Abstract
Eukaryotic Subcellular Localization DataBase collects the annotations of subcellular localization of eukaryotic proteomes. So far five proteomes have been processed and stored: Homo sapiens, Mus musculus, Caenorhabditis elegans, Saccharomyces cerevisiae and Arabidopsis thaliana. For each sequence, the database lists localization obtained adopting three different approaches: (i) experimentally determined (when available); (ii) homology-based (when possible); and (iii) predicted. The latter is computed with a suite of machine learning based methods, developed in house. All the data are available at our website and can be searched by sequence, by protein code and/or by protein description. Furthermore, a more complex search can be performed combining different search fields and keys. All the data contained in the database can be freely downloaded in flat file format. The database is available at http://gpcr.biocomp.unibo.it/esldb/.
INTRODUCTION
Subcellular localization is a key feature for characterizing physiological functions of proteins: in eukaryotes compartmentalization finalizes the sets of possible interacting molecules and therefore the biological process(es) in which a protein is involved. Experimental determination of localization is however an expensive and time-consuming procedure. To date it has been carried out only for a narrow subset of known proteins.
Presently only unicellular species have been extensively analyzed by means of high-throughput experiments, such as S.cerevisiae (1,2). Different approaches, such as green fluorescent protein (GFP)-tagging (2) and immunoluminescence (1), agree only on 75% of the annotations and this is mainly due to experimental limitations and possible interference of the tagging procedures on the normal protein trafficking (3,4). Although it is difficult to scale these techniques to more complex organisms, a partial map for mouse liver cells was recently produced (5).
Curated annotations of the subcellular localization, although probably not covering all the available experimental knowledge, are contained in the SwissProt database. The amount of proteins with an experimental annotation listed in SwissProt is different for different species and reaches at the most half of the total amount of proteins in a genome (see below).
The question remains, however, as to how one can obtain a reliable annotation of subcellular localization for the rest of the proteins. A first approach is based on similarity search. Although in principle a change in few residues could result in a change of the localization of a protein, in practice with very few exceptions natural proteins with a sequence identity >30% share the same localization (6–8). In the most successful cases, about two-thirds of a genome can be annotated both by experimental results and similarity search.
The remaining proteins can be annotated only by computational approaches. Many predictors have been developed recently [(4,7–11), a list is available at www.psort.org]. A prerequisite for a predictive method is its capability of well performing when the query sequence shares very low sequence similarity to known proteins. It is therefore important to implement and adopt predictors tested with a rigorous cross-validation procedure on sets of proteins <30% identical with respect to the sequences used for the training. Another important feature to be considered in adopting a predictor is how the relative abundance of localization among different sub-compartments was treated during the training phase. Indeed, with the available data it is difficult to estimate the real proportions between the proteins targeted into the different subcellular localizations; most of the predictive methods tend in fact to overestimate localization types for which more examples are known (8). Moreover the mentioned proportions are likely to be different in different species. It is therefore necessary to adopt predictive methods that attempt to correct bias towards one or more localization classes.
Databases storing information on subcellular localization have been previously described. They include (i) the results of large-scale experiments for the determination of subcellular localization in specific organisms [YGFP (1), for yeast; and ORMDB (5), for mouse]; (ii) the annotations of proteins to be found in organelles [plprot (12), for proteins from plastids; OrganelleDB (13), for proteins in different organelles; and MitoP2 (14), for mitochondrial proteins]. A database collecting all the annotations listed in SwissProt is also available [DBSubLoc (15)]. Finally databases that implement predictors of subcellular localization based on different methods have been reported [LOCtarget (16) and PA-GOSUB (17)]. LOCtarget (16) is specific for structural genomics targets and lists some 50 000 proteins from different organisms. PA-GOSUB (17) contains the annotations of eukaryotic subcellular localization and protein function of different genomes and is based on homology search and Bayesian artificial networks for prediction.
We recently developed BaCelLo, a well-performing balanced method for the prediction of subcellular localization, outperforming previously existing methods for the same task (8). We adopt BaCelLo to annotate whole genomes in association with methods specifically implemented for the prediction of the topology of integral membrane proteins.
In this paper we present eukaryotic Subcellular Localization DataBase (eSLDB), a database of protein subcellular localization which provides an annotation for the entire proteomes of eukaryotic organisms. For each sequence our database contains the experimental localization, when available, the homology-based annotation, when feasible, and the predicted localization computed with the in-house developed machine learning based methods. By this the new database provides more features than other existing databases. To date, five proteomes were fully processed: Homo sapiens, Mus musculus, Caenorhabditis elegans, Saccharomyces cerevisiae and Arabidopsis thaliana.
