Abstract
Phosphorylation is the most common protein post-translational modification. Phosphorylated residues (serine, threonine and tyrosine) play critical roles in the regulation of many cellular processes. Since the amount of data produced by screening assays is growing continuously, the development of computational tools for collecting and analysing experimental data has become a pivotal task for unravelling the complex network of interactions regulating eukaryotic cell life. Here we present Phospho3D, http://cbm.bio.uniroma2.it/phospho3d, a database of 3D structures of phosphorylation sites, which stores information retrieved from the phospho.ELM database and is enriched with structural information and annotations at the residue level. The database also collects the results of a large-scale structural comparison procedure providing clues for the identification of new putative phosphorylation sites.
INTRODUCTION
The phosphorylation of specific protein residues is a crucial event in the regulation of several cellular processes, operating on activation, deactivation or recognition of the target protein. A great deal of eukaryotic proteins (∼30% in the human genome) undergo this reversible post-translational modification (1). Phosphorylation on serine/threonine or tyrosine residues is accomplished by protein kinases (PKs), one of the largest protein families, comprising 1.5–2.5% of all eukaryotic genes (2).
Although the amount of data produced in various screening assays is steadily growing (3–6), experimental identification of phosphoproteins and the determination of individual phosphorylation sites remains a difficult and time-consuming task. Hence, the implementation of computational tools proves to be very useful for collecting and analysing experimental data.
Several sequence-based methods to predict phosphorylation sites were developed using different computational approaches such as regular expressions with context-based rules (7), position-specific scoring matrices (PSSMs) (8), artificial neural networks (9,10), support vector machines (SVMs) (11,12), hidden Markov models (13) and iterative statistical methods (14). All these methods are based on the hypothesis that the sequence surrounding the phosphorylated residue represents the main determinant for kinase specificity. They are reasonably accurate and work well with a number of specific kinases. However, the specificity determinants and rules remain elusive for a large number of protein kinases that display a number of substrates sharing little or no sequence similarity in the known phosphopeptides. We propose that, at least in some cases, the rules of kinase specificity may reside in the presence of structural determinants which only occasionally overlap with sequence consensi and which might be independent of the residue order in protein sequences.
Here we describe Phospho3D, a database of 3D structure of phosphorylation sites. It collects information retrieved from the phospho.ELM database (15) and is enriched with structural information and diverse annotations at the residue level. In addition, the database stores the results of a large-scale local structural comparison which suggest functional annotation of phosphorylation sites by 3D similarity. Cases of significant structural similarity between phosphorylation sites may indicate that they are phosphorylated by the same kinase.
DATABASE CONSTRUCTION AND CONTENT
The Phospho3D database was constructed by collecting data from the phospho.ELM database which gathers experimentally verified phosphorylation sites manually extracted from the literature. The phospho.ELM dataset used in this work (version 4.0) contains 5314 phosphorylation sites, or instances, belonging to 1805 different sequences.
The correspondence between phospho.ELM sequences and the Protein Data Bank (PDB) chains was established via the Seq2Struct resource (16), an exhaustive collection of annotated links between SwissProt-TrEMBL and PDB sequences. Links are based on sequence alignment using pre-established highly reliable thresholds. From a list of 4530 sequence–structure links (for further details see website documentation), only the ones having the phosphorylable residue in the alignment region were retained, this resulting in 2726 instances (166 unique phospho.ELM instances on 1219 protein chains).
The basic information stored in Phospho3D consists of the instance, its flanking sequence (10 residues) and any residue whose distance from the instance does not exceed 12 Å thus defining a 3D neighbourhood which we define as zone.
For each zone, annotation at the residue level is provided, namely solvent accessibility supplied by the NACCESS program (17), secondary structure assignment given by the DSSP program (18) and residue conservation as from the Consurf-HSSP database (19).
Users can also retrieve information extracted from the phospho.ELM dataset; for instance, the Medline reference PMID and, when available, the kinase(s) that phosphorylate(s) the given site.
In addition, for each zone the results of a large-scale local structural comparison versus a representative dataset of PDB (20) protein chains from eukaryotic organisms are also given. The comparison was carried out using the Query3D sequence/fold independent algorithm (21). Structural matches are assessed by two criteria: structural similarity and biochemical similarity. The structural similarity demands that matching residues have a root mean square deviation (r.m.s.d.) lower than a given threshold, whereas the biochemical similarity is evaluated using a Dayhoff substitution matrix (22). The score of the match is the number of matching residues which fulfil the similarity criteria. The significance of the score is evaluated by calculating the Z-score over the score distribution of the query zone comparison to the whole dataset.
