Abstract
The Candida Genome Database (CGD, http://www.candidagenome.org/) contains a curated collection of genomic information and community resources for researchers who are interested in the molecular biology of the opportunistic pathogen Candida albicans. With the recent release of a new assembly of the C.albicans genome, Assembly 20, C.albicans genomics has entered a new era. Although the C.albicans genome assembly continues to undergo refinement, multiple assemblies and gene nomenclatures will remain in widespread use by the research community. CGD has now taken on the responsibility of maintaining the most up-to-date version of the genome sequence by providing the data from this new assembly alongside the data from the previous assemblies, as well as any future corrections and refinements. In this database update, we describe the sequence information available for C.albicans, the sequence information contained in CGD, and the tools for sequence retrieval, analysis and comparison that CGD provides. CGD is freely accessible at http://www.candidagenome.org/ and CGD curators may be contacted by email at candida-curator@genome.stanford.edu.
INTRODUCTION
Candida albicans is a serious human health concern, especially for the growing population of immunocompromised patients (1,2). C.albicans is a common fungal organism that can live as a commensal in its mammalian host, and it does so in a large fraction of the human population (3). It can also enter a pathogenic state, causing painful mucosal infections (e.g. oral thrush and vaginitis) or deadly systemic disease (4,5).
The Candida Genome Database (CGD, http://www.candidagenome.org/) grew out of the Candida research community's need for a centralized resource for genomic sequence and gene annotation, and for improved access to the wealth of published experimental data about Candida molecular biology. CGD is based on the framework of the Saccharomyces Genome Database (SGD, available at http://www.yeastgenome.org/) (6). CGD became available online in August 2004. In the past 2 years CGD has grown in many respects, with greatly expanded curation of the experimental literature (see Table 1). Results of large-scale studies have been incorporated into or linked from CGD in several ways: the Gene Ontology terms assigned by the Candida Annotation Working Group have been added to CGD GO Annotations (7); an archive of publicly available large-scale datasets has been created; and gene-specific Locus pages in CGD have been linked to experimentally determined transcription modules (8). Additional community resources have also been added, including a Candida labs directory and a listing of Candida-related postdoctoral positions and other job opportunities.
Table 1.
Candida Genome Database curation statistics. September 2006 and March 2005 statistics are presented
| CGD curation statistics | March 2005 | September 2006 | References cited 2006 |
|---|---|---|---|
| Curated gene descriptions | 1455 | 3988 | 829 |
| Genes with phenotype annotations | 652 | 867 | — |
| Total phenotype annotations | 2393 | 3864 | 587 |
| Genes with GO annotations | 3673 | 4066 | — |
| Total gene ontology annotations | 14 121 | 16 920 | — |
| Total literature-based GO annotations | 2617 | 4439 | 940 |
The ‘Total GO annotations’ figure includes the similarity-based predictions generated by the Annotation Working Group, whereas the ‘Total literature-based GO annotations’ figure excludes this set.
This update highlights the sequence resources added to CGD over the past 2 years, a period in which there have been significant advances in the assembly, annotation and availability of the C.albicans genome sequence. A large-scale community-based revision of the C.albicans genome assembly was completed and has been recently released to the public (7,9). During this time, CGD has incorporated extensive DNA and protein sequence information from multiple versions of the C.albicans genome assembly, including Assembly 20, along with tools for accessing and comparing the sequences.
C.albicans sequence assemblies
The latest C.albicans genome assembly, Assembly 20, is a collaborative effort of groups at the Biotechnology Research Institute of the National Research Council of Canada, the University of Minnesota, and Chiba University of Japan (9). Assembly 20 follows Assembly 19, which was released by the Stanford Genome Technology Center in 2004 (10), and improves upon it in a number of ways. Most notably, Assembly 20 is a chromosome-level assembly; the 266 Assembly 19 contigs that span the haploid complement of the genome have been mapped onto the eight C.albicans chromosomes and joined into nearly contiguous sequence. Numerous corrections to the sequence and to the predicted boundaries of ORFs have also been made as part of this effort (9).
