Abstract
The University of California, Santa Cruz Genome Browser Database contains, as of September 2006, sequence and annotation data for the genomes of 13 vertebrate and 19 invertebrate species. The Genome Browser displays a wide variety of annotations at all scales from the single nucleotide level up to a full chromosome and includes assembly data, genes and gene predictions, mRNA and EST alignments, and comparative genomics, regulation, expression and variation data. The database is optimized for fast interactive performance with web tools that provide powerful visualization and querying capabilities for mining the data. In the past year, 22 new assemblies and several new sets of human variation annotation have been released. New features include VisiGene, a fully integrated in situ hybridization image browser; phyloGif, for drawing evolutionary tree diagrams; a redesigned Custom Track feature; an expanded SNP annotation track; and many new display options. The Genome Browser, other tools, downloadable data files and links to documentation and other information can be found at http://genome.ucsc.edu/.
INTRODUCTION
The University of California, Santa Cruz (UCSC) Genome Browser (1,2) is a web-based interface that provides access to the finished human genome sequence assembly and the draft assemblies of other species and their annotations. As of September 2006, the Browser supports 13 vertebrate species including model organisms such as chimp, mouse and rat. Also presented are 19 non-vertebrate organisms, including 11 Drosophila species and yeast. Extensive cross-species pairwise and multiple alignments are presented for all assemblies. This publication focuses on changes introduced since September 2005, the last published update (2).
Each assembly displayed in the Browser graphic view consists of multiple data ‘tracks’ collected into several groups with shared characteristics, such as Gene Expression. Typically, a main table, keyed to genomic sequence coordinates, is used to draw the annotation in the graphic display, while one or more supporting tables may be used to supply additional information on a ‘details page’ for each item, which can be accessed by clicking on the graphic display. Tables in a variety of formats may be used to support browser tracks, as described in http://genome.ucsc.edu/FAQ/FAQformat.
In the ever-changing landscape of genome assemblies, the archiving activities of the UCSC Browser group provide stability for publications that refer to a specific data freeze. Several of the latest updates per species are maintained on the public site, with the superseded assemblies supported on a separate archive server, http://genome-archive.cse.ucsc.edu.
The Browser toolset includes BLAT (3), In Silico PCR, the Gene Sorter (4) and the Proteome Browser (5) and now, VisiGene. The Table Browser (6) provides access to all database tables and supports advanced queries, filters and table intersection. New users may find assistance through online help and in training materials offered by OpenHelix (http://www.openhelix.com/ucscmaterials.shtml). A recent publication (7) details use of the Browser in comparative genomics.
NEW DATA
New assemblies
In the past year (2), the number of genome assemblies supported by the Genome Browser has increased more than 60% with 22 new sequence assembly databases added, including five new species: the purple sea urchin (Strongylocentrotus pupuratus), and four Drosophila species: erecta, grimshawii, persimilis and sechellia.
The Genome Browser now supports a total of 59 assemblies for 32 species. New assemblies have been released for 15 species previously supported, including human, mouse, rat, chimp, rhesus, opossum, chicken, frog (Xenopus tropicalis), zebrafish, dog, sea squirt (Ciona intestinalis) and four Drosophilids. The sequencing centers, organizations and individuals who contributed to the assemblies and annotations are acknowledged at http://genome.ucsc.edu/goldenPath/credits.html.
Each new assembly contains at a minimum, assembly data, comparative genomics tracks and automatically updated tracks of RefSeq Genes and GenBank mRNA and EST alignments (8). For assemblies with small numbers of native RefSeq alignments and significant evolutionary distance from human (e.g. cow, chicken, opossum and Xenopus), we assist gene-finding with a track, Human Proteins, which maps human exons using tBLASTn.
Human genome browsers
The genome browsers on the human assemblies have the richest set of annotations: the latest assembly, hg18 (NCBI Build 36), was released in 2006. Collaborators continue to submit data for both hg17 and hg18. Human assembly annotations include assembly details, FISH-clone mapping information, fosmid and BAC end pairs, SNPs, sno/miRNAs, numerous gene-prediction tracks, splice-variant data, microarrays and other gene expression data, promoter prediction, microsatellites, segmental duplications and other repeats, and comparative genomics data.
Additionally, the hg16 and hg17 browsers present dozens of tracks as the data portal for the Encyclopedia of DNA Elements (ENCODE) project (9). This project provides deep coverage of 1% of the genome, mapping a variety of characteristics, including DNase hypersensitivity, histone methylation and acetylation, microarray-detected gene expression, transcription initiation sites, replication origins and gene predictions.
Known genes
The search capability for finding Known Genes (10) has been upgraded for speed, and now includes keywords from all descriptive fields in the GO (11), UniProt (12) and RefSeq records as well as gene aliases from a variety of sources. The UCSC Known Genes track includes link-outs to a wide variety of databases, with each Known Gene serving as a portal to a rich store of information, linking to the relevant pages in UniProt, Entrez Gene, PubMed, OMIM (13), GeneLynx (14), GeneCards (15), HGNC (16), CGAP (17), HPRD (18), Stanford Source (19), Ensembl (20), ExonPrimer, MGD (21), H-INV (22), GO, KEGG (23), BioCyc, Reactome and Allen Brain Atlas (ABA) (24).
MAMMALIAN GENE COLLECTION
The Mammalian Gene Collection (MGC) (25) at the National Institutes of Health is building a collection of full-length open-reading-frame clones for human, mouse, rat and cow genes. The MGC Genes track on the UCSC Genome Browser displays alignments for these clones and now supports a redesigned details page. Data for individual clones are displayed with details from MGC, including tissue source, identity of the gene product, clone library information, coding sequence coordinates and modification date. As with other alignment tracks in the Genome Browser, links are provided to the sequences of the DNA, RNA and protein, for both the clone and the reference assembly. Link-outs are provided to multiple resources, including the GenBank record, IMAGE Consortium page, and the MGC Clone database. Additionally, it is now possible to order MGC clones directly from the appropriate supplier via a link on the UCSC MGC Gene track details page.
Conservation tracks
Sequence conservation tracks using multiz and phastCons (26,27) are presented for 10 of the 12 new vertebrate assemblies, including 17-species comparatives on the most recent human and mouse assemblies, hg18 and mm8. These tracks allow direct visualization of homology to all the other species, providing intuitive detection of cross-species relationships in the DNA