Abstract
xanthusBase (http://www.xanthusbase.org) is the official model organism database (MOD) for the social bacterium Myxococcus xanthus. In many respects, M.xanthus represents the pioneer model organism (MO) for studying the genetic, biochemical, and mechanistic basis of prokaryotic multicellularity, a topic that has garnered considerable attention due to the significance of biofilms in both basic and applied microbiology research. To facilitate its utility, the design of xanthusBase incorporates open-source software, leveraging the cumulative experience made available through the Generic Model Organism Database (GMOD) project, MediaWiki (http://www.mediawiki.org), and dictyBase (http://www.dictybase.org), to create a MOD that is both highly useful and easily navigable. In addition, we have incorporated a unique Wikipedia-style curation model which exploits the internet's inherent interactivity, thus enabling M.xanthus and other myxobacterial researchers to contribute directly toward the ongoing genome annotation.
INTRODUCTION
In the era of post-genomics, model organism databases (MODs) have become an integral component of research for the molecular life sciences. MODs now serve several important functions: as a medium for learning (1), for coordinating resource sharing (2) and, perhaps most importantly, for knowledge discovery (3). While the number of MODs is increasing, their default design and implementation have stabilized, evolving into an internet-based annotation centered on a core genome sequence. Additional genomic information, such as microarray data and strain collection libraries, are all integrated to work in concert with the genome core. Within this context, MODs can function as an ordered repository, an interactive forum, and a catalyst for data integration. Because a genome's annotation is never truly ‘finished’, a MOD represents a continually evolving annotation document.
The xanthusBase core is the 9.14 Mb Myxococcus xanthus genome sequence, recently released by the Monsanto Company and The Institute for Genomic Research (TIGR). This genome sequence, and its annotation, represents a wealth of information for both basic and applied research. For example, the myxobacteria in general, and M.xanthus in particular, have a rich secondary metabolism (4) that has already been exploited to develop therapeutic agents, such Myxovirescen A, an antibacterial compound (5), and Prolyl Endopetidase, an oral treatment for Celiac Sprue (6). M.xanthus has a large number of polyketide synthases, on the order of Streptomyces ceolicolor. Future research will be greatly enhanced by genome-scale analysis of the M.xanthus polyketide synthase clusters, as well as the other genes involved in the production of secondary metabolites.
In addition to its potential utility as a source of pharmaceutical agents, the primary focus of M.xanthus research is as a topic of basic scientific interest to discover the behavioral genomics of emergent behavior and the evolutionary underpinnings of multicellularity (7). Although each M.xanthus cell is autonomous with respect to both metabolism and reproduction, it exists within a homogeneous predatory biofilm called a swarm, which moves and feeds as a single entity. In response to starvation-stress, the millions of cells that comprise a swarm display an ordered series of emergent behaviors called development, during which they execute a complex behavioral genetic program to self-organize into fruiting bodies (8,9). A fruiting body is a macroscopic (∼0.1 mm) roughly spherical structure of ∼1 × 105 cells that is composed of a sticky peptidoglycan layer surrounding a core bolus of metabolically quiescent and environmentally resistant spores. The completed fruiting body is raised upon a stalk, and it is sticky. The cooperative feeding behavior of M.xanthus betrays the purpose of fruiting prior to sporulation; if a moving object, such as a leg of an insect, comes in contact with a fruiting body, all of the component spores will stick to the leg and be translocated as a unit. This way, if carried to a new food source, the thousands of spores can emerge as an ‘instant’ swarm, rather than having to re-establish a swarm from a single spore (10,11).