However, Assembly 20 does not yet fully replace Assembly 19. Assembly 19 is a diploid assembly, which for many regions of the genome includes two contigs, corresponding to polymorphic allelic regions that show significant sequence differences. In contrast, Assembly 20 is a haploid assembly, and in generation of Assembly 20, updates to Assembly 19 have been made in only one allele of each pair. It should also be noted that Assembly 20 does not represent a haploid set of chromosomes from the sequenced strain per se, but instead is a mosaic of the two haplotypes. Thus, it is important that access to Assembly 19 sequences be maintained even though Assembly 20 is considered to be a more complete assembly. In addition, earlier assemblies of the genomic sequence and their associated gene nomenclatures are in widespread use in the research community and in the scientific literature. CGD continues to provide easy access to all versions of the genome sequence (including the outdated ones) while minimizing confusion, despite the necessarily complex nature of the situation.
In CGD, the primary sequence for each gene is the sequence that is specified in Assembly 20. Where the sequence and annotation of individual genes has changed, it is essential for the research community to have access to the different versions, not only to be able to understand exactly what has changed, but also to facilitate a critical evaluation of the sequence and to facilitate the continuing refinement process. To this end, CGD makes multiple versions of the sequence available to the user community. The Assembly 20 and 19 sequences of each gene (and of both alleles, if available) are readily available from each gene's Locus page, and the genomic context on the chromosome (from Assembly 20) or the contig(s) (from Assembly 19) can be explored using the GBrowse genome browser (11). Sequences from all of the assemblies, including the archived Assemblies 4 and 6, are available for query by BLAST and for bulk download, as described in more detail below.
Gene names and identifiers
Frequently, users need to retrieve sequence information for a single gene, or for a small number of genes. The most direct route to this sequence information is through the Locus page. CGD is gene-centric in nature; the website is organized around Locus pages that provide access to all available information about each gene, including the literature-derived information about the function, role and localization of the gene product, the phenotypes caused by mutation of the gene, the experimental literature that describes the gene, and, of course, its sequence. To make it possible for users to easily access sequence and other information about any given gene, CGD collects all of the names by which each gene has been called, and makes all of these gene name synonyms, or ‘aliases’, searchable. The gene names in CGD come from several sources. The genetic-style names of format ‘ABC1’ have been manually collected by CGD curators from the published literature (12). Numerically based names (e.g. Contig4-3061_0012, orf6.8002 and orf19.5007) were assigned to the predicted protein-coding genes during the annotation of Assemblies 4, 6 and 19, respectively (7,10). At CGD, sequence comparisons were used to determine the mapping between the ORFs from Assemblies 4 and 6 and the corresponding genes in Assembly 19, and the appropriate aliases were added to each Locus page. The ORFs in Assembly 20 share the names of format ‘orf19.#’ that were assigned during production of Assembly 19. Names of format ‘IPF.#’ (e.g. IPF22272.1 and IPF1819.1) and CA# (e.g. CA5255) were assigned by CandidaDB (13). Additionally, names of format ‘CaJ7.#’ have been assigned to ORFs on Chromosome 7 (14). Cases in which confusion exists regarding the assignment of a give gene name are documented in CGD Nomenclature Notes, which appear prominently on the Locus page so that users are alerted to the situation.
In the future, it may be desirable to adopt a chromosomal position-based systematic gene nomenclature, similar to the one used in the Saccharomyces cerevisiae community. If the Candida research community decides to develop such a system, CGD will facilitate the process and will adopt the new nomenclature as the primary systematic gene identifiers.
Sequence visualization and retrieval at CGD
CGD provides numerous ways to retrieve sequence data. The CGD Sequence Retrieval Tool is accessible via the ‘Get Sequence’ link, located on the left-hand sidebar on the Home Page, under Search Options. The tool retrieves sequences specified by gene name, alias, systematic ORF name or allele name. It also allows the user to define how much of the upstream and downstream flanking sequences to retrieve, to select only the coding sequence (without introns), or to translate the sequence to protein. The output is available in either FASTA or GCG format. The tool is also able to retrieve an entire chromosome, contig, or a particular region of a chromosome or contig identified by starting and ending coordinates.
Several types of sequence may also be retrieved from individual CGD Locus pages using options on the ‘Retrieve Sequences’ pull-down menu. The options include the genomic DNA (with introns), the coding sequence (with introns removed), the genomic DNA with 1 kb upstream and downstream flanking regions, and the predicted protein sequence (ORF translation). The sequences in Assembly 20 and Assembly 19 are available from this menu. In cases where the two diploid alleles are known to differ in sequence in Assembly 19, both of these sequences are available. The selected sequence is displayed in a browser window in FASTA format, and may be copied and pasted into another application.