In summary, eSLDB is, to our knowledge, the first database containing the available experimental and similarity-based annotations for eukaryotic proteomes listing for each protein sequence also the predicted subcellular localization.
DATABASE CONSTRUCTION AND CONTENT
Five different genomes were downloaded as specified: H.sapiens (ENSEMBL NCBI36), M.musculus (ENSEMBL NCBIM36), S.cerevisiae (ENSEMBL SGD1), C.elegans (ENSEMBL CEL150) and A.thaliana (TAIR6).
For each protein the corresponding SwissProt entry in release 50 was found, when existing, searching for exactly matching sequences. The amount of genomic sequences that is deposited in the SwissProt database ranges from 13% for both A.thaliana and C.elegans to 79% for S.cerevisiae (Table 1).
Table 1.
Number of proteins with an experimental or a similarity-based annotation of the subcellular localization
| No. of sequences in the genome | No. of sequences with a SwissProt entry | No. of sequences experimentally annotated | No. of sequences annotated by similarity | |
|---|---|---|---|---|
| Homo sapiens | 48 926 | 12 927 (26%) | 9341 (19%) | 33 225 (68%) |
| Mus musculus | 31 302 | 6228 (20%) | 4669 (15%) | 20 764 (66%) |
| Caenorhabditis elegans | 25 714 | 3327 (13%) | 1612 (6%) | 11 222 (44%) |
| Saccharomyces cerevisiae | 6680 | 5296 (79%) | 3106 (46%) | 3503 (52%) |
| Arabidopsis thaliana | 30 600 | 4030 (13%) | 2645 (9%) | 10 121 (33%) |
The sequences experimentally annotated are included among those annotated by similarity.
For these proteins the experimental annotation was extracted by parsing the ‘Subcellular localization’ section of the COMMENT field of the SwissProt file. Entries annotated as ‘probable’, ‘possible’ or ‘by similarity’ were not considered. The annotations directly or implicitly referring to one of the following 17 classes were taken into account: Nucleus, Cytoplasm, Mitochondrion, Plastid, Golgi, Endoplasmic reticulum, Lysosome, Endosome, Vesicles, Peroxisome, Vacuole, Cell wall, Secretory pathway, Extracellular, Cytoskeleton, Membrane and Transmembrane (Table 1 in Supplementary Data lists the keywords that have been considered for assigning the localization). Only 22% of all the SwissProt entries for the five considered species record the experimental subcellular localization. The rate of experimental annotation ranges from 46% of the S.cerevisiae proteome to <10% for A.thaliana and C.elegans (Table 1).
The ‘Experimental annotation’ column in Table 2 lists the amount of proteins experimentally annotated in each one of the 17 types of considered localization. It is worth mentioning that the same sequence can be annotated in SwissProt with two (or rarely more) different localizations. For example, this happens for proteins that shuttle between the nucleus and the cytoplasm. In these cases the same entry counts two (or more) times in Table 2. It is evident that the amount of proteins in the different localizations spans two orders of magnitude.
Table 2.
Number of sequences in the 17 different subcellular localizations as derived with experimental and similarity-based annotations
| Subcellular localization | Experimental annotation | Similarity-based annotation |
|---|---|---|
| Cell wall | 73 | 404 |
| Cytoplasm | 4468 | 23 492 |
| Cytoskeleton | 519 | 4099 |
| Endoplasmic reticulum | 1058 | 4115 |
| Endosome | 202 | 1091 |
| Extracellular | 340 | 1719 |
| Golgi | 806 | 3231 |
| Lysosome | 208 | 959 |
| Membrane | 1913 | 9956 |
| Mitochondrion | 1829 | 4490 |
| Nucleus | 3413 | 25 812 |
| Peroxisome | 112 | 718 |
| Plastid | 429 | 1525 |
| Secretory pathway | 2157 | 7262 |
| Transmembrane | 6773 | 19 586 |
| Vacuole | 179 | 506 |
| Vesicles | 390 | 2289 |
The sequences experimentally annotated are included among those annotated by similarity.
The best way to annotate the remaining proteins is to search for experimentally annotated sequences sharing high identity (6–8). Since the three eukaryotic kingdoms (Metazoa, Viridiplantae and Fungi) differ in number and types of possible localizations, three kingdom-specific datasets of annotated proteins were extracted from SwissProt. These dataset contains 26 192 sequences for Metazoa, 6370 sequences for Viridiplantae and 4023 sequences for Fungi. All the sequences of the five considered genomes were searched for similar sequences in the appropriate dataset using BLAST (18). When matches were found with an E-value <104 (roughly corresponding to an identity level >30%) the annotation of the best-scoring match was transferred to the query sequence. When multiple matches are found with the same best scoring E-value, all of them are reported in the database. This procedure assigns localization to 55% of the proteins in the database. This rate ranges from 33% of A.thaliana up to 68% for H.sapiens (Table 1).
