Abstract
The SUPERFAMILY database provides protein domain assignments, at the SCOP ‘superfamily’ level, for the predicted protein sequences in over 400 completed genomes. A superfamily groups together domains of different families which have a common evolutionary ancestor based on structural, functional and sequence data. SUPERFAMILY domain assignments are generated using an expert curated set of profile hidden Markov models. All models and structural assignments are available for browsing and download from http://supfam.org. The web interface includes services such as domain architectures and alignment details for all protein assignments, searchable domain combinations, domain occurrence network visualization, detection of over- or under-represented superfamilies for a given genome by comparison with other genomes, assignment of manually submitted sequences and keyword searches. In this update we describe the SUPERFAMILY database and outline two major developments: (i) incorporation of family level assignments and (ii) a superfamily-level functional annotation. The SUPERFAMILY database can be used for general protein evolution and superfamily-specific studies, genomic annotation, and structural genomics target suggestion and assessment.
INTRODUCTION
The SUPERFAMILY database (1) provides predictions of the protein domains in amino acid sequences. The classification of these domains is built on the Structural Classification of Proteins (SCOP) database (2) which groups domains of known structure hierarchically into classes, folds, superfamilies and families. The SCOP superfamily level groups together the most distantly related domains; and this level is what SUPERFAMILY is primarily based upon.
The SUPERFAMILY database contains domain assignments to ∼60% of the sequences of completely sequenced genomes, i.e. currently 64 eukaryotes, 327 bacteria (including 89 isolates) and 24 archae. New genomes are constantly being added. Our database also includes assignments for several sequence collections, i.e. UniProt (SwissProt and TrEMBL) (3) and the PDB (4) chains. We strongly encourage sequence submissions from the community.
Underlying the assignments is an expert curated library of profile hidden Markov models (HMMs) (5). HMMs (6) are profiles based on multiple sequence alignments designed to represent a domain superfamily (or family). Each of the models has a web page with a figure showing its amino acid composition, strongly conserved sites, hydrophobicity and regions in which insertions and deletions occur.
Since domains of a common superfamily are often diverged beyond easily detectable sequence similarity, the assignment of domain superfamilies is a non-trivial problem of remote homology detection. We use the Sequence Alignment and Modeling (SAM) HMM software package (7), as it is one of the best tools for remote homology detection (8,9), to build the model library, score the protein sequences and search the database for homologues. A program to produce domain assignments and the model library [in SAM, HMMER (6) and PSI-BLAST (10) formats] are available for download. SUPERFAMILY uses 10 894 models to represent the 1539 superfamilies in SCOP 1.69. The use of multiple models to represent a superfamily improves results (5). The model building procedure is described in (5).
The SUPERFAMILY web site, at http://supfam.org, offers a variety of methods for navigating and analysing genome assignments. Figure 1 gives an overview of the functionality and results that are available through the website. For example, the user can perform keyword searches for sequence identifiers, organism names, SCOP superfamily names, SCOP superfamily identifiers, SCOP unique identifiers, SUPERFAMILY model numbers and PDB identifiers. Further, up to 20 sequences can be submitted in the FASTA format for domain assignment. These sequences are first passed through a BLAST filter to find any assignments already stored in the database. If no hits are found in our pre-computed database, the sequences are then scored against the model library. If this does not produce any significant hits the sequences are scored using PRC (http://supfam.org/PRC/), which is an extremely sensitive profile–profile comparison method. Larger numbers of sequences can be accommodated by contacting the authors.
Figure 1.
Summary of the functionality and results that are available as part of the SUPERFAMILY analysis framework via the web interface.
For each genome, the website displays a list of predicted superfamilies including links to the individual protein assignments. In addition, undirected domain occurrence networks for all superfamilies are available in two graphical representations, produced by graphviz (11) and VCN (12). Nodes in the domain occurrence networks represent genomes and connections between genomes represent domain architectures which are common to both genomes. It is also possible to detect over- or under-represented superfamilies by comparing the domain composition of the given genome with a selectable list of other genomes.
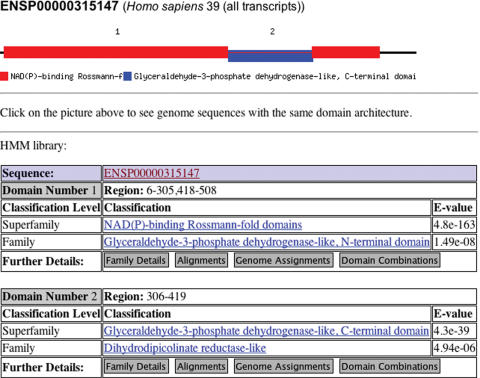
Domain architectures can be examined for each protein assignment. Figure 2 shows an example of a SUPERFAMILY assignment page which links to separate pages with the alignment between the sequence and the HMM which matched the domain. Further, one can enter a query that returns all examples of a given domain's combinations from all the genomes in SUPERFAMILY.