The published annotation of the M.xanthus genome (12) is a single circular chromosome with 9 139 763 bases of GC rich DNA (69%) predicted to encode 7331 coding sequences (CDS)—a number that rivals lower eukaryotes such as Saccharomyces cerevisiae. As the first myxobacteria to be completely sequenced, this predicted annotation supports the hypothesis that the complex behavior of M.xanthus is controlled through multiple signaling cascades evolved by gene duplication and diversification. To understand the complex genetics through which M.xanthus interacts with its environment and controls spatial patterning, the genome must be recapitulated as a series of interconnected functional networks. There is an obvious need for an open and interactive MOD—a need that xanthusBase strives to fulfill. A MOD for M.xanthus was first conceived at the 32rd International Conference on the Biology of the Myxobacteria where it was officially endorsed by the community. With this enthusiastic user base it was felt that a MOD based on community annotations was worth attempting.
A MOD FOR M.xanthus
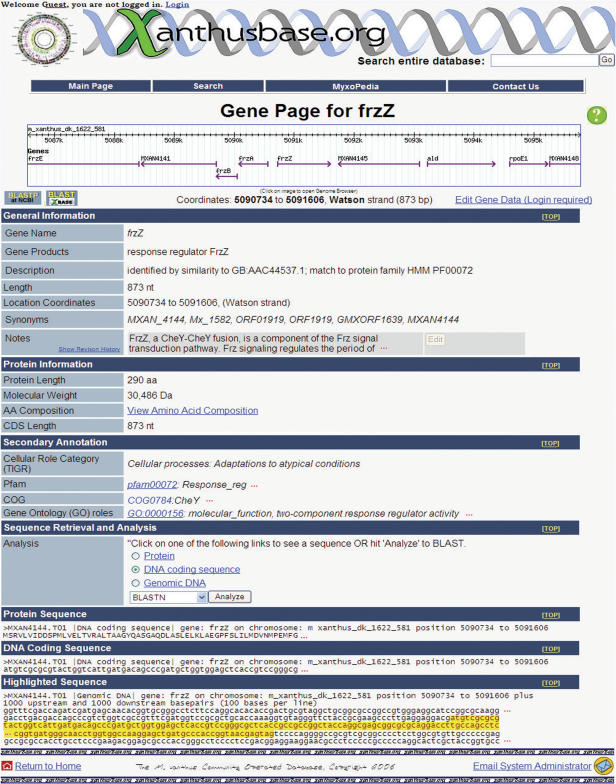
Annotations for each predicted CDS in the M.xanthus genome are accessible through a gene page as shown in Figure 1. The majority of this content is editable by clicking on the ‘Edit Gene Data’ link located at the top of each CDS page. Any field that has received user revisions to its content will display a pull down link showing the revision history of that field (see Discussion below). Each gene page provides basic information for a predicted CDS beginning with a GBrowse (13) implemented graphical representation of the CDS on the chromosome. This visualization presents the orientation of the CDS, the chromosomal coordinates, and the relative positions of nearby CDSs. Located immediately below this graphic is a General Information section which provides a basic description of the CDS, as originally annotated by TIGR. This section includes the gene name, the gene product, a short description of the CDS, the sequence length of the CDS, the exact coordinates of the CDS on the chromosome, the orientation of the CDS, and a list of synonyms that describe other nomenclatures associated with the CDS. In addition, this section also contains a free text notes field for users to supply additional general information about each CDS. The Protein Information section contains the length of the translated protein, the molecular weight of this protein, and a link which displays the amino acid composition.
Figure 1.
A representative xanthusBase gene page. Some sections have been condensed for display purposes.
Further annotations of the CDS are located in the Secondary Annotation section beginning with links to existing pages. Additional secondary annotations in the form of Enzyme Commission (EC) numbers, Cellular Role Categories as designated by TIGR, Gene Ontology (14), Pfam (15), COG (16) and KEGG (17) are also presented in this section. Clicking on each annotation's respective links will display the specific annotation as maintained by each annotation's online database. This section also contains a list of any relevant publications associated with the CDS.
