebrates | ||||
| Jawless fishes | 26 | 17 (16) | 1.3–4.6 | 2.3 |
| Cartilaginous fishes | 183 | 130 (13) | 2.5–17.1 | 5.7 |
| Lungfishes | 14 | 4 (66) | 50–133 | 90.4 |
| Chondrostean fishes | 38 | 22 (42) | 1.2–7.3 | 3.5 |
| Teleost fishes | 1761 | 1354 (5) | 0.4–4.4 | 1.2 |
| Amphibians | 870 | 463 (9) | 0.95–120.1 | 16.7 |
| Reptiles | 406 | 309 (4) | 1.1–5.4 | 2.3 |
| Birds | 274 | 205 (2) | 1.0–2.2 | 1.5 |
| Mammals | 600 | 432 (9) | 1.7–8.4 | 3.5 |
| Invertebrates | ||||
| Insects | 512 | 433 (0.05) | 0.1–16.9 | 1.6 |
| Crustaceans | 259 | 227 (0.6) | 0.16–38.0 | 3.1 |
| Arachnids | 120 | 117 (0.2) | 0.08–7.5 | 2.4 |
| Molluscs | 202 | 183 (0.2) | 0.4–5.9 | 1.8 |
| Echinoderms | 46 | 46 (0.9) | 0.5–4.4 | 1.3 |
| Annelids | 128 | 126 (1) | 0.06–7.6 | 1.5 |
| Flatworms | 65 | 61 (0.3) | 0.06–20.5 | 2.1 |
| Nematodes | 52 | 41 (0.3) | 0.03–2.5 | 0.2 |
| Miscellaneous other | 121 | 106 | — | — |
| Total | 5677 | 4276 | ||
The nearly 3:1 bias in favor of vertebrates and the discrepancy between total records and species coverage for many groups indicate that available animal genome size data are derived from an unrepresentative subsample of animal diversity and that considerable work remains to be performed to correct this. This table does not include known but unpublished data that are not yet listed in the database [cf. (3)].
Animal genome size data are accessed through either browse or search functions. The browse function allows users to select an entire group of animals (e.g. mammals, insects), or to select subsections of the database using progressive pull-down menus ranging in specificity from phylum to species. The advanced search feature allows a variety of queries, including genus, species or common name, as well as options to select genome sizes equal to, less than/greater than or between user-specified values. Finally, it is also possible to retrieve all records generated using a given method, standard species or cell type.
Data are returned in customizable dynamic tables, with users specifying the number of records displayed per page (100, 250, 500 or All). The default results page includes taxonomic details (Phylum/Subphylum, Class, Order, Family, Genus, Species, common name), C-value in pg, chromosome number (where available), and the method, cell type and standard species used in the analysis. The source is given as a numbered reference with a hotlink to the full citation. Two courses of action are possible from this results table: (i) the data can be downloaded and can be viewed using Excel (with the spreadsheet following the same customized format as the dynamic tables), or (ii) users can click on species names to enter individual species pages. The latter option provides a detailed record for the species of choice, including taxonomic and methodological details, the C-value estimate from the chosen record as well as links to other available records for the same species, chromosome number, the full source citation and both internal links (e.g. to call up data for all members of the genus, family, order, etc.) and external links [e.g. to NCBI, image searches and both general (e.g. the Integrated Taxonomic Information Service) and specific (e.g. FishBase, AmphibiaWeb) taxonomic databases as applicable]. There are no limitations on browsing or searching the database, but downloading data to Excel requires users to input a name and valid email address as a digital signature of a data sharing agreement. A randomized and limited-duration link to the compiled spreadsheet is then emailed to the input address as a means of protecting intellectual property without hindering access to information.
Release 2.0 of the Animal Genome Size Database also provides users with up-to-the-minute summary statistics for the entire database and each major taxonomic group and subgroup therein, number of species covered, min/max, mean ± standard error, a breakdown of methods, cell types, standards used for all records in the given group, and a brief summary of the major patterns and correlates reported to date for the taxon in question. Other features available to users include a real-time Flash-based graphical summary of the total dataset, relevant announcements and a list of the 10 most recently added records on the main page, as well as a fully searchable reference list, an FAQ, author contact information, links to related sites and a genome size discussion forum.