The ‘similarity-based annotation’ column in Table 2 contains the number of proteins annotated with the above described procedure in each localization (including the sequences experimentally annotated). Also in this case, sequences that end up with a multiple annotation are counted several times.
It appears that a large portion of the sequences, ranging from 32% in H.sapiens up to 67% in A.thaliana, is not endowed with similar counterparts with an annotated localization. In this case subcellular localization can be predicted with specifically suited methods.
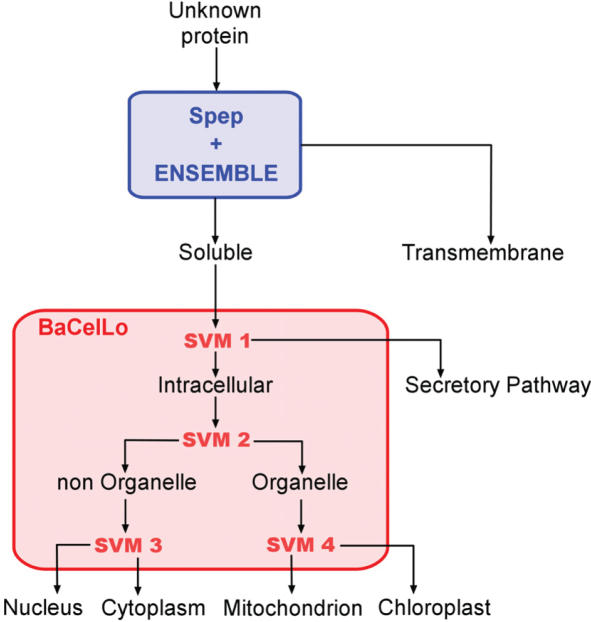
For generating our annotation system we developed a pipeline that comprises previously described methods, all based on machine learning tools and that are proved to outperform most of the available predictors for the same task when rigorous cross-validation procedures are adopted (8). The pipeline is shown in Figure 1. First of all, membrane proteins are discriminated with Spep (19) and ENSEMBLE (20): the former is a neural network based method for predicting the presence of signal peptide while the latter is a method based on neural networks and hidden Markov models for the prediction of the topology of all-alpha transmembrane proteins. When a signal peptide is predicted, it is cleaved from the sequence before predicting the presence and the location of the transmembrane helices. If no transmembrane helix is found, the uncleaved sequence is analyzed using BaCelLo (8), a recently developed tool for predicting the subcellular location of eukaryotic proteins. This is based on a decision tree of support vector machines and it discriminates four localizations in Metazoa and Fungi (cytoplasm, nucleus, extracellular and mitochondrion) and five localizations in Viridiplantae (the same as before plus chloroplast).
Figure 1.
Flow chart of the predicting pipeline adopted in eSLDB. SVM, support vector machine. BaCelLo, Spep and ENSEMBLE are predictive methods described previously (8,17,18).
At the end of the pipeline up to five localizations can be discriminated in Metazoa and Fungi and up to six in Viridiplantae. Although the possible types of localization are 17 (see above), the actual reduction in the number of discriminated localization is due to the lack of an adequate number of non-redundant examples for training. A novelty of BaCelLo is that first takes into consideration that the actual proportion of proteins targeted towards each compartment remains unknown by adopting an equiprobability hypothesis and a balancing procedure (8).
The structure of the predictive system allows annotating the subcellular localization in a hierarchical way. First, all membrane proteins are separated from soluble ones; the latter are then divided into intracellular and secreted. Intracellular proteins are separated in nucleocytoplasmic and organellar; the former are then separated in cytoplasmic and nuclear while the latter, in the case of Viridiplantae, are further divided into mitochondrial and chloroplastic.
The topology of the decision tree and the balancing procedure were adopted for maximizing the prediction performances as evaluated on testing sets independent of the training sets. The best scoring binary decisions are at the top of the tree, the worst-scoring at the bottom. This procedure minimizes the propagation of the errors through the hierarchy of the tree. The predictions are stored in the database along with the hierarchy of the decisions in the pipeline.
All the proteins of a genome, also when experimental and/or homology-based annotation are possible, are annotated by means of the predictive method. In Table 3 the number of proteins predicted in each class is listed.
Table 3.