Sequence information may be retrieved in bulk from the CGD website. The downloadable files are indexed on the CGD Downloads page (http://www.candidagenome.org/DownloadContents.shtml). The raw sequences of C.albicans chromosomes from Assembly 20 are available, as well as an archive of contigs from previous assemblies (Assemblies 4, 5, 6 and 19) from the Stanford Genome Technology Center (10). For Assemblies 19 and 20, files with processed sequences are also available: DNA sequences of all coding regions, with or without introns, and with and without 1 kb flanking regions. The protein sequences derived from ORF translation are available for the Assemblies 20, 19 and 6. The DNA sequence of the mitochondrial genome is also available for download (10), as are the sequences of the tRNA genes predicted at CGD using the tRNAscan-SE algorithm (15).
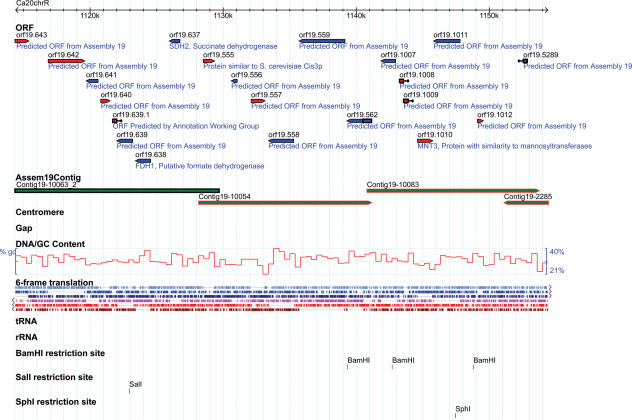
CGD also makes C.albicans sequence information and gene annotation available via the GBrowse genome browser, which is a sequence visualization and analysis tool developed as part of the Generic Model Organism Database Project (11). The tool displays a user-selected fragment of the genome (contig, chromosome, ORF, etc.) in the form of an interactive map that can be zoomed in and out, or scrolled along the selected region. The sequence of the region within the browser's window can be saved to a file at any time. The content displayed on the map is also user-selectable, using ‘tracks’. The user may elect to display (or hide) tracks containing ORFs, tRNA genes, restriction enzyme sites, six-frame translation, and DNA sequence (at high resolution) or G-C content (at low resolution). At this time, users may choose to browse either the Assembly 20 chromosomes or the Assembly 19 contigs. The Assembly 20 view includes a track containing the Assembly 19 contigs, to facilitate comparison between the two assemblies (Figure 1). In the future, ‘historical’ tracks containing the contigs and ORFs from Assemblies 6 and 4 will be added to the browser. Upon selecting these tracks, users will have a graphical view of the changes between the genome assemblies; for example, it will be clear where contigs have been assembled with each other, or where ORFs have been added, merged, or deleted from one version of the genome assembly to the next.
Figure 1.
GBrowse view of data from Assembly 20 of the C.albicans genome sequence. The view may be scrolled along the Assembly 20 chromosome, or zoomed in on a particular region. Selectable tracks of data include ORFs, Contig 19s, centromeric regions, and sequence gap regions.
All the features displayed in the genome browser are hyperlinked to their respective Locus pages, so that all the other pieces of information (experimental literature, mutant phenotypes, Gene Ontology assignments) are just a mouse click away. On each Locus page, GBrowse-generated thumbnails show schematic diagrams of the chromosomal surroundings and serve as a link to the full-featured genome browser, so that switching back and forth between the Locus page and GBrowse windows is rapid and straightforward.
Comparative sequence analysis at CGD
For sequence analysis, CGD provides a BLAST tool accessible via a link shown in the banner at the top of almost every page on the CGD website (http://www.candidagenome.org/cgi-bin/nph-blast). The tool allows comparison of any query sequence to one of several C.albicans sequence datasets. The full suite of BLAST programs is available [BLASTN, BLASTP, BLASTX, TBLASTX and TBLASTN (16)]. The sequence databases that can be searched include the complete sequence of Assembly 20 chromosomes, Assembly 19 supercontigs, Assembly 6 contigs or Assembly 4 contigs; the coding sequences of ORFs from Assemblies 20 or 19 with or without introns; the translation products of ORFs from Assemblies 20, 19 or 6; the predicted tRNA genes from Assembly 19, or the mitochondrial genome sequence.
