Abstract
The Gene Expression Omnibus (GEO) repository at the National Center for Biotechnology Information (NCBI) archives and freely disseminates microarray and other forms of high-throughput data generated by the scientific community. The database has a minimum information about a microarray experiment (MIAME)-compliant infrastructure that captures fully annotated raw and processed data. Several data deposit options and formats are supported, including web forms, spreadsheets, XML and Simple Omnibus Format in Text (SOFT). In addition to data storage, a collection of user-friendly web-based interfaces and applications are available to help users effectively explore, visualize and download the thousands of experiments and tens of millions of gene expression patterns stored in GEO. This paper provides a summary of the GEO database structure and user facilities, and describes recent enhancements to database design, performance, submission format options, data query and retrieval utilities. GEO is accessible at http://www.ncbi.nlm.nih.gov/geo/
INTRODUCTION
Microarray and other high-throughput technologies have led to an explosion in the rate of molecular abundance data generated in the last decade. For the last seven years the Gene Expression Omnibus (GEO) database has served as a central hub for these data, operating primarily as a public archive and distribution center, but also providing flexible mining tools that enable users to easily query, filter, inspect and download data in the context of their specific interests (1,2).
GEO is currently the largest fully public gene expression resource. Since its inception, the database has grown exponentially each year. As of September 2006, the database holds over 120 000 samples, representing over 3.2 billion individual measurements, spanning over 200 organisms, and addressing a wide variety of biological phenomena. These data have been deposited by >2000 laboratories from around the world. All data are freely available online and via bulk FTP download.
GEO supports minimum information about a microarray experiment (MIAME)-compliant data submissions. MIAME is a data content standard developed by the microarray gene expression data (MGED) society to outline what information should be provided when describing a microarray experiment (3). Making microarray data public in a MIAME-compliant manner has become a precondition for publication for many journals. Publishing original data and protocols facilitates independent evaluation of results and reanalysis, and is in keeping with the spirit of open-access (4). Consequently, most of the data in GEO have been submitted by the research community in fulfillment of journal requirements.
DATABASE STRUCTURE AND DATA FLOW
The GEO database architecture is designed for the efficient capture, storage and retrieval of large-scale functional genomic data. The diverse and complex nature of such data presents considerable challenges in data handling and querying. There are many different types of high-throughput methodologies and researchers use a wide variety of hardware and software to generate and process data. Thus, data come in many different formats and comprise varying content. Furthermore, technologies and processing strategies continue to rapidly evolve. In light of these considerations, GEO was designed with a flexible structure that can accommodate diverse styles of data. This flexibility is largely attributed to the fact that tabular data are not fully granulated in the core database but instead are treated as plain text, tab-delimited tables that may contain any number of rows or columns. Although the primary database has no knowledge and applies no restrictions on these tab-delimited tables, some columns reserve special meanings and data from selected fields are extracted to secondary databases and used in downstream query and analysis applications. Accompanying supplementary and native file types are linked from each record and stored on an FTP server.
Expression data can be rendered meaningless unless accompanied by the contextual biological and processing details under which they were generated. To address this, GEO has a MIAME-compliant infrastructure that supports fully annotated records. Biological and other descriptive metadata are stored in designated fields with proper relations or restrictions within database tables.
Submitter-supplied data
The overall structure of the core GEO database remains as described previously (1,2). Briefly, data submitted to GEO are stored in a relational MSSQL database partitioned into three entity types:
Platform
Includes a summary description of the array and a data table defining the array template. Each row in the table corresponds to a single feature, and includes sequence annotation and tracking information as provided by the submitter. The table may contain any number of columns allowing thorough annotation of the array.
Sample
Includes a description of the biological material and the experimental protocols to which it was subjected, and a data table containing hybridization measurements for each feature on the corresponding platform. The table may contain any number of columns in which to comprehensively present hybridization results. The metadata fields may hold very large volumes of text to allow elaborate descriptions of the biological source and protocols.