THE WEB INTERFACE
The Phospho3D database can be searched by kinase name, by PDB identification code or keyword. A browsing function has been also implemented.
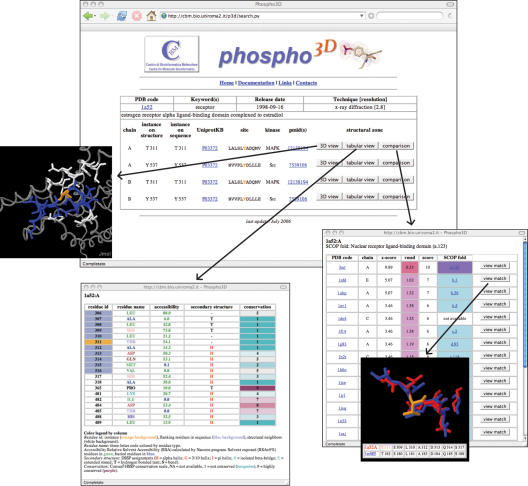
The information returned to the user consists of a brief description of the PDB structure(s) which fulfil the search criterion and of a list of instances presented along with associated information (Figure 1). For each instance, the user can select three options related to the surrounding structural zone: a graphical view using the Jmol Java Applet (http://www.jmol.org); a tabular view reporting the zone annotation at the residue level; a list of 3D matches identified by local structural comparison. Each match can be visualized using Jmol. A tabular view of the matching residues is also presented (Figure 1).
Figure 1.
In the central panel a list of instances for the PDB file 1A52 is shown. For each of them, users can visualize the corresponding zone via the Jmol viewer, the annotation at the residue level and the results of the large-scale local structural comparison. For each structural match the score, the Z-score, and the rmsd are reported along with the SCOP fold (27) of the matching PDB files.
CONCLUSION AND FUTURE PERSPECTIVES
The Phospho3D database is a useful tool for the analysis of the structural features of experimentally verified phosphorylation sites. Moreover, it provides the results of a large-scale local structural comparison between the zones and a representative set of eukaryotic protein chains. The results of such a comparison identify new putative phosphorylation sites and suggest the kinase(s) responsible for phosphorylation.
Phospho3D will be regularly updated as soon as the new Phospho.ELM datasets are released. The annotations will be integrated as a feature in the pdbFun server (23). We are also planning to identify and annotate those sites which are recognized by protein phosphatases and phosphoresidues-binding modules (24–26).
The Phospho3D dataset (annotations at the residue level and structural comparison results) is available upon request.
Acknowledgments
The authors would like to thank Francesca Diella for support and suggestions and for providing the Phospho.ELM dataset. We acknowledge the support of AIRC, Telethon (GGP04273), a PNR 2001–2003 (FIRB art.8) and a PNR 2003–2007 (FIRB art.8). Funding to pay the Open Access publication charges for this article was provided by AIRC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cohen P. The origins of protein phosphorylation. Nature Cell. Biol. 2002;4:E127–E130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- 2.Manning G., Plowman G.D., Hunter T., Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 2002;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- 3.Salomon A.R., Ficarro S.B., Brill L.M., Brinker A., Phung Q.T., Ericson C., Sauer K., Brock A., Horn D.M., Schultz P.G., et al. Profiling of tyrosine phosphorylation pathways in human cells using mass spectrometry. Proc. Natl Acad. Sci. USA. 2003;100:443–448. doi: 10.1073/pnas.2436191100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shu H., Chen S., Bi Q., Mumby M., Brekken D.L. Identification of phosphoproteins and their phosphorylation sites in the WEHI-231 B lymphoma cell line. Mol. Cell. Proteomics. 2004;3:279–286. doi: 10.1074/mcp.D300003-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Brill L.M., Salomon A.R., Ficarro S.B., Mukherji M., Stettler-Gill M., Peters E.C. Robust phosphoproteomic profiling of tyrosine phosphorylation sites from human T cells using immobilized metal affinity chromatography and tandem mass spectrometry. Anal. Chem. 2004;76:2763–2772. doi: 10.1021/ac035352d. [DOI] [PubMed] [Google Scholar]