Figure 2.
Domain architecture and assignment details for the Ensembl protein ENSP00000315147 from human. Shown are the superfamily and family classification and associated E-values for two domains. Links to further family details, alignments between the SUPERFAMILY model and the protein, assignments for the human genome and domain combinations in which the superfamily domain occur in are included for each domain.
All the domain assignments and information on domain combinations is available for download in the form of a MySQL database dump.
The domain assignments are also available through a protein Distributed Annotation Server (DAS). The DAS protocol is primarily used to combine annotation data from multiple online sources. This facility enables high traffic genomic servers, such as Ensembl (13), to easily stay up to date with the live data in SUPERFAMILY. A machine readable list of SUPERFAMILY data sources (genomes) can be viewed at http://supfam.org/SUPERFAMILY/cgi-bin/das/dsn. Further details on the DAS protocol can be found at http://biodas.org.
In the following sections, we describe two major SUPERFAMILY developments since the previous publication (1): family level assignments and functional annotation of superfamilies. We conclude with an overview of future improvements to SUPERFAMILY.
FAMILY LEVEL ASSIGNMENT
In the hierarchical organization of SCOP, each domain superfamily comprises one or more domain families. For example, the superfamily of NAD(P)-binding Rossmann-fold domains consist of 12 families, e.g. Tyrosine-dependent oxidoreductases or CoA-binding domains. Sequence identity between members of a domain family is higher than between families of one superfamily. Most databases of protein domain assignments aggregate data at a single structural level. The Pfam database (14) includes data from the functionally related family level. The newest release of SUPERFAMILY now includes family level assignment in addition to the superfamily assignments, predicting 2845 families. The family level in the SUPERFAMILY and Pfam databases are similar, but not equivalent. The family level in SUPERFAMILY reflects the SCOP classification which is independent of Pfam. The family level provides a wealth of functional information that is of use to structural, computational and bench biologists.
We developed an algorithm (15) predicting family association for a domain, given its known association with a SCOP superfamily. From a statistical viewpoint, we test the hypothesis that assuming a query domain is in superfamily A, the query domain is in family A1. The corresponding null hypothesis is: the query domain is not in family A1.
The general method to solve the superfamily to family level mapping can be considered to be a hybrid pairwise-profile method. The query domain and the member sequences of a family are each separately aligned to the SUPERFAMILY model. These alignments provide the profile aspects of the method. HMMs usually give a many-against-one (profile) score. We infer the pairwise alignment between the query domain and a family sequence by comparing the following two profile alignments: (i) query domain to SUPERFAMILY model and (ii) family sequence to SUPERFAMILY model. Residues from each sequence are aligned to each other if they align to the same position in the model. A pairwise score can be calculated from the inferred alignment using the BLOSUM 62 substitution matrix and affine gap penalties. A ‘gap open’ penalty of 3 and ‘gap extend’ penalty of 0.8 were found to give optimal results. The low gap open penalty, relative to BLAST which uses a value of 5, is due to the fact that the alignment comes from the SUPERFAMILY model. The HMM is aware of other sequences in the superfamily and so is locally less refined on the query domain-family sequence pair. The hybrid method does not require any family level models.
Conserved sites in the HMM contribute more to the profile score than variable sites. For normal SUPERFAMILY model scoring a relative weighting factor takes the importance of the model position into account. However, performance of the hybrid method was found to be best when the position-specific weights were not used. When the weights are included the hybrid method performs similarly to the original HMM score (15).
To test the hypothesis, we calculate the pairwise score between the query domain and the best profile scoring sequence from family A1. From this result we subtract, the pairwise score between the query domain and the family sequence not in A1 which gives the best profile score. If a query domain has a strong score to one family and weak scores to the remaining families then discrimination between families is strong. Conversely, if a query domain has comparable scores to more than one family, the distinction is weak.
Alignments and scores between every SCOP family member and each of the SUPERFAMILY models are pre-calculated and stored in the SUPERFAMILY database. Only the alignment and score between the query domain and the SUPERFAMILY model must be calculated. These are calculated as part of the superfamily classification process. They are used as the query side of the hybrid method. There is little additional computational cost involved in applying the family level sub-classification method.
We consider family level domain classifications with E-values below 0.0001 to be strong classifications. This threshold was chosen to minimize the error rate. The family level E-value function being used is
where K, λ and τ are coefficients derived from a benchmark dataset (15), n is the length of the sequence in amino acids and S is the raw score. The 1 subscript refers to the hit with the highest score and subscript 2 denotes the second highest scoring hit. The second highest scoring hit must occur outside the family with the highest scoring hit. In the cases where there is no second highest scoring hit n2 is set to the average domain length (180) and because there is no alignment S2 is set to zero.