A summary of the genomic and annotation data in xanthusBase is presented in Table 1. The Sequence Retrieval and Analysis section provides the user with the ability to perform various alignments of the CDS using different programs of BLAST (18) against a local database containing all of the predicted CDSs in M.xanthus. In addition, the ability to perform an alignment of the CDS using BLASTP against the National Center for Biotechnology Information's (NCBI) non-redundant database (19) is also provided. The DNA Coding Sequence and Protein Coding Sequence sections display the nucleotide and protein sequences of the CDS in FASTA format, respectively. Finally, the Highlighted Sequence section contains the nucleotide sequence of the CDS highlighted in yellow, with both adjacent 1000 bp upstream and downstream regions.
Table 1.
Genomic data and annotations found in xanthusBase as of August 2006
| Predicted coding sequences (CDSs) | 7331 |
| Predicted ribosomal RNA genes | 12 |
| Predicted transfer RNA genes | 67 |
| Gene products | 982 |
| GenBank records | 286 |
| PubMed references | 874 |
| GO annotations | 2940 |
| Pfam annotations | 3818 |
| COG annotations | 4318 |
| KEGG pathways | 586 CDSs in 114 pathways |
| CDSs with enzyme commission numbers | 744 |
| Colleagues | 40 laboratories |
MYXOPEDIA: A WIKI FOR M.xanthus
MyxoPedia is an implementation of the MediaWiki software (http://www.mediawiki.org) implemented on xanthusBase. The MyxoPedia pages provide a free text forum for discussion about a wide variety of topics on M.xanthus and allows for the incorporation of images, time-lapse microscopy videos, and protocols into the database. Gene related data is generally not stored in MyxoPedia, but is rather stored in the main database where wiki-editing principles have been superimposed onto the database to allow the editing of its standardized fields. A discussion of the application of these ‘wiki-editing’ methods to the main genomic content of the database follows.
XANTHUSBASE: A WIKIPEDIA-STYLE MODEL FOR MODS
The most prevalent curation method for a MOD is the museum model (20), which is performed at one or more central locations by professional curators who annotate an MO genome through the interpretation of primary scientific literature. To benefit more directly from the expertise of the scientific community, distributed annotation systems have been described that allow for the periodic exchange of data maintained by individual labs (21), or that allow community members to make online modifications that are used as suggestions for a central curator (22). The Wikipedia (http://www.wikipedia.com) model is the single most successful community-driven internet database format. Unlike many other models, Wikipedia has a proven track record, and a body of literature analyzing the reasons for its success and the impressive accuracy of its annotations (23). Recent analysis of Wikipedia has revealed that the motivating force behind user participation is very similar to the motivation described by academic scientists, an amorphous and non-quantifiable merit system termed ‘credibility’ (24). We believe that this strong similarity indicates that MODs can be adapted to the Wikipedia model. The potential benefits of such a system—an open and interactive annotation that is driven and curated by the same scientific experts that generate data—are immeasurable.
Frustrating any attempt at a ‘finished’ annotation is the fact that, for the molecular life sciences, all data remains open to reanalysis and reinterpretation. In this way, the museum MOD curation schema does not fully leverage the interactive potential of the internet, because a virtual firewall exists between a MOD and the community of scientific experts who generate the data. xanthusBase is a new model for MODs in which Wikipedia-editing principles are super-imposed onto standardized genome database fields.
Two essential paradigms of this approach are distributed curation and the maintenance of annotation history. The distributed curation paradigm assigns annotation responsibility to a voluntary community of experts, rather than a group of professional annotators. Each member of the community is given the right to edit database entries, with editorial access controlled through individual login accounts. Data considered to be core genome components, which include DNA and protein sequences, CDS predictions, and GenBank assigned ID numbers, will not be modifiable. All other components, however, are editable by community members as outlined in Table 2. Changes to editable fields are accomplished in a manner similar to the process employed by Wikipedia. In xanthusBase, common tasks, including minor corrections and additions, are designed to be easy for any user to perform. All modifications are updated and displayed instantaneously to the public. Since curation, in this model, is provided by members of the community with expert knowledge in specific areas, their contributions, in some cases, will be more accurate than the annotations that a group of professional annotators could provide. With respect to the metric of credibility (24), the content of xanthusBase will be viewed as a manuscript in progress, and active curators will be viewed as contributing authors.