Number of sequences in the six predicted subcellular localizations
| Subcellular localization | H.sapiens | M.musculus | C.elegans | S.cerevisiae | A.thaliana |
|---|---|---|---|---|---|
| Transmembrane | 10 229 | 7750 | 6593 | 1657 | 8079 |
| Secretory | 7816 | 4971 | 5172 | 348 | 3001 |
| Nucleus | 12 358 | 6820 | 4733 | 1717 | 7649 |
| Cytoplasm | 14 720 | 9356 | 6280 | 1710 | 6033 |
| Mitochondrion | 3630 | 2326 | 1454 | 1112 | 963 |
| Chloroplast | — | — | — | — | 4875 |
Table 4 contains the evaluation of the coverage and the accuracy of the prediction for the proteins of H.sapiens, as compared with both the experimental and the similarity derived annotations. We considered 6444 unique proteins experimentally annotated and 25 134 unique sequences for which a similarity-based annotation is available. Table 4 also lists the distribution of these proteins among the different classes. The coverage is computed as the fraction of correctly predicted sequences in each class over the number of proteins belonging to the class. The accuracy is the fraction of correctly predicted proteins over the total number of proteins predicted in the class. The agreement between the annotations and the prediction is good, especially when predictions are compared with the experimental annotations and the higher levels of the hierarchical prediction are considered.
Table 4.
Performance of the prediction pipeline as compared with the experimental and the similarity-based annotations
| Subcellular localization | With respect to the experimental annotations | With respect to the similarity-based annotations | ||||
|---|---|---|---|---|---|---|
| No. of proteins | Coverage (%) | Accuracy (%) | No. of proteins | Coverage (%) | Accuracy (%) | |
| Transmembrane | 2244 | 87.7 | 93.0 | 6660 | 76.5 | 82.3 |
| Soluble | 4200 | 92.4 | 86.6 | 18 474 | 92.3 | 83.0 |
| Secretory pathway | 865 | 82.3 | 60.6 | 2844 | 68.8 | 48.3 |
| Intracellular | 3364 | 89.0 | 90.5 | 15 776 | 87.2 | 83.5 |
| Nucleus or cytoplasm | 3013 | 88.2 | 90.7 | 14 788 | 83.0 | 82.6 |
| Nucleus | 2107 | 66.5 | 85.1 | 8973 | 54.5 | 69.6 |
| Cytoplasm | 1410 | 56.7 | 62.4 | 8779 | 51.0 | 57.2 |
| Organelle (mitochondrion) | 398 | 58.0 | 60.9 | 1230 | 42.3 | 31.9 |
The indentation of the subcellular localization names reflects the hierarchy of the prediction (see Figure 1). Coverage = (no. of proteins of class i predicted as class i)/(total no. of proteins in class i). Accuracy = (no. of proteins of class i predicted as class i)/(total no. of proteins predicted as class i).
DATABASE ACCESSION
All the data are available at the website (http://gpcr.biocomp.unibo.it/esldb) and can be accessed by protein code, as derived from the original and above-mentioned versions of the genomic databases, or by protein description, as derived from the SwissProt entries. Alternatively the sequence of interest can be submitted and, by means of the MD5 encoding, the match engine searches for identical sequences deposited in the database. Moreover, complex searches can be performed combining the different annotation methods and the different localizations. The use of Boolean connectors can improve the complexity of the queries.
The entries matching the query keys can either be displayed or downloaded in a tabular format that contains the experimental and the similarity-derived annotation and the localization prediction for each match. Pages containing more details about each one of the proteins (Figure 2) are linked to the protein codes and contain the sequence, the description and the link to the SwissProt file when an experimental annotation exists.
Figure 2.
Output page of eSLDB for a single protein chain.
When the sequence can be annotated by similarity, the SwissProt entry corresponding to the most similar sequence is reported together with the E-value as computed by BLAST (18). In both the tabular and the detailed pages, the results of the prediction are given together with the complete path through the decision tree. This information can be useful since, as we commented in the previous section, the accuracy of the prediction lowers as the number of discriminated classes increases. This means that annotations in the macro-classes are endowed with a higher reliability.
All the data contained in the database can be freely downloaded in flat file format. The database resides in a PostgreSQL server and the web interface has been implemented using Python, HTML 4.0 and CSS 2.0 languages.