Series
Defines a set of related samples considered to be part of a study, and describes the overall study aim and design. Series may also incorporate tabular summary tables pertaining to the experiment as a whole.
Each of these objects is essentially under the submitter's editorial control and is assigned a stable and unique accession number that may be used to cite and retrieve the records. The accession consists of a number and a letter prefix indicating whether the record is a GEO Platform (GPL), GEO Sample (GSM), or GEO Series (GSE).
In addition to the user-submitted objects described above, GEO defines and creates a number of related data objects to facilitate data mining, visual rendering and transposition of submitted data into alternative structures. The principal object used for this purpose is the DataSet object.
GEO DataSets
Despite the variety of style and content of the data received, submissions have a common core set of elements:
sequence identity tracking information of each feature on the array
normalized hybridization measurements
a description of the biological source used in each hybridization
Using a combination of automated data extraction and manual curation, this information is taken from the submitter-supplied records and organized into an upper-level object called a GEO DataSet. A DataSet represents a collection of similarly-processed experimentally-related samples, summarized and categorized according to experimental variables. DataSets allow for the transformation of diverse styles of incoming data from multiple unrelated projects, into a relatively standardized format upon which downstream data analysis and data display tools are based.
DataSets provide two discrete renderings of the data (Figure 1):
An experiment-centered representation that encapsulates the entire study. This information is presented as a DataSet record which comprises a synopsis of the experiment, a breakdown of the experimental variables, access to auxiliary objects, several data display and analysis tools, and download options.
A gene-centered representation that presents quantitative gene expression measurements for one gene across a DataSet. This information is presented as a GEO Profile which comprises gene identity annotation, DataSet title, links to auxiliary information and a chart depicting the expression level and rank of that gene across each sample in the DataSet. Gene annotation is derived from querying sequence identifiers (e.g. GenBank accessions, clone IDs) with the latest Entrez Gene and UniGene databases, an important point given the dynamic nature of gene annotation.
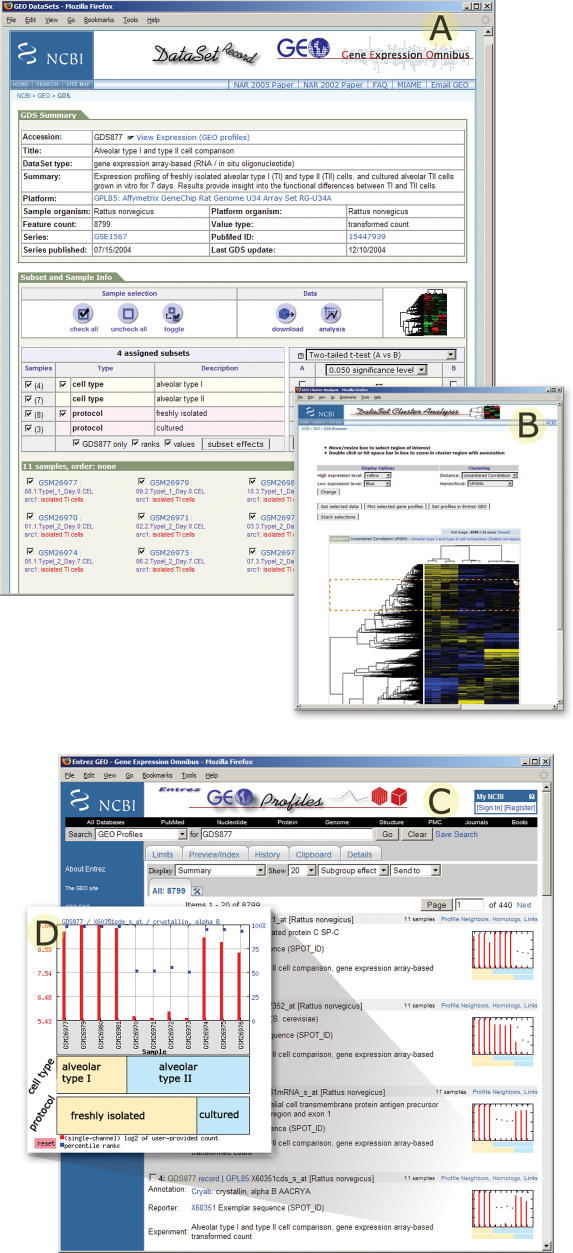
Figure 1.