Table 2.
Summary table of all editable and non-editable gene page components in xanthusbase.org
| Non-editable core components | Editable Wiki components |
|---|---|
| Sequence length | Gene Name |
| Coordinates | Gene Product |
| Synonyms | Gene Description |
| Protein length | MyxoPedia Pages |
| Molecular weight | Relevant Publications |
| DNA coding sequence | Enzyme Commission Number* |
| Protein sequence | TIGR Role Categories* |
| Highlighted sequence | COG* |
| KEGG* | |
| GO* | |
| Pfam* |
Non-editable components indicate those annotations that users are not permitted to modify. Editable components indicate those annotations that users are allowed to modify by deleting, adding, or editing. An asterisk (*) indicates editable components where the original annotations can not be deleted or edited, but new annotations can be added.
The maintenance of an annotation history is a second key feature of the xanthusBase model. All modifications are permanently recorded, with changes to each field tracked via both the user login and time-stamp. This feature is essential to the distributed nature of this model because it provides an assurance that each community member's contributions are always credited and accessible. In addition, the maintenance of an annotation history is an important safeguard against unforeseen events, such as accidental deletions or unintentional modifications; the database can revert to a previous annotation state at any given time. The implementation of an annotation history for each entry changes the database from a collection of static, single-value fields into a dynamic and evolving knowledge-base. Importantly, the annotation history of each entry can be displayed, providing insight into the ‘evolution’ of an annotation as scientific research progresses.
CONCLUSIONS AND FUTURE DIRECTIONS
The xanthusBase model is a viable alternative curation schema that, in combination with open-source database software, will enable smaller research communities, like the one for M.xanthus, to support a MOD. This would provide the community with a central resource for learning and contributing information about the MO, so that the knowledge base would evolve as the community grows. Information can be extracted from such MODs and used to contribute to comprehensive resources, such as GenBank, in the same way many of the large MODs contribute their information. Genome annotation represents an iterative continuum, and any concept of a ‘finished’ annotation is a long way off.
IMPLEMENTING NEW MOD FEATURES
New features will be developed for xanthusBase as needed. For example, there will be instances when community members are in long-term disagreement as to the correct annotation for certain CDSs. A feature is being added to xanthusBase that will enable users to create a branched annotation, so that all community members can contribute while the disagreement is being resolved. Each branch will evolve individually via the xanthusBase model.
DEVELOPING A SOURCEFORGE CODE BASE
Through a collaborative effort with dictyBase (2), we will be establishing an open source resource on sourceForge (http://sourceforge.net) with the entire code base behind both xanthusBase and dictyBase. Where xanthusBase and dictyBase have chosen disparate implementations, both in terms of suitability towards an organism type (i.e. prokaryote versus eukaryote) and MOD schema (i.e. museum versus xanthusBase curation), there will be alternative modules available for an administrator to choose from when deploying a new MOD. This tool kit will allow smaller communities to more easily implement a MOD for their MO.
UNDERGRADUATE EDUCATION SUPPORT SCHEMA
An education support model is currently in development to allow undergraduate students to contribute to the xanthusBase annotation as part of a classroom curriculum. Annotations performed by students will be maintained in the MOD, separate from community member annotations. This will allow students to freely make their contributions without risking the integrity of the xanthusBase. Community members will have access to view student annotations and, when appropriate, apply them to xanthusBase.