Usage of the database
Traffic at the Animal Genome Size Database has increased steadily since its launch in 2001, and the main page now receives 50–100 unique visitors per day. Records regarding individual queries are not kept, but a typical data download includes all data for one or more entire groups of animals (i.e. up to several hundred species for a particular vertebrate group). The database has been cited in ∼90 publications since 2001.
FUNGAL GENOME SIZE DATABASE
Development and coverage of the Fungal Genome Size Database, Release 1.0
In a discussion of the plant and animal genome size databases penned in mid-2004, it was noted that ‘unfortunately, equivalent databases have not yet been compiled for fungi or "protists", although this would clearly be a worthy project for experts in those groups to undertake' (3). On March 20, 2005, a major portion of this gap had been filled with the launch of the Fungal Genome Size Database (6).
Numerous relative genome sizes (i.e. in arbitrary units) had been estimated in the late 1980s and early 1990s by researchers at the University of Regensburg in Germany using a classical cytophotometry technique, including 287 records for Basidiomycetes (20,21) and 743 for Ascomycetes (22). Using the same method as well as flow cytometry and image cytometry, and by employing an internal standard (Saccharomyces cerevisiae), it became possible to convert these estimates from arbitrary units into far more informative absolute genome sizes in Mb (23–25). These converted data formed the basis of the Fungal Genome Size Database, which has since been expanded to include 1298 records covering 739 species and 335 genera from 40 orders (Table 4) based on the taxonomy of the Index Fungorum Partnership (www.indexfungorum.org) (26).
Table 4.
Number of records in fungal genome size database
| Phylum order | Number of records |
|---|---|
| Ascomycota | 911 |
| Ascosphaerales | 1 |
| Chaetothyriales | 1 |
| Diaporthales | 3 |
| Elaphomycetales | 1 |
| Eurotiales | 23 |
| Helotiales | 523 |
| Hypocreales | 47 |
| Hysteriales | 5 |
| Lecanorales | 5 |
| Mycocaliciales | 1 |
| Mycosphaerellales | 3 |
| Onygenales | 24 |
| Ophiostomatales | 17 |
| Ostropales | 1 |
| Pezizales | 153 |
| Pleosporales | 9 |
| Pneumocystidales | 2 |
| Rhytismatales | 6 |
| Saccharomycetales | 33 |
| Schizosaccharomycetales | 4 |
| Sordariales | 11 |
| Teloschistales | 1 |
| Xylariales | 12 |
| Incertae sedis | 25 |
| Basidiomycota | 358 |
| Agaricales | 78 |
| Boletales | 261 |
| Filobasidiales | 6 |
| Hymenochaetales | 1 |
| Microbotryales | 1 |
| Phallales | 1 |
| Polyporales | 2 |
| Sporidiobolales | 1 |
| Tremellales | 1 |
| Uredinales | 4 |
| Ustilaginales | 2 |
| Chytridiomycota | 5 |
| Blastocladiales | 1 |
| Chytridiales | 3 |
| Neocallimastigales | 1 |
| Glomeromycota | 11 |
| Diversisporales | 3 |
| Glomerales | 8 |
| Zygomycota | 13 |
| Mucorales | 13 |
Numerals in boldface indicate records within phyla and numerals in roman indicate records within orders.
Data from the Fungal Genome Size Database are made available through queries (PHP, HTML) of a MySQL database. The user and administrative interfaces for the database are generated by a CMS system developed by Trump Trading Ltd (TTCMS). The data can be queried by different taxonomic levels (phylum, order, genus, species epithet, variety) as well as by ploidy level, chromosome number, chromosome size range, method of genome size estimation, standard specimens used, cell type analyzed and source reference. Responses to queries are presented as HTML tables, with detailed information about given records (e.g. herbarium index, original reference and additional remarks) provided in a separate pop-up window accessed by clicking on a given genus or species name in the main table.