Sequence comparisons with species other than C.albicans are available through the SGD (http://www.yeastgenome.org). SGD currently provides a Fungal BLAST tool [http://seq.yeastgenome.org/cgi-bin/blast-fungal.pl; (17)] that is updated regularly with all fungal sequences available in GenBank; it allows users to BLAST against all fungal sequences or against any desired combination of species. Rather than re-creating this tool, CGD links to it from the CGD BLAST search page, such that users may readily input any sequence for comparison to myriad fungal species.
Since S.cerevisiae proteins have in general been more widely and deeply characterized than those of C.albicans, it is useful for CGD users to be able to access quickly any published information about the putative S.cerevisiae ortholog of a C.albicans protein. Therefore, for each C.albicans protein with a predicted S.cerevisiae ortholog, we have added a prominent link to the Locus Summary page of the ortholog in the SGD (http://www.yeastgenome.org). We have also added a search feature whereby the name of an S.cerevisiae gene may be used to search for the C.albicans ortholog in CGD. Orthologs were mapped using the InParanoid software (version 1.35) developed at the Karolinska Institutet (18). The haploid complement of C.albicans proteins from CGD was compared to S.cerevisiae proteins from SGD, using the wormpep 145 set of C.elegans proteins from the Sanger Institute as an outgroup. Using relatively stringent cutoffs (BLOSUM80 matrix and an InParanoid score of 100%), 3594 ortholog mappings met these criteria when the analysis was performed in 2005 using Assembly 19 of the C.albicans genome sequence. The analysis will be repeated using Assembly 20, and also periodically in the future when the sequence is updated significantly.
DISCUSSION
Future comparative genomics tools at CGD
Annotated genome sequences are now available for multiple fungal species related to C.albicans [http://cbi.labri.fr/Genolevures/index.php, (19); http://www.sanger.ac.uk/sequencing/Candida/dubliniensis/; http://www.broad.mit.edu/annotation/fungi/fgi/]. To facilitate CGD users' access to these data, which are stored at diverse locations, we will collect DNA and protein sequences and make the files available for download. In the near future, we will implement two tools that allow comparison of closely related genomes with regard to conservation of gene order and of orthologous sequences. The Synteny Viewer, which is already in use at SGD (20), is an interactive display of aligned chromosomal segments from different species, with genes color-coded by degree of similarity and hyperlinked to sequence or additional information. This tool will be used to display regions of synteny, or conserved gene order, between C.albicans and the other Candida species. The Fungal Alignment Viewer, also used by SGD (20), facilitates comparison among orthologous genes as well as their 5′ and 3′ flanking sequences by generating custom color-coded ClustalW alignments between nucleotide or protein sequences.
The degree of similarity between a pathogen drug target and host molecules is an important consideration in predicting drug toxicity and side effects. In the future, CGD will implement a new BLAST tool containing sequences from pathogenic and non-pathogenic fungi plus sequences from mammalian hosts, so that users may evaluate conservation between possible drug targets and any host homologs.
Future developments in C.albicans sequence information
With the summer 2006 release of Assembly 20, the CGD team has now taken on the responsibility of maintaining the ‘official’ copy of the C.albicans genome sequence, and the task of incorporating corrections and updates as the sequence data are refined. The most significant challenge remaining in refinement of the C.albicans sequence will be the completion of an updated diploid assembly. The adjustment of the boundaries of allelic ORFs and, eventually, accurate representation of haplotype information (as far as haplotype data are available) are challenging tasks for the future. As the genomic sequence assembly continues to evolve, CGD looks forward to providing the Candida research community with the latest DNA and protein sequence, and with the tools necessary to navigate and analyze this wealth of information.
CGD curators welcome your suggestions and comments. Please email CGD at candida-curator@genome.stanford.edu.