One of the benefits of our hybrid method is the ability to infer the presence of new families from negative results. This ability has been used to make improvements to the SCOP hierarchy and should be of great utility to the structural genomics projects. In addition to the family level classification the method can for a given sequence suggest a closest homologue with a known 3D structure. This closest homologue is the PDB structure of the SCOP family member with the highest score.
An example of a family level assignment can be seen in Figure 2. The family level data added to SUPERFAMILY is being used for the functional annotation of genomes, individual sequence family studies, predictions of new family level targets for structural genomics projects, suggesting the most closely related structures for homology modeling, working with functional sets of domains, e.g. transcription factors (16) and further development of the Gene Ontology (GO) (17) for SCOP.
The hybrid method produces superior results to either profile or pairwise techniques (15). No updates to sub-level models, phylogenetic trees, neural networks etc, are required when the database is updated. The method scales up to the most complex genomes and has been applied to over 400 genomes and sequence collections. The results are available for browsing and download on the SUPERFAMILY web site.
The aim of the hybrid method is to create a statistical technique for classifying domains into a pre-existing biological classification system. Given an existing profile library with genomic assignments for one level of a classification scheme, sub-level assignments come at next to no computational cost compared to alternative methods.
The hybrid method is general, so it could be applied directly to other databases such as Gene3D (18), or indirectly to Pfam. It is anticipated that the method will be of great use in future versions of SCOP which are expected to include a more fluid hierarchy. The authors welcome feedback and collaborations on this major new SUPERFAMILY development.
FUNCTIONAL ANNOTATION OF DOMAIN SUPERFAMILIES
The definition of the function of a protein domain is still a matter of debate and can vary depending on the actual context of the biological problem being addressed. We developed a domain-centric scheme for functional annotation instead of the widely used whole-gene (17) or whole-protein (19) annotation. Thus a particular protein with two domains can be assigned to two different functional categories, e.g. a protein could consist of a kinase and a small-molecule binding domain.
Our definition of domain function is a combination of the ‘biological process’ and ‘molecular function’ used in the GO (17) and was modeled after the scheme used in the Cluster of Orthologous Groups (COGs) database (19).
Our scheme is comprised of 50 detailed functional categories which map to seven general categories. These seven general functional categories are as follows: (i) Information: storage, maintenance of the genetic code; DNA replication/repair; general transcription/translation; (ii) Regulation: regulation of gene expression and protein activity; information processing in response to environmental input; signal transduction; general regulatory or receptor activity; (iii) Metabolism: anabolic and catabolic processes; cell maintenance/homeostasis; secondary metabolism; (iv) Intra-cellular processes: cell motility/division; cell death; intra-cellular transport; secretion; (v) Extra-cellular processes: inter-, extra-cellular processes, e.g. cell adhesion; organismal processes, e.g. blood clotting, immune system; (vi) General: general and multiple functions; interactions with proteins, ions, lipids or small molecules; and (vii) Other/Unknown: unknown function, viral proteins/toxins. Supplementary Data S1 describe the superfamily distribution in the 50 functional categories over all SCOP classes.
We annotated domain superfamilies manually to ensure high quality. The annotation describes the superfamily's dominant molecular function, i.e. function as part of the protein, or dominant biological process, i.e. their role in the cell. Each domain superfamily is associated with only one function, even though some superfamilies, especially large ones, may have a variety of functions. One example are Immunoglobulin domains which are assigned to ‘cell adhesion’ for common function in cell surface receptors, although in vertebrates this domain superfamily is also involved in the immune system. In cases where no one function was obviously dominant, the domain function was classified as ‘General or several functions’.
We based our annotation on information from SCOP (2), InterPro (20), Pfam (14), Swiss-Prot (3) and literature. For validation, we used the existing annotations of GO biological process and GO molecular function for Pfam domains in InterPro, mapping them to SCOP superfamilies based on sequence similarity. This procedure largely confirmed our annotations for 647 and 667 domain superfamilies of known GO process and GO function, respectively.
As further validation, we extracted the largest superfamilies (∼300), i.e. which have at least 25 members in one of the completely sequenced eukaryotes (21), and re-examined their annotations, consulting co-workers as an independent source of information. Based on our experience, we estimate error rates of <10% for abundant superfamilies and <20% for other superfamilies.
In total, we annotated all domain superfamilies of the SCOP classes 1–11, with an emphasis on the 1261 domain superfamilies of the primary classes 1–7. The SCOP classes are— (1) all alpha proteins, (2) all beta proteins, (3) alpha and beta proteins (a/b), (4) alpha and beta proteins (a+b), (5) multi-domain proteins (alpha and beta), (6) membrane and cell surface proteins and peptides, (7) small proteins, (8) coiled coils proteins, (9) low-resolution structures, (10) peptides and (11) designed proteins.