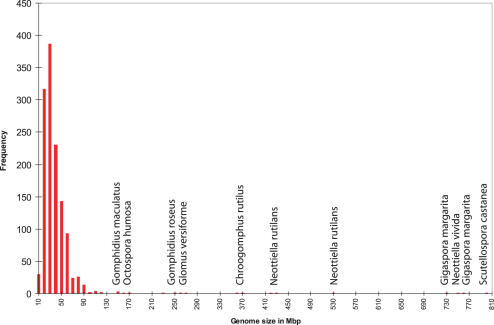
Compared with plants and animals, fungi display very small genomes: ∼90% of the available fungal data lie within the range of 1C = 10–60 Mb, with an average of ∼37 Mb and a median of 28 Mb (Figure 1). The largest fungal genome size reported to date, that of Scutellospora castanea (Diversisporales) is a mere 795 Mb (0.81 pg) (27), whereas the smallest, 6.5 Mb (0.007 pg) in Pneumocystis carinii f. sp. muris (Pneumocystidales), is far more miniscule than even the most streamlined animal or non-algal plant genomes (www.broad.mit.edu/annotation/fungi/fgi/FGI_01_whitepaper_2002.pdf) (28).
Figure 1.
Histogram presenting fungal genome sizes (Mb) in the database. A majority of genome size estimates cover the range from 10 to 60 Mb. The odd values are labeled with species names.
As with plants (and to a far lesser but not insignificant degree with animals), ploidy level variability is an important consideration in fungi. Ploidy level (x) has been estimated for 1036 (80%) of the records in the database, and varies from 1x to 50x. Diploidy (2x) is the single most commonly observed level (36% of records), although haploidy (1x) is also common; a level of 50x has been reported for only one species, Neottiella rutilans (22). Chromosome numbers have been reported for 81 of the species included in the database, ranging from n = 3 in Schizosaccharomyces pombe (Schizosaccharomycetales) (29) to n = 20 in Ustilago hordei (Ustilaginales) and Batrachochytrium dendrobatidis (Chytridiales) (28,30).
In both plants and animals, the majority of variation among estimates for individual species is attributed to experimental error (3,14,15). In fungi, however, it remains unclear to what extent apparent intraspecific variation is non-artifactual as data regarding heteroploidy in this group remain controversial (20,31,32). There is evidence that interspecific hybrids may occur in most fungal phyla, with both sexual and asexual origins evident among the growing list of apparent fungal hybrids (33). Hybrids may be diploid or maintain the dikaryotic state, they may undergo karyogamy and normal meiosis to reconstitute the euploid state, or they may undergo abnormal meiosis to yield a heteroploid hybrid. During vegetative growth, chromosomes and chromosome segments can be lost at random, which would generate legitimate variation in estimated genome sizes.
Electrophoretic karyotyping has shown that variation in chromosome number and size is a rule rather than an exception for many, mostly asexual, species (32). This method indicated that genome size in Pleurotus ostreatus (Agaricales) ranges from 20.8 to 35.1 Mb (0.021–0.036 pg, a relative difference of >60%) and chromosome number ranges from 6 to 11 (34,35). Using flow cytometry, genome size in the same species appears to range from 18.5 to 28.7 Mb (0.019–0.021 pg, a 55% difference) (B. Kullman, unpublished data), whereas microfluorometric measurements resulted in a reported range of 24.0–27.53 Mb (0.025–0.028 pg, a 15% difference) (21). It bears noting, however, that even small absolute differences among estimates that might be considered within the margin of measurement error in plants or animals (e.g. 0.01 pg) translate into substantial relative differences in species with such tiny genomes.
Usage of the database
At this early stage, the database receives ∼10–20 unique hits per day, and at the time of this writing has been visited by >9000 visitors from around the world.
FUTURE PROSPECTS
Taken together, the three eukaryotic genome size databases represent some of the broadest genetic datasets available, covering >10 000 species. In relative terms, however, this comprises a very small minority of eukaryotic diversity. It is therefore a primary objective of modern genome size research to greatly increase the coverage of taxa in all three kingdoms. Perhaps the least well studied of all, however, are the members of the extremely diverse (and paraphyletic) assemblage commonly known as ‘protists’. The construction of a database of genome sizes for this group, and subsequent efforts to fill the gaps therein, represents an equivalently high priority. Overall, the release of these databases has proved to be a boon for the advancement of knowledge about eukaryotic genome structure and evolution, and has made it possible for the first time to identify the key areas still in need of intensive study.
