Abstract
Estrogen has a profound impact on human physiology affecting transcription of numerous genes. To decipher functional characteristics of estrogen responsive genes, we developed KnowledgeBase for Estrogen Responsive Genes (KBERG). Genes in KBERG were derived from Estrogen Responsive Gene Database (ERGDB) and were analyzed from multiple aspects. We explored the possible transcription regulation mechanism by capturing highly conserved promoter motifs across orthologous genes, using promoter regions that cover the range of [−1200, +500] relative to the transcription start sites. The motif detection is based on ab initio discovery of common cis-elements from the orthologous gene cluster from human, mouse and rat, thus reflecting a degree of promoter sequence preservation during evolution. The identified motifs are linked to transcription factor binding sites based on the TRANSFAC database. In addition, KBERG uses two established ontology systems, GO and eVOC, to associate genes with their function. Users may assess gene functionality through the description terms in GO. Alternatively, they can gain gene co-expression information through evidence from human EST libraries via eVOC. KBERG is a user-friendly system that provides links to other relevant resources such as ERGDB, UniGene, Entrez Gene, HomoloGene, GO, eVOC and GenBank, and thus offers a platform for functional exploration and potential annotation of genes responsive to estrogen. KBERG database can be accessed at http://research.i2r.a-star.edu.sg/kberg.
INTRODUCTION
Estrogen affects many aspects of body functioning through influence of numerous genes. The mechanisms of gene reaction to estrogen are complex and multiple factors are involved. The changes of gene expression level after estrogen stimulation could be the result of hormone–receptor interaction via estrogen response elements (EREs), activation of the target gene transcription through interaction with co-activators or over non-genomic pathways (1,2). A number of genes whose transcription is affected by estrogen have been well documented in manually curated estrogen responsive genes database (ERGDB) (3). Although ERGDB provides information about genes responsive to estrogen proved in experiments, it does not give any clue about the way these genes are regulated and what functional properties these genes may have. Thus, to facilitate functional analysis of estrogen responsive genes we do need a different framework not available in ERGDB. The new KnowledgeBase for Estrogen Responsive Genes (KBERG) provides users with such options to explore some of the genes' functional properties and complements the experimental-information-rich ERGDB.Since the set of estrogen responsive genes covers a broad spectrum of functionality, belongs to many different pathways and may be related to many diseases, it is thus a significant challenge to characterize functions of these genes and to provide clues on how these genes are regulated and what potential effects we should expect. As a part of functional analysis, transcription regulation of these genes is an important step to unravel parts of the underlying estrogen driven signal pathways and to help develop insights of estrogen effects that can bring us closer to efficient therapies for estrogen-related diseases.
The binding of transcription factors (TFs) to specific regulatory elements is a primary mechanism for controlling gene transcription. Promoter region is the control center for such transcription regulation, where multiple transcription factor binding sites (TFBSs) are clustered forming specific regulatory modules (4). A comparative genome study found that the complexity of the organism is not correlated to the number of genes (5) but to the complexity of gene regulation systems. Recent studies (6,7) document that the richness of transcript variants stemming from multiple transcription start sites (TSSs) in the vast majority of mammalian gene loci are controlled in various ways at the promoter level. In brief, dynamic gene regulation is achieved through multi-level manipulation in which transcription level control is an essential step. The sophisticated gene regulatory modes characterized by a set of TFBSs determine when, where and how gene will express based on the integration of internal and external signals. Broad gene groups are subjected to different regulatory programs as shown in (8). Thus, identification of the structure and organization of TFBSs in promoters is the primary step towards gaining understanding of the gene's transcription regulation. However, detection of functional TFBSs is challenging due to features such as short motif length, high degeneration and flexible distance location. Position weight matrix (PWM) approach has been widely used in TFBS detection although high false positive rate is the main drawback (9). Comparative genomic approach that searches for highly preserved motifs across different species has been applied to improve the accuracy of prediction (10). The method has been corroborated in a genome-wide screen for high-affinity ERE in human and mouse (11). Through analyzing remarkably consistent patterns in orthologous promoters, we aim to provide highly selective promoter motifs that could be involved in the control of transcription of genes responsive to estrogen.