More available eukaryotic proteomes are currently under process and will be added to the database. Moreover we plan to regularly update the database as new versions of SwissProt or new releases of the considered proteomes will be available.
eSLDB is publicly available at http://gpcr.biocomp.unibo.it/esldb/.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
R.C. acknowledges the receipt of the following grants: PNR 2001–2003 (FIRB art.8) for a project on Bioinformatics for Genomics and Proteomics, a FIRB 2003 LIBI-International Laboratory of Bioinformatics and the support to the Bologna node of the Biosapiens Network of Excellence project within the European Union's VI Framework Programme (contract number LSHG-CT-2003-503265). P.F. acknowledges MIUR for a grant on Proteases. A.P. and P.L.M. are supported by a FIRB 2003-LIBI grant. Funding to pay the Open Access publication charges for this article was provided by FIRB 2003/Biosapiens.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kumar A., Agarwal S., Heyman J.A., Matson S., Heidtman M., Piccirillo S., Umansky L., Drawid A., Jansen R., Liu Y., et al. Subcellular localization of the yeast proteome. Genes Dev. 2002;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 3.Simpson J.C., Pepperkok R. Localizing the proteome. Genome Biol. 2003;4:240. doi: 10.1186/gb-2003-4-12-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nair R., Rost B. Mimicking cellular sorting improves prediction of subcellular localization. J. Mol. Biol. 2005;348:85–100. doi: 10.1016/j.jmb.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 5.Forster L.J., de Hoog C.L., Zhang Y., Xie X., Mootha V.K., Mann M. A Mammalian organelle map by protein correlation profile. Cell. 2006;125:187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 6.Nair R., Rost B. Sequence conserved for subcellular localization. Protein Sci. 2002;11:2836–2847. doi: 10.1110/ps.0207402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu C.S., Chen Y.C., Lu C.H., Hwang J.K. Prediction of protein subcellular localization. Protein. 2006;64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- 8.Pierleoni A., Martelli P.L., Fariselli P., Casadio R. BaCelLo: a Balanced subCellular Localization predictor. Bioinformatics. 2006;22:e408–e416. doi: 10.1093/bioinformatics/btl222. [DOI] [PubMed] [Google Scholar]
- 9.Nakai K., Horton P. PSORT: a program for detecting the sortine signals of proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–35. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 10.Bhasin M., Raghava G.P.S. ESLpred: SVM-based method for subcellular localization of eukaryotic proteins using dipeptide composition and PSI-BLAST. Nucleic Acids Res. 2004;32:W414–W419. doi: 10.1093/nar/gkh350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guda C., Subramaniam S. pTARGET: a new method for predicting protein subcellular localization in eukaryotes. Bioinformatics. 2005;21:3963–3969. doi: 10.1093/bioinformatics/bti650. [DOI] [PubMed] [Google Scholar]
- 12.Kleffmann T., Hirsch-Hoffmann M., Gruissem W., Baginsky S. plprot: a comprehensive proteome database for different plastid types. Plant Cell Physiol. 2006;47:432–436. doi: 10.1093/pcp/pcj005. [DOI] [PubMed] [Google Scholar]
- 13.Wiwatwattana N., Kumar A. OrganelleDB: a cross-species database of protein localization and function. Nucleic Acids Res. 2005;33:D598–D604. doi: 10.1093/nar/gki071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prokisch H., Andreoli C., Ahting U., Heiss K., Ruepp A., Scharfe C., Meitinger T. MitoP2: the mitochondrial proteome database-now including mouse data. Nucleic Acids Res. 2006;34:D705–D711. doi: 10.1093/nar/gkj127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo T., Hua S., Ji X., Sun Z. DBSubLoc: database of protein subcellular localization. Nucleic Acids Res. 2004;32:D122–D124. doi: 10.1093/nar/gkh109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair R., Rost B. LOCnet and LOCtarget: sub-cellular localization for structural genomics targets. Nucleic Acids Res. 2004;32:W517–W521. doi: 10.1093/nar/gkh441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu P., Szafron D., Greiner R., Wishart D.S., Fyshe A., Pearcy B., Poulin B., Eisner R., Ngo D., Lamb N. PA-GOSUB: a searchable database of model organism protein sequences with their predicted Gene Ontology molecular function and subcellular localization. Nucleic Acids Res. 2005;33:D147–D153. doi: 10.1093/nar/gki120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Fariselli P., Finocchiaro G., Casadio R. SPEPlip: the detection of signal peptide and lipoprotein cleavage sites. Bioinformatics. 2003;19:2498–2499. doi: 10.1093/bioinformatics/btg360. [DOI] [PubMed] [Google Scholar]
- 20.Martelli P.L., Fariselli P., Casadio R. An ENSEMBLE machine learning approach for the prediction of all-alpha membrane proteins. Bioinformatics. 2003;19:i205–i211. doi: 10.1093/bioinformatics/btg1027. [DOI] [PubMed] [Google Scholar]