Acknowledgments
We would like to thank the Candida Annotation Working Group, in particular Andre Nantel and Marco van het Hoog, for providing sequence and annotation files to CGD and helping to check and reconcile versions of the sequence; and Mike Cherry and the SGD Group for helpful advice and discussions. CGD is supported by NIH grant R01 DE015873 from the NIDCR at the NIH. Funding to pay the Open Access publication charges for this article was provided by grant R01 DE015873 from the NIDCR at the NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mavor A.L., Thewes S., Hube B. Systemic fungal infections caused by Candida species: epidemiology, infection process and virulence attributes. Curr. Drug Targets. 2005;6:863–874. doi: 10.2174/138945005774912735. [DOI] [PubMed] [Google Scholar]
- 2.Richardson M.D. Changing patterns and trends in systemic fungal infections. J. Antimicrob. Chemother. 2005;56(Suppl. 1):i5–i11. doi: 10.1093/jac/dki218. [DOI] [PubMed] [Google Scholar]
- 3.Soll D.R. Candida commensalism and virulence: the evolution of phenotypic plasticity. Acta Trop. 2002;81:101–110. doi: 10.1016/s0001-706x(01)00200-5. [DOI] [PubMed] [Google Scholar]
- 4.Calderone R.A. Introduction and historical perspectives. In: Calderone R.A., editor. Candida and Candidiasis. ASM Press, Washington DC; 2002. pp. 3–13. [Google Scholar]
- 5.Vazquez J.A., Sobel J.D. Mucosal candidiasis. Infect. Dis. Clin. North Am. 2002;16:793–820. doi: 10.1016/s0891-5520(02)00042-9. [DOI] [PubMed] [Google Scholar]
- 6.Hirschman J.E., Balakrishnan R., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., Fisk D.G., Hong E.L., Livstone M.S., Nash R., et al. Genome Snapshot: a new resource at the Saccharomyces Genome Database (SGD) presenting an overview of the Saccharomyces cerevisiae genome. Nucleic Acids Res. 2006;34:D442–D445. doi: 10.1093/nar/gkj117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun B.R., van Het Hoog M., d'Enfert C., Martchenko M., Dungan J., Kuo A., Inglis D.O., Uhl M.A., Hogues H., Berriman M., et al. A human-curated annotation of the Candida albicans genome. PLoS Genet. 2005;1:36–57. doi: 10.1371/journal.pgen.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ihmels J., Bergmann S., Berman J., Barkai N. Comparative gene expression analysis by differential clustering approach: application to the Candida albicans transcription program. PLoS Genet. 2005;1:e39. doi: 10.1371/journal.pgen.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nantel A. The long hard road to a completed Candida albicans genome. Fungal Genet. Biol. 2006;43:311–315. doi: 10.1016/j.fgb.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Jones T., Federspiel N.A., Chibana H., Dungan J., Kalman S., Magee B.B., Newport G., Thorstenson Y.R., Agabian N., Magee P.T., et al. The diploid genome sequence of Candida albicans. Proc. Natl Acad. Sci. USA. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnaud M.B., Costanzo M.C., Skrzypek M.S., Binkley G., Lane C., Miyasato S.R., Sherlock G. The Candida Genome Database (CGD), a community resource for Candida albicans gene and protein information. Nucleic Acids Res. 2005;33:D358–D363. doi: 10.1093/nar/gki003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.d'Enfert C., Goyard S., Rodriguez-Arnaveilhe S., Frangeul L., Jones L., Tekaia F., Bader O., Albrecht A., Castillo L., Dominguez A., et al. CandidaDB: a genome database for Candida albicans pathogenomics. Nucleic Acids Res. 2005;33:D353–D357. doi: 10.1093/nar/gki124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chibana H., Oka N., Nakayama H., Aoyama T., Magee B.B., Magee P.T., Mikami Y. Sequence finishing and gene mapping for Candida albicans chromosome 7 and syntenic analysis against the Saccharomyces cerevisiae genome. Genetics. 2005;170:1525–1537. doi: 10.1534/genetics.104.034652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Balakrishnan R., Christie K.R., Costanzo M.C., Dolinski K., Dwight S.S., Engel S.R., Fisk D.G., Hirschman J.E., Hong E.L., Nash R., et al. Fungal BLAST and Model Organism BLASTP Best Hits: new comparison resources at the Saccharomyces Genome Database (SGD) Nucleic Acids Res. 2005;33:D374–D377. doi: 10.1093/nar/gki023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Remm M., Storm C.E., Sonnhammer E.L. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 19.Souciet J., Aigle M., Artiguenave F., Blandin G., Bolotin-Fukuhara M., Bon E., Brottier P., Casaregola S., de Montigny J., Dujon B., et al. Genomic exploration of the hemiascomycetous yeasts: 1. A set of yeast species for molecular evolution studies. FEBS Lett. 2000;487:3–12. doi: 10.1016/s0014-5793(00)02272-9. [DOI] [PubMed] [Google Scholar]
- 20.Christie K.R., Weng S., Balakrishnan R., Costanzo M.C., Dolinski K., Dwight S.S., Engel S.R., Feierbach B., Fisk D.G., Hirschman J.E., et al. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32:D311–D314. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]