In addition to the detection of conserved motifs in promoter regions, mapping these motifs to known TFBS provides hints of the interaction with TFs and the formation of transcription initiation complex. As typical TFs, estrogen receptors (ERs) influence their target genes by either directly binding to ERE or indirectly interacting with a wide range of co-regulators forming complexes that bind to promoters (12). The availability of co-regulators is tissue specific and development stage specific. However, the experimental detection of co-regulators is time and labor consuming. In KBERG, we provided partial solution regarding these issues by matching the highly preserved motifs in promoters to the TRANSFAC database. This allowed identification of TFBSs that potentially bind identified motifs. As a result, we found that SP1-binding site is the most frequent TFBS in promoter regions in our collection. This prediction result is in good concordance with promoter deletion studies that specifies the essential role of ER/SP1 complex for gene's response to estrogen at transcription level (13,14). The top 10 most frequently found TFBSs are given on the KBERG website under the section ‘Statistic’.
Systematic assessment of gene functionality can help us to understand the complexity of living cells. It can be achieved by studying genes that share similarities in sequence and expression. Co-expression is a biological property that reflects functional relationship among a group of genes. Co-expressed genes tend to share certain regulatory mechanism by using similar sets of TFs. The phenomenon of co-expression can also be a result of the biological dependency among a group of proteins. It has been observed that proteins working closely in the same interaction network are also likely to be co-expressed under the same biological condition. Sets of NF-κB target genes were overexpressed in aggressive breast cancer comparing with non-aggressive breast cancer (15). Thus, clustering genes based on their expression profile is one method to potentially deduce gene functional characteristics.
Another systematic approach to cluster gene for functional annotation is to group them based on Gene Ontology (GO) (16) categories by using well-defined terminology to describe biological process, molecular function and cellular component where genes exert their activity. Owing to the important impact of estrogen in human physiology and its involvement in numerous diseases, we believe that functionally characterizing estrogen responsive genes can benefit both basic research and clinical applications. However, up to date, there is no supporting system that facilitates such functional exploration.
To complement currently available estrogen-related database and to provide researchers with comprehensive information related to potential functionality of estrogen responsive genes, we developed KBERG. Users can obtain integrated information covering the following aspects: orthologous gene information from human, mouse and rat; ab initio determined DNA motifs preserved in promoter regions; possible relation between motifs and known TFs based on the TRANSFAC database (17). The system also helps user to deduce functional association of gene groups annotated by the same GO term or possessing the same expression characteristics. Links to UniGene, Entrez Gene, HomoloGene, GenBank, GO (16), eVOC (18) and ERGDB (3) make KBERG a unique site for exploring estrogen responsive genes. At current stage, KBERG contains information of 1362 homolog groups and 1526 estrogen responsive genes from human, mouse and rat. The detail statistic data are available in the website. Although KBERG is specialized for estrogen responsive gene, the framework can be adapted to other gene groups for their functional exploration and annotation. KBERG is free for academic and non-profit users and can be accessed at http://research.i2r.a-star.edu.sg/kberg.
DATABASE CONSTRUCTION
We have created a comprehensive system that presents functional properties of estrogen responsive genes from multiple viewpoints. All genes in KBERG are experimentally proved to respond to estrogen—their expression levels are significantly altered by estrogen. These genes were derived from a manually curated estrogen-specific database, ERGDB (3).
KBERG is constructed through several modules. Each module performs specialized functions that provide users a convenient way to find desired information. The modules are described in the following sections.
Promoter analysis and motif detection module
The goal of this module is to identify highly conserved promoter motifs from groups of orthologous genes. To facilitate promoter analysis, we built an in-house promoter database. Using genes derived from ERGDB as a primary list, orthologous genes from human (Homo sapiens), mouse (Mus musculus) and rat (Rattus norvegicus) genomes are downloaded from the NCBI HomoloGene site (ftp://ftp.ncbi.nlm.nih.gov/pub/HomoloGene/).