A selection of GEO screenshots from a typical experiment (GEO DataSet GDS877; 16). (A) DataSet record includes experiment summary information, DataSet subset classifications, and access to data mining features such as hierarchical cluster heat map and ‘Query subset A versus B’ tool. (B) DataSet hierarchical cluster heat map calculated by un-centered correlation coefficient/average linkage option. Regions of interest can be selected using the red image cropper box, then either expanded to view sample and gene annotation, downloaded, charted as line plots, or linked directly to corresponding Entrez GEO profiles records. (C) GEO profiles retrieval results; each entity includes sequence identifier and DataSet information, and a thumbnail profile image. (D) Expanded profile chart depicts expression value information for the crystallin gene across each sample in DataSet GDS877. Experimental subset groupings are reflected in labels at foot of chart.
SUBMISSION PROCEDURES, FORMATS AND STANDARDS
We endeavor to make data deposit procedures as straightforward as possible. Submitters have several options for data submission; selecting which method to use depends on the amount and type of data to be submitted, and what format the data are already in. Regardless of the deposit method chosen, the final GEO records will look similar and contain equivalent information. Each format captures all components of the MIAME checklist, as well as any additional information that the submitter wants to provide.
Upload options and formats
Web deposit
The web submission process is designed for quick and easy deposit of individual records by occasional submitters, or for smaller experiments. This route consists of a set of interactive web forms that provide a simple step-by-step procedure for deposit of data tables and accompanying descriptive information.
SOFT format
Simple Omnibus Format in Text (SOFT) is a simple, line-based, tab-delimited format designed for rapid batch deposit. A single SOFT file can hold both data tables and accompanying descriptive information for multiple platforms, samples and series records. The simplicity of SOFT allows it to be readily generated from commonly-used database and spreadsheet applications. Conveniently, two versions of SOFT are available:
SOFTtext
SOFT-formatted data are organized as concatenated records.
SOFTmatrix
SOFT-formatted data are organized side-by-side as a matrix table, usually in an Excel spreadsheet.
MINiML format
MIAME Notation in Markup Language, (MINiML, pronounced ‘minimal’) is a recent addition to GEO's upload/download options. MINiML is effectively an XML rendering of SOFT format, and is similarly designed for rapid batch submission and upload of data. The MINiML XML schema definition and a detailed description are available at the GEO website.
MAGE-ML format
MicroArray Gene Expression Markup Language (MAGE-ML) is an XML format devised by the MGED consortium (5) and a direct derivation from the corresponding MAGE object model. GEO is not based on the MAGE object model and cannot receive these files directly. Nonetheless, parsers have been written to extract data from some of the various flavors of MAGE-ML and reformat according to GEO schema. It is worth noting here that having data formatted as MAGE-ML does not in any way imply MIAME-compliance. MIAME is a data content standard, not a format standard. MIAME-compliant data may be submitted in many formats.
Detailed documentation and examples of submission options and formats are available on the GEO website. However, if submitters have questions or require assistance with submission procedures they are encouraged to contact GEO curation staff at geo@ncbi.nlm.nih.gov for prompt support.
Submitters may keep their records private until a manuscript describing the data is published. Submitters may generate read-only passwords that give reviewers and collaborators confidential access to their private data.