Acknowledgments
The authors wish to thank their many colleagues and collaborators for assistance with various aspects of the construction and maintenance of the genome size databases. Work on the Animal Genome Size Database has been supported by the Natural Sciences and Engineering Research Council of Canada in the form of several scholarships, fellowships and grants to T.R.G. Research leading to the development of the Fungal Genome Size Database was supported by Estonian Science Foundation grant number 4989 to B.K. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bennett M.D., Leitch I.J. Genome size evolution in plants. In: Gregory T.R., editor. The Evolution of the Genome. San Diego, CA: Elsevier; 2005. pp. 89–162. [Google Scholar]
- 2.Gregory T.R. Synergy between sequence and size in large-scale genomics. Nature Rev. Genet. 2005;6:699–708. doi: 10.1038/nrg1674. [DOI] [PubMed] [Google Scholar]
- 3.Gregory T.R. Genome size evolution in animals. In: Gregory T.R., editor. The Evolution of the Genome. San Diego, CA: Elsevier; 2005. pp. 3–87. [Google Scholar]
- 4.Bennett M.D., Leitch I.J. Plant DNA C-values Database. 2005 doi: 10.1007/978-1-0716-3389-2_9. [DOI] [PubMed] [Google Scholar]
- 5.Gregory T.R. Animal Genome Size Database. 2005 [Google Scholar]
- 6.Kullman B., Tamm H., Kullman K. Fungal Genome Size Database. 2005 [Google Scholar]
- 7.Dolezel J., Bartos J., Voglmayr H., Greilhuber J. Nuclear DNA content and genome size of trout and human. Cytometry. 2003;51A:127–128. doi: 10.1002/cyto.a.10013. [DOI] [PubMed] [Google Scholar]
- 8.Sparrow A.H., Price H.J., Underbink A.G. A survey of DNA content per cell and per chromosome of prokaryotic and eukaryotic organisms: some evolutionary considerations. In: Smith H.H., editor. Evolution of Genetic Systems. New York: Gordon and Breach; 1972. pp. 451–494. [PubMed] [Google Scholar]
- 9.Tiersch T.R., Wachtel S.S. On the evolution of genome size of birds. J. Hered. 1991;82:363–368. doi: 10.1093/oxfordjournals.jhered.a111105. [DOI] [PubMed] [Google Scholar]
- 10.Bennett M.D., Smith J.B. Nuclear DNA amounts in angiosperms. Philos. Trans. R. Soc. Lond. Ser. B. 1976;274:227–274. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- 11.Bennett M.D., Smith J.B., Heslop-Harrison J.S. Nuclear DNA amounts in angiosperms. Proc. R. Soc. Lond. B. 1982;216:179–199. [Google Scholar]
- 12.Bennett M.D., Smith J.B. Nuclear DNA amounts in angiosperms. Philos. Trans. R. Soc. Lond. Ser. B. 1991;334:309–345. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- 13.Bennett M.D., Leitch I.J. Nuclear DNA amounts in angiosperms. Ann. Bot. 1995;76:113–176. [Google Scholar]
- 14.Greilhuber J. Intraspecific variation in genome size: a critical reassessment. Ann. Bot. 1998;82(Suppl. A):27–35. [Google Scholar]
- 15.Greilhuber J. Intraspecific variation in genome size in angiosperms—identifying its existence. Ann. Bot. 2005;95:91–98. doi: 10.1093/aob/mci004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dolezel J., Bartos J. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. 2005;95:99–110. doi: 10.1093/aob/mci005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Angiosperm Phylogeny Group II. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants. Bot. J. Linnean Soc. 2003;141:399–436. [Google Scholar]
- 18.Gregory T.R. Nucleotypic effects without nuclei: genome size and erythrocyte size in mammals. Genome. 2000;43:895–901. doi: 10.1139/g00-069. [DOI] [PubMed] [Google Scholar]