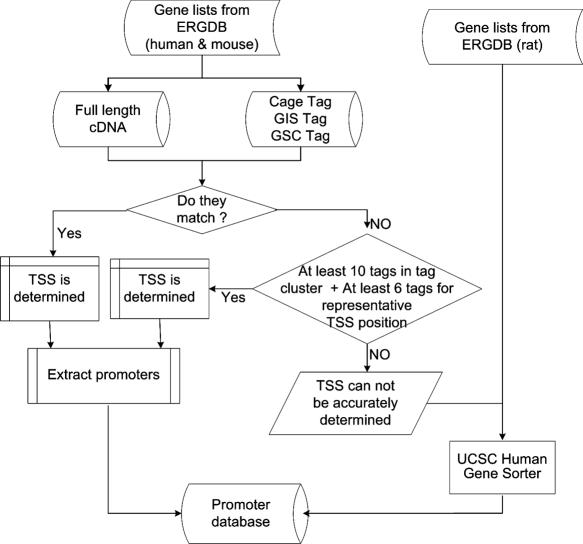
Recent genome comparison study (7) indicates that the availability of precise TSSs increases the accuracy of finding conserved motifs by focusing on phylogenetically relevant promoter sequences. To accurately analyze functional motifs from selected promoters, we first identified TSSs with solid experimental evidence. Each mouse and human TSS is identified through multiple selection steps. Supporting evidences include (i) full-length cDNA from FANTOM3 database (http://fantom.gsc.riken.jp); (ii) CAGE tags from RIKEN tag-cluster database (http://fantom.gsc.riken.jp/); and (iii) 5′-UTR sequences based on UCSC Human Gene Sorter database (http://genome.ucsc.edu/cgi-bin/hgNear/). Details of the promoter processing are illustrated in Figure 1.
Figure 1.
Flow chart for promoter extraction.
We limited search for conserved motifs to the region [−1200, +500] relative to TSS as most functional TFBSs are located in these region, although some can be found several thousand base pairs away from the TSS. The shorter promoter sequence also reduces the noise in motif identification. Dragon Motif Builder (19,20) was used to identify ab initio DNA motifs in each orthologous promoter sequence group. The highly preserved motifs were detected without any presumption of what may be TFs that control activation of the respective genes. However, sets of motifs within close mutual distance and in preserved orders in orthologous sequences are considered embedding more reliable and more significant information for transcriptional regulation. We expect some of these predicted motifs will turn out to be functional TFBSs, although further experimental validation is needed.
In the report page, we provide both graphic overview of the motif arrangements and details of the motif sequence information, which enable users to further investigate these motifs experimentally, or to compare them to the known TFBS through other data sources. The length of motifs we considered was 9 bp. In each orthologous promoter group, five motifs were selected based on their sequence preservation, motif combination order and maximum allowed distance between motifs (50 bp). Details of motif family consensus, matrix, positions, strands and rough graphic distribution in the orthologous promoter group are provided in the report page. We also matched each motif to the TRANSFAC Professional database, version 9.2, to suggest possible TFBSs that these motifs contain or overlap with. We allowed one mismatch in 9 bp motif using the Patch program (TRANSFAC) and the similarity score was given in the report page. Although the evaluation of motif prediction is beyond the scope of this paper, we would like to suggest the user to take full advantage of the information in the report page. The matching score of potential TFBS with TRANSFAC, the total information content and position distribution, the degree of sequence preservation and the combination pattern with neighboring motifs are valuable indicators to estimate the accuracy of prediction.
Gene Ontology grouping module
This module enables users to group estrogen responsive genes that share common GO category annotation. In this way genes in KBERG can be clustered into functionally related groups. Using GO terms we associate wide range of biological knowledge to groups of gene. By examining the frequency of the used GO terms for the gene group, a user can estimate the significant features shared by the group of genes. The ‘Browse Gene Ontology’ function in KBERG allows user to select estrogen responsive genes that share the same GO annotation term and the statistic information about the term. For example, there are 101 genes under the GO term ‘receptor activity’ (GO:0004872) and only 22 genes belong to ‘steroid hormone receptor activity’ (GO:0003707). Users can easily obtain two relevant but not identical lists of genes to study receptors with specific annotation and with broad annotation. We believe that clustering genes according to GO terms is a complementary way to associate genes with their functions and a logical way to assess genes. Allowing user to download the list of classified genes is one of the helpful features of the system.