Most researchers submit to GEO to support data discussed in a journal manuscript, so it is important to present the data as it was processed in the manuscript. However, over the past two years, greater emphasis has been placed on provision of raw, unmanipulated native data files to accompany the processed data within GEO records. Such files include, e.g. Affymetrix CEL or GenePix GPR scan files. Recent modifications to submission procedures now make it more convenient for submitters to supply these raw files: the web deposit route specifically requests supplementary files; the batch deposit routes allow for raw data files to be zipped/tarred together with bulk submissions. Provision of raw data not only enables other researchers to faithfully reproduce the data selection, transformation and analysis steps that are the basis of a publication, but also maximizes the long-term value of submissions, enabling recycling of the data into repeated rounds of analysis.
All submitted data undergo syntactic validation and are inspected by curators for content integrity. When content or format problems are identified, curators work with the submitter until the issue is resolved. However, given the huge diversity of biological themes, technology types, processing techniques, and statistical transformations applied to microarray data, it is impractical for curators to decisively determine the accuracy, validity or score the degree of MIAME-compliance of submitted data. Thus, researchers are ultimately responsible for the completeness, quality and accuracy of their submissions. This validation process can benefit from feedback by journal editorial reviewers or funding agency enforcement. Through their GEO accounts, researchers retain full editorial control of their records and can update or edit their records at any time.
In addition to satisfying possible journal requirements for publication, there are other significant benefits to depositing data with GEO. Data receive long term archiving at a centralized repository, integration with other NCBI resources which afford greatly increased usability and visibility, as well as possible links back to submitters' own project websites.
TOOLS TO RETRIEVE, EXPLORE AND VIZUALIZE DATA
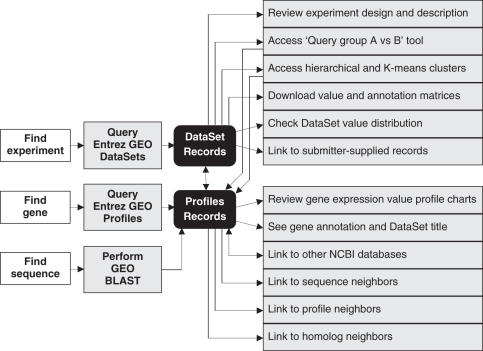
To maximize the utility and value of the massive volumes of data in GEO, a selection of intuitive tools and features has been developed to assist researchers to quickly locate, analyze and visualize data relevant to their interests. These features incorporate traditional data reduction techniques and concise displays designed for human scanning, helping the user identify and categorize gene and sample relationships. Figure 2 depicts a schematic overview of the query workflow and how the various features and tools are interlinked. A summary of where the main features are located and their purpose is provided in Table 1. Query approaches include standard text-based searches, sequence-based searches, mining based on expression behavior characteristics or combinations of these factors.
Figure 2.
A schematic overview of query workflow, and how various features and tools are interlinked. A description of the location and purpose of these features is provided in Table 1.
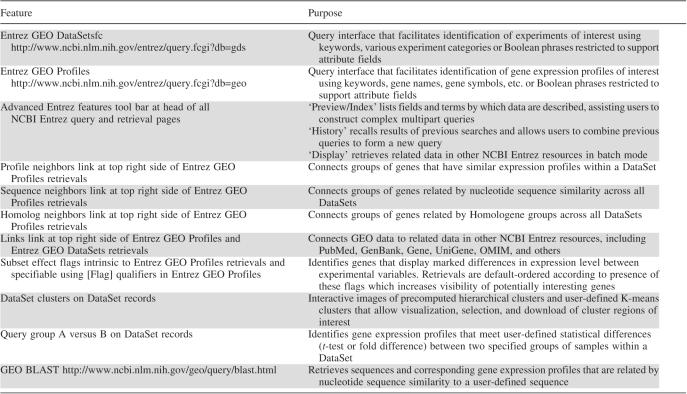
Table 1.
Summary of location and purpose of various GEO data mining tools and features
These tools do not require specialized knowledge of microarray analysis methods, nor do they require time-consuming download or processing of large data sets. However, it should be stated that the analysis features are not primarily intended for robust systematic data mining. The diverse nature of the data in GEO restricts to some extent the statistical tools that can be developed. All data are treated similarly; criteria such as scaling factors, filter parameters, and number of repeats are not considered. Despite these issues, these tools are extremely useful for quick and easy identification of relevant and noteworthy data.