Acknowledgments
The authors would like to thank members of the M.xanthus community for their support on this project. The authors would also like to thank Steve A. Arshinoff, Barry S. Goldman, Rion G. Taylor, and members of the Welch Lab for their encouragement, suggestions, and advice. The authors also thank Brian Calhoun-Bryant for technical assistance. Funding to pay the Open Access publication charges for this article was provided by Syracuse University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Goodner B.W., Wheeler C.A., Hall P.J., Slater S.C. Massively Parallel Undergraduates for Bacterial Genome Finishing. ASM News. 2003;69:584–585. [Google Scholar]
- 2.Chisholm R.L., Gaudet P., Just E.M., Pilcher K.E., Fey P., Merchant S.N., Kibbe W.A. dictyBase, the model organism database for Dictyostelium discoideum. Nucleic Acids Res. 2006;34:D423–D427. doi: 10.1093/nar/gkj090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blake J.A., Bult C.J. Beyond the data deluge: data integration and bio-ontologies. J. Biomed. Inform. 2006;39:314–320. doi: 10.1016/j.jbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Reichenbach H., Hofle G. Biologically active secondary metabolites from myxobacteria. Biotechnol. Adv. 1993;11:219–277. doi: 10.1016/0734-9750(93)90042-l. [DOI] [PubMed] [Google Scholar]
- 5.Simunovic V., Zapp J., Rachid S., Krug D., Meiser P., Muller R. Myxovirescin A biosynthesis is directed by hybrid polyketide synthases/nonribosomal peptide synthetase, 3-hydroxy-3-methylglutaryl-CoA synthases, and trans-acting acyltransferases. Chembiochem. 2006;7:1206–1220. doi: 10.1002/cbic.200600075. [DOI] [PubMed] [Google Scholar]
- 6.Gass J., Ehren J., Strohmeier G., Isaacs I., Khosla C. Fermentation, purification, formulation, and pharmacological evaluation of a prolyl endopeptidase from Myxococcus xanthus: implications for Celiac Sprue therapy. Biotechnol. Bioeng. 2005;92:674–684. doi: 10.1002/bit.20643. [DOI] [PubMed] [Google Scholar]
- 7.Kaiser D. Control of multicellular development: Dictyostelium and Myxococcus. Annu. Rev. Genet. 1986;20:539–566. doi: 10.1146/annurev.ge.20.120186.002543. [DOI] [PubMed] [Google Scholar]
- 8.Kaiser D. Coupling cell movement to multicellular development in myxobacteria. Nature Rev. Microbiol. 2003;1:45–54. doi: 10.1038/nrmicro733. [DOI] [PubMed] [Google Scholar]
- 9.Kaiser D. Signaling in myxobacteria. Annu. Rev. Microbiol. 2004;58:75–98. doi: 10.1146/annurev.micro.58.030603.123620. [DOI] [PubMed] [Google Scholar]
- 10.Dworkin M., Kaiser D. Myxobacteria II. Washington,D.C: American Society for Microbiology Press; 1993. [Google Scholar]
- 11.Shimkets L.J. Social and developmental biology of the myxobacteria. Microbiol. Rev. 1990;54:473–501. doi: 10.1128/mr.54.4.473-501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldman B.S., Nierman W.C., Kaiser D., Slater S.C., Durkin A.S., Eisen J., Ronning C.M., Barbazuk W.B., Blanchard M., Field C., et al. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl Acad. Sci. USA. 2006;103:15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finn R.D., Mistry J., Schuster-Bockler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tatusov R.L., Koonin E.V., Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 17.Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K.F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein L. Genome annotation: from sequence to biology. Nature Rev. Genet. 2001;2:493–503. doi: 10.1038/35080529. [DOI] [PubMed] [Google Scholar]
- 21.Dowell R.D., Jokerst R.M., Day A., Eddy S.R., Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tripathy S., Pandey V.N., Fang B., Salas F., Tyler B.M. VMD: a community annotation database for oomycetes and microbial genomes. Nucleic Acids Res. 2006;34:D379–D381. doi: 10.1093/nar/gkj042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giles J. Internet encyclopaedias go head to head. Nature. 2006;438:900–901. doi: 10.1038/438900a. [DOI] [PubMed] [Google Scholar]
- 24.Forte A., Bruckman A. Group 2005 Conference; Sanibel Island, Florida, USA. 2005. [Google Scholar]