eVOC module
eVOC (18) associates human gene expression data with controlled ontology terms in sets of hierarchical vocabularies, covering categories such as anatomical system, cell type, pathology, developmental stage, etc. We use eVOC ontology in our system to provide an alternative approach to cluster gene for functional assessment. The expression data provide extensive information about the gene co-expression under various biological conditions, which, although coarse, becomes essential indicators for gene co-regulation and interaction. Expression results also represent important gene behaviors that should not be missed out in gene functional characterization. In KBERG, we integrated eVOC (version 2.7) to reveal gene expression in terms of human organ, cell type, disease and age. Thus, a user can easily find out some of the associated information for gene expression (organ and cell type, whether in normal or diseased tissues and in the age group). The downloadable list of classified genes is provided and could be useful for user's further exploration.
SUMMARY
Designed for estrogen-related research, KBERG provides a wide range of information about estrogen responsive genes through promoter motifs and functional associations via well-defined vocabularies from GO and eVOC. We summarized the main functions of the system in Table 1. In brief, through searching this system, within a short period of time, a user can gain valuable information related to either individual gene or a group of genes that share the same ontology description. However, there is limitation of the system that user needs to bear in mind. The promoter motif prediction is a pure computational estimation and needs to be verified by experiments. Also, classifying gene based on ontology categories may not always be the best way to reveal its biological function. Thus, the result of interpretation relies on the user's knowledge.
Table 1.
Summarized functions of KBERG
| Web page | What you can get? |
|---|---|
| Search page | (i) Search can be done by Gene Symbol, Gene ID, accession number, GO or anatomic terms |
| (ii) Examples for each searching method enable user to learn how to use the system | |
| Browse Gene | (i) Alphabetic list of gene names |
| (ii) Links to UniGene, Entrez Gene and Explore page | |
| Explore Gene | (i) Gene information with gene name and description |
| (ii) Otholog gene information with links to ortholog genes and their sequences (human, mouse and rat only) | |
| (iii) Promoter analysis | |
| (iv) Expression information | |
| (v) Gene Ontology | |
| Promoter Analysis | (i) Promoter sequences |
| (ii) Ab initio detected motifs | |
| Ab initio motif detection | (i) Prediction and graphic presentation of motifs in promoter regions [−1200, +500] bp |
| (ii) Details of motif position, matrix, information content and consensus | |
| (iii) Possible TF site suggested by TRANSFAC with score | |
| Expression Info from eVOC | (i) List of eVOC terms associated with the gene in each category |
| (ii) Links to a group of genes sharing the same eVOC term | |
| Gene Ontology Info | (i) List of GO terms describing the gene |
| (ii) Links of each term to GO for genes sharing the same term in KBERG | |
| Browse GO | (i) Allows browsing in the three GO categories (biological process, molecular function, cellular component) |
| (ii) List of GO terms and the number of genes that share annotation by that term | |
| (iii) Both terms and number of gene allow sorting | |
| (iv) Downloadable gene list for the selected GO term | |
| Browse eVOC | (i) List of terms used in eVOC anatomic categories and the number of genes sharing that annotation term |
| (ii) Sorting of both terms and number of genes | |
| (iii) Downloadable gene list for the selected organ/tissue | |
| Link to ERGDB | (i) Gene name and sequences |
| (ii) Experimental information, reference papers and link to PubMed | |
| (iii) ERE prediction in gene promoter region [−4500, +500] bp |