NCBI's Entrez search system serves as the basis for most queries. Entrez GEO DataSets contains experiment-centered data and Entrez GEO Profiles contains gene-centered data. Most biologists are familiar with Entrez, using it routinely to search other NCBI databases like PubMed and GenBank (6,7). It has a straightforward interface where users can locate relevant material by simply typing in keywords or Boolean phrases restricted to supported attribute fields. Examples of typical queries and query fields are provided at http://www.ncbi.nlm.nih.gov/projects/geo/info/qqtutorial.html.
Full use is made of Entrez's powerful linking capabilities. Intra-database links connect genes related by expression pattern or sequence. Where possible, reciprocal inter-database links connect GEO data with related data in other NCBI resources such as PubMed, GenBank, Gene, UniGene, MapViewer, OMIM and others. Advanced Entrez features allow generation of complex multipart queries or combination of multiple queries that find common intersections in retrievals. GEO's Entrez query facilities were recently further enhanced by implementation of a spell-check function, as well as automatic term mapping using MeSH translation tables.
Graphics are an important tool to aid visualization and interpretation of high-dimensional expression data. The expression pattern of each gene within a DataSet is represented as a profile chart (Figure 1D). A breakdown of the experimental design is provided along the bottom of the chart, helping the user to quickly assess whether expression levels are shifting with experimental variables. Thumbnail chart images provided on batch profile retrievals are useful for rapid batch profile scanning and comparison. Value distribution charts are provided on DataSets records, providing at-a-glance indication of how well normalized the data are within a DataSet. Precomputed interactive hierarchical cluster heat map images are available on each DataSet record, providing suggestions for groups of coordinately regulated genes within entire DataSets.
Within the last year, the back-end structure of the Profiles, DataSets and annotation databases was completely redesigned. These changes allow more flexibility on the front-end user interfaces and will permit development of more advanced query, analysis and download tools, including enhanced Entrez utilities user-scripting options. These changes also help to streamline internal indexing procedures, enabling more frequent release of new DataSets and profiles.
For users who prefer to use their own analysis software or want to perform more robust analyses, all GEO data are available for bulk download via anonymous FTP at ftp://ftp.ncbi.nih.gov/pub/geo/DATA/. Files include SOFT- and MINiML-formatted Platform and Series families, SOFT-formatted DataSets and original supplementary data types. Various software packages have been developed by the community to handle GEO data formats, including the GEOquery R/BioConductor package, http://bioconductor.org/packages/release/bioc/html/GEOquery.html.
CONCLUSIONS
GEO currently represents the largest single resource for public gene expression data. Beyond archiving and making data freely-available for peer review and download, the GEO repository also provides an extensive complement of utilities and strategies that enable effective data mining on either a small or large scale.
The data in GEO gain value as they accumulate. Pooling masses of expression data into common formats at a single location affords researchers the opportunity to distill disparate data sets and identify common gene expression trends, dissect regulatory networks and predict functions of uncharacterized genes. Increasingly, GEO data are used and cited by third parties as evidence to support and complement their own studies, selected examples include (8–15).
Having GEO data cross-annotated with extensive sequence, mapping and bibliographic resources via the NCBI Entrez system of interlinked databases imparts further value and context to the data. This diverse integrated data environment leverages multiple types of information and enables traditional disciplinary boundaries to be crossed, ultimately accelerating systems-level hypothesis formation and scientific discovery.
Future plans for GEO include continued development of data retrieval and mining features, and enhancing novice user experience. We also plan to improve rendering and representation of the non-gene-expression data types that GEO accepts, which include chromatin-immunoprecipitation on arrays (ChIP-chip) studies, array comparative genomic hybridization (aCGH), SNP arrays and some proteomic data.