Superfamilies of metabolism, e.g. enzymes, are the most abundant category. Close to half of all superfamilies (533) have metabolism related functions. Each of the other categories comprise <15% of the domain superfamilies. Around 10% of the superfamilies (200) have unknown functions. Members of some superfamilies, particularly the large ones, may have a variety of functions.
To date, we applied the functional annotation to several problems involving analyses of statistically over-represented domain combinations (21), domain recombination (22) and protein family expansions in relation to biological complexity (23). The annotation could also be used in superfamily-specific studies, improvements to the GO annotation of SCOP and as part of the SUPERFAMILY assignments. The general and detailed annotations for all superfamily domains in SCOP classes 1–7 are available for download from http://sufpam.org/SUPERFAMILY/function.html. Users of the functional annotation are encouraged to contact us with questions.
FUTURE DIRECTIONS
A large part of the development of SUPERFAMILY will be dictated by the changes to the SCOP hierarchy which is expected to become more fluid. The hybrid method outlined above will be invaluable for dealing with these changes to appear with release 1.73.
We currently study a domain architecture approach to the phylogenetic distribution of superfamilies which we expect to produce large amounts of data useful for evolutionary studies. With respect to the model building process we currently investigate how to incorporate information on the most common domain combinations to improve assignment accuracy and coverage.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We gratefully acknowledge comments on the manuscript from Madan Babu Mohan. C. V. acknowledges support by the Boehringer Ingelheim Fonds, the Medical Research Council and the International Human Frontier of Science Program. Funding to pay the Open Access publication charges for this article was provided by the Medical Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Madera M., Vogel C., Kummerfeld S.K., Chothia C., Gough J. The superfamily database in 2004: additions and improvements. Nucleic Acids Res. 2004;32:D235–D239. doi: 10.1093/nar/gkh117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreeva A., Howorth D., Brenner S.E., Hubbard T.J., Chothia C., Murzin A.G. Scop database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu C.H., Apweiler R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., et al. The universal protein resource (uniprot): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deshpande N., Addess K.J., Bluhm W.F., Merino-Ott J.C., Townsend-Merino W., Zhang Q., Knezevich C., Xie L., Chen L., Feng Z., et al. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gough J., Karplus K., Hughey R., Chothia C. Assignment of homology to genome sequences using a library of hidden markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 6.Eddy S.R. Profile hidden markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 7.Karplus K., Barrett C., Hughey R. Hidden Markov models for detecting remote protein homologies. Bioinformatics. 1998;14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- 8.Madera M., Gough J. A comparison of profile hidden markov model procedures for remote homology detection. Nucleic Acids Res. 2002;30:4321–4328. doi: 10.1093/nar/gkf544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wistrand M., Sonnhammer E.L. Improved profile HMM performance by assessment of critical algorithmic features in SAM and HMMER. BMC Bioinformatics. 2005;6:99–109. doi: 10.1186/1471-2105-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gansner E.R., North S.C. An open graph visualization system and its applications to software engineering. Softw. Pract. Exp. 1999;30:1203–1233. [Google Scholar]
- 12.Batada N.N. CNplot: visualizing pre-clustered networks. Bioinformatics. 2004;20:1455–1456. doi: 10.1093/bioinformatics/bth080. [DOI] [PubMed] [Google Scholar]
- 13.Birney E., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finn R.D., Mistry J., Schuster-Böckler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gough J. Genomic scale sub-family assignment of protein domains. Nucleic Acids Res. 2006;34:3625–3633. doi: 10.1093/nar/gkl484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kummerfeld S.K., Teichmann S.A. DBD: a transcription factor prediction database. Nucleic Acids Res. 2006;34:D74–D81. doi: 10.1093/nar/gkj131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeats C., Maibaum M., Marsden R., Dibley M., Lee D., Addou S., Orengo C.A. Gene3D: modeling protein structure, function and evolution. Nucleic Acids Res. 2006;34:D281–D284. doi: 10.1093/nar/gkj057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41–55. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. Interpro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogel C., Berzuini C., Bashton M., Gough J., Teichmann S.A. Supra-domains: evolutionary units larger than single protein domains. J. Mol. Biol. 2004;336:809–823. doi: 10.1016/j.jmb.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 22.Vogel C., Teichmann S.A., Pereira-Leal J. The relationship between domain duplication and recombination. J. Mol. Biol. 2005;346:355–365. doi: 10.1016/j.jmb.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 23.Vogel C., Chothia C. Protein family expansions and biological complexity. PLoS Comput. Biol. 2006;2:e48. doi: 10.1371/journal.pcbi.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]