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by Institute for Infocomm Research, Singapore.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nilsson S., Gustafsson J.A. Estrogen receptor transcription and transactivation: basic aspects of estrogen action. Breast Cancer Res. 2000;2:360–366. doi: 10.1186/bcr81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrington W.R., Kim S.H., Funk C.C., Madak-Erdogan Z., Schiff R., Katzenellenbogen J.A., Katzenellenbogen B.S. Estrogen dendrimer conjugates that preferentially activate extranuclear, nongenomic versus genomic pathways of estrogen action. Mol. Endocrinol. 2006;20:491–502. doi: 10.1210/me.2005-0186. [DOI] [PubMed] [Google Scholar]
- 3.Tang S., Han H., Bajic V.B. ERGDB: Estrogen Responsive Genes DataBase. Nucleic Acids Res. 2004;32:D533–D536. doi: 10.1093/nar/gkh083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Werner T., Fessele S., Maier H., Nelson P.J. Computer modeling of promoter organization as a tool to study transcriptional coregulation. FASEB J. 2003;17:1228–1237. doi: 10.1096/fj.02-0955rev. [DOI] [PubMed] [Google Scholar]
- 5.Claverie J.M. Gene number. What if there are only 30,000 human genes? Science. 2001;291:1255–1257. doi: 10.1126/science.1058969. [DOI] [PubMed] [Google Scholar]
- 6.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 7.Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nature Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 8.Bajic V.B., Tan S.L., Christoffels A., Schonbach C., Lipovich L., Yang L., Hofmann O., Kruger A., Hide W., Kai C., et al. Mice and men: their promoter properties. PLoS Genet. 2006;2:e54. doi: 10.1371/journal.pgen.0020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Q., Liu J.S. Modeling within-motif dependence for transcription factor binding site predictions. Bioinformatics. 2004;20:909–916. doi: 10.1093/bioinformatics/bth006. [DOI] [PubMed] [Google Scholar]
- 10.Blanchette M., Schwikowski B., Tompa M. Algorithms for phylogenetic footprinting. J. Comput. Biol. 2002;9:211–223. doi: 10.1089/10665270252935421. [DOI] [PubMed] [Google Scholar]
- 11.Bourdeau V., Deschenes J., Metivier R., Nagai Y., Nguyen D., Bretschneider N., Gannon F., White J.H., Mader S. Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol. Endocrinol. 2004;18:1411–1427. doi: 10.1210/me.2003-0441. [DOI] [PubMed] [Google Scholar]
- 12.Shao W., Brown M. Advances in estrogen receptor biology: prospects for improvements in targeted breast cancer therapy. Breast Cancer Res. 2004;6:39–52. doi: 10.1186/bcr742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleming J.G., Spencer T.E., Safe S.H., Bazer F.W. Estrogen regulates transcription of the ovine oxytocin receptor gene through GC-rich SP1 promoter elements. Endocrinology. 2006;147:899–911. doi: 10.1210/en.2005-1120. [DOI] [PubMed] [Google Scholar]
- 14.Khan S., Abdelrahim M., Samudio I., Safe S. Estrogen receptor/Sp1 complexes are required for induction of cad gene expression by 17β-estradiol in breast cancer cells. Endocrinology. 2003;144:2325–2335. doi: 10.1210/en.2002-0149. [DOI] [PubMed] [Google Scholar]
- 15.Van Laere S.J., Auwera I., Eynden G.G., Elst H.J., Weyler J., Harris A.L., van Dam P., Van Marck E.A., Vermeulen P.B., Dirix L.Y. Nuclear factor-κB signature of inflammatory breast cancer by cDNA microarray validated by quantitative real-time reverse transcription–PCR, immunohistochemistry, and nuclear factor-κB DNA-binding. Clin. Cancer Res. 2006;12:3249–3256. doi: 10.1158/1078-0432.CCR-05-2800. [DOI] [PubMed] [Google Scholar]
- 16.Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matys V., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelso J., Visagie J., Theiler G., Christoffels A., Bardien S., Smedley D., Otgaar D., Greyling G., Jongeneel C.V., McCarthy M.I., et al. eVOC: a controlled vocabulary for unifying gene expression data. Genome Res. 2003;13:1222–1230. doi: 10.1101/gr.985203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L., Huang E., Bajic V.B. Some implementation issues of heuristic methods for motif extraction from DNA sequences. Int. J. Comp. Syst. Signals. 2005;6:3–12. [Google Scholar]
- 20.Huang E., Yang L., Chowdhary R., Kassim A., Bajic V.B. In Proceedings of the Information Processing and Living Systems. Singapore: World Scientific Press; 2005. An algorithm for ab initio DNA motif detection; pp. 611–614. [Google Scholar]