- 6.Beausoleil S.A., Jedrychowski M., Schwartz D., Elias J.E., Villen J., Li J., Cohn M.A., Cantley L.C., Gygi S.P. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl Acad. Sci. USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puntervoll P., Linding R., Gemund C., Chabanis-Davidson S., Mattingsdal M., Cameron S., Martin D.M., Ausiello G., Brannetti B., Costantini A., et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Obenauer J.C., Cantley L.C., Yaffe M.B. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blom N., Gammeltoft S., Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 10.Iakoucheva L.M., Radivojac P., Brown C.J., O'Connor T.R., Sikes J.G., Obradovic Z., Dunker A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plewczynski D., Tkacz A., Godzik A., Rychlewski L. A support vector machine approach to the identification of phosphorylation sites. Cell. Mol. Biol. Lett. 2005;10:73–89. [PubMed] [Google Scholar]
- 12.Kim J.H., Lee J., Oh B., Kimm K., Koh I. Prediction of phosphorylation sites using SVMs. Bioinformatics. 2004;20:3179–3184. doi: 10.1093/bioinformatics/bth382. [DOI] [PubMed] [Google Scholar]
- 13.Huang H.D., Lee T.Y., Tzeng S.W., Wu L.C., Horng J.T., Tsou A.P., Huang K.T. Incorporating hidden Markov models for identifying protein kinase-specific phosphorylation sites. J. Comput. Chem. 2005;26:1032–1041. doi: 10.1002/jcc.20235. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz D., Gygi S.P. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 2005;23:1391–12398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 15.Diella F., Cameron S., Gemund C., Linding R., Via A., Kuster B., Sicheritz-Ponten T., Blom N., Gibson T.J. Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics. 2005;5:79. doi: 10.1186/1471-2105-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Via A., Zanzoni A., Helmer-Citterich M. Seq2Struct: a resource for establishing sequence-structure links. Bioinformatics. 2005;21:551–553. doi: 10.1093/bioinformatics/bti049. [DOI] [PubMed] [Google Scholar]
- 17.Hubbard S., Thornton J.M. NACCESS Computer Program. 1993. Department of Biochemistry and Molecular Biology, University College, London. [Google Scholar]
- 18.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 19.Glaser F., Rosenberg Y., Kessel A., Pupko T., Ben-Tal N. The ConSurf-HSSP database: the mapping of evolutionary conservation among homologs onto PDB structures. Proteins. 2005;58:610–617. doi: 10.1002/prot.20305. [DOI] [PubMed] [Google Scholar]
- 20.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ausiello G., Via A., Helmer-Citterich M. Query3d: a new method for high-throughput analysis of functional residues in protein structures. BMC Bioinformatics. 2005;4:S5. doi: 10.1186/1471-2105-6-S4-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dayhoff M.O., Schwartz R.M., Orcutt B.C. A model of evolutionary change in proteins. Atlas Prot. Seq. Struct. 1978;5:345–352. [Google Scholar]
- 23.Ausiello G., Zanzoni A., Peluso D., Via A., Helmer-Citterich M. pdbFun: mass selection and fast comparison of annotated PDB residues. Nucleic Acids Res. 2005;33:W133–W137. doi: 10.1093/nar/gki499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Espanel X., Huguenin-Reggiani M., Van Huijsduijnen R.H. The SPOT technique as a tool for studying protein tyrosine phosphatase substrate specificities. Protein Sci. 1998;11:2326–2334. doi: 10.1110/ps.0213402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walchli S., Espanel X., Harrenga A., Rossi M., Cesareni G., van Huijsduijnen R.H. Probing protein-tyrosine phosphatase substrate specificity using a phosphotyrosine-containing phage library. J. Biol. Chem. 2004;279:311–318. doi: 10.1074/jbc.M307617200. [DOI] [PubMed] [Google Scholar]
- 26.Yaffe M.B., Smerdon S.J. The use of in vitro peptide-library screens in the analysis of phosphoserine/threonine-binding domain structure and function. Annu. Rev. Biophys. Biomol. Struct. 2004;33:225–244. doi: 10.1146/annurev.biophys.33.110502.133346. [DOI] [PubMed] [Google Scholar]
- 27.Murzin A.G., Brenner S.E., Hubbard T., Chothia C. SCOP: a Structural Classification Of Proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]