Acknowledgments
This research, and funding to pay the Open Access publication charges for this article, were supported by the Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Barrett T., Suzek T.O., Troup D.B., Wilhite S.E., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brazma A., Hingamp P., Quackenbush J., Sherlock G., Spellman P., Stoeckert C., Aach J., Ansorge W., Ball C.A., Causton H.C., et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 4.Ball C., Brazma A., Causton H., Chervitz S., Edgar R., Hingamp P., Matese J.C., Parkinson H., Quackenbush J., Ringwald M., et al. Standards for microarray data: an open letter. Environ. Health Perspect. 2004;112:A666–A667. doi: 10.1289/ehp.112-1277123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spellman P.T., Miller M., Stewart J., Troup C., Sarkans U., Chervitz S., Bernhart D., Sherlock G., Ball C., Lepage M., et al. Design and implementation of microarray gene expression markup language (MAGE-ML) Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-9-research0046. RESEARCH0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuler G.D., Epstein J.A., Ohkawa H., Kans J.A. Entrez: molecular biology database and retrieval system. Methods Enzymol. 1996;266:141–162. doi: 10.1016/s0076-6879(96)66012-1. [DOI] [PubMed] [Google Scholar]
- 8.Yuan Z., Tie A., Tarnopolsky M., Bakovic M. Genomic organization, promoter activity, and expression of the human choline transporter-like protein 1. Physiol. Genomics. 2006;26:76–90. doi: 10.1152/physiolgenomics.00107.2005. [DOI] [PubMed] [Google Scholar]
- 9.Byrnes J.K., Morris G.P., Li W.H. Reorganization of adjacent gene relationships in yeast genomes by whole-genome duplication and gene deletion. Mol. Biol. Evol. 2006;23:1136–1143. doi: 10.1093/molbev/msj121. [DOI] [PubMed] [Google Scholar]
- 10.Siddiqui A.S., Delaney A.D., Schnerch A., Griffith O.L., Jones S.J., Marra M.A. Sequence biases in large scale gene expression profiling data. Nucleic Acids Res. 2006;34:e83. doi: 10.1093/nar/gkl404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norris A.W., Kahn C.R. Analysis of gene expression in pathophysiological states: balancing false discovery and false negative rates. Proc. Natl Acad. Sci. USA. 2006;103:649–653. doi: 10.1073/pnas.0510115103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham R.R., Kozyrev S.V., Baechler E.C., Reddy M.V., Plenge R.M., Bauer J.W., Ortmann W.A., Koeuth T., Gonzalez Escribano M.F., Pons-Estel B., et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nature Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 13.Calvo S., Jain M., Xie X., Sheth S.A., Chang B., Goldberger O.A., Spinazzola A., Zeviani M., Carr S.A., Mootha V.K. Systematic identification of human mitochondrial disease genes through integrative genomics. Nature Genet. 2006;38:576–582. doi: 10.1038/ng1776. [DOI] [PubMed] [Google Scholar]
- 14.Zhou X.J., Kao M.C., Huang H., Wong A., Nunez-Iglesias J., Primig M., Aparicio O.M., Finch C.E., Morgan T.E., Wong W.H. Functional annotation and network reconstruction through cross-platform integration of microarray data. Nat. Biotechnol. 2005;23:238–243. doi: 10.1038/nbt1058. [DOI] [PubMed] [Google Scholar]
- 15.Griffith O.L., Pleasance E.D., Fulton D.L., Oveisi M., Ester M., Siddiqui A.S., Jones S.J. Assessment and integration of publicly available SAGE, cDNA microarray, and oligonucleotide microarray expression data for global coexpression analyses. Genomics. 2005;86:476–488. doi: 10.1016/j.ygeno.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez R., Yang Y.H., Griffin C., Allen L., Tigue Z., Dobbs L. Freshly isolated rat alveolar type I cells, type II cells, and cultured type II cells have distinct molecular phenotypes. Am. J. Physiol. Lung Cell Mol. Physiol. 2005;288:L179–L189. doi: 10.1152/ajplung.00272.2004. [DOI] [PubMed] [Google Scholar]