- 19.Gregory T.R. A bird's-eye view of the C-value enigma: genome size, cell size, and metabolic rate in the class Aves. Evolution. 2002;56:121–130. doi: 10.1111/j.0014-3820.2002.tb00854.x. [DOI] [PubMed] [Google Scholar]
- 20.Bresinsky A., Wittmann-Meixner B., Weber E., Fischer M. Karyologische Untersuchungen an Pilzen mittels Fluoreszenzmikroskopie. Z. Mykol. 1987;53:303–318. [Google Scholar]
- 21.Wittmann-Meixner B. Polyploidie bei Pilzen. Biblioth. Mycol. 1989;131:1–163. [Google Scholar]
- 22.Weber E. Untersuchungen zu Fortpflanzung und Ploidie verschiedener Ascomyceten. Biblioth. Mycol. 1992;140:1–186. [Google Scholar]
- 23.Kullman B. Application of flow cytometry for measurement of nuclear DNA content in fungi. Folia Cryptog. Estonica. 2000;36:31–46. [Google Scholar]
- 24.Kullman B. Nuclear DNA content, life cycle and ploidy in two Neottiella species (Pezizales, Ascomycetes) Persoonia. 2002;18:103–115. [Google Scholar]
- 25.Kullman B., Teterin W. Estimation of fungal genome size: comparison of image cytometry and photometric cytometry. Folia Cryptog. Estonica. 2006;42:43–56. [Google Scholar]
- 26.Index Fungorum Partnership. 2004. Index Fungorum. Custodians CABI Bioscience, CBS and Landcare Research. [Google Scholar]
- 27.Hijri M., Sanders J.R. Low gene copy number shows that arbuscular mycorrhizal fungi inherit genetically different nuclei. Nature. 2005;433:160–163. doi: 10.1038/nature03069. [DOI] [PubMed] [Google Scholar]
- 28.Birren B., Fink G., Lander E. Fungal Genome Initiative White Paper. 2002 [Google Scholar]
- 29.Wood V., Gwilliam R., Rajandream M.A. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 30.McCluskey K., Mills D. Identification and characterization of chromosome length polymorphisms among strains representing fourteen races of Ustilago hordei. Mol. Plant- Micr. Interact. 1990;3:336–373. [Google Scholar]
- 31.Tolmsoff W.J. Heteroploidy as a mechanism of variability among fungi. Annu. Rev. Phytopathol. 1983;21:317–340. [Google Scholar]
- 32.Beadle J., Wright M., McNeely L., Bennett J.W. Electrophoretic karyotype analysis in fungi. Adv. Appl. Microbiol. 2003;53:243–270. doi: 10.1016/s0065-2164(03)53007-6. [DOI] [PubMed] [Google Scholar]
- 33.Schardl C.L., Craven K.D. Interspecific hybridization in plant-associated fungi and oomycetes: a review. Mol. Ecol. 2003;12:2861–2873. doi: 10.1046/j.1365-294x.2003.01965.x. [DOI] [PubMed] [Google Scholar]
- 34.Sagawa I., Nagata Y. Analysis of chromosomal DNA of mushrooms in genus Pleurotus by pulsed field gel electrophoresis. J. Gen. Appl. Microbiol. 1992;38:47–52. [Google Scholar]
- 35.Ramírez L., Larraya L.M., Pisabarro A.G. Molecular tools for breeding basidiomycetes. Int. Microbiol. 2000;3:147–152. [PubMed] [Google Scholar]
- 36.Kapraun D.F. Nuclear DNA content estimates in multicellular eukaryotic green, red and brown algae: phylogenetic considerations. Ann. Bot. 2005;95:7–44. doi: 10.1093/aob/mci002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu Y.L., Palmer J.D. Phylogeny of early land plants: insights from genes and genomes. Trends. Plant Sci. 1999;4:26–30. doi: 10.1016/s1360-1385(98)01361-2. [DOI] [PubMed] [Google Scholar]
- 38.Murray B.G., Leitch I.J., Bennett M.D. 2001. Gymnosperm DNA C-values Database. [Google Scholar]
- 39.Greilhuber J., Borsch T., Müller K., Worberg A., Porembski S., Barthlott W. Smallest angiosperm genomes found in Lentibulariaceae with chromosomes of bacterial size. Plant Biol. 2006 doi: 10.1055/s-2006-924101. in press. [DOI] [PubMed] [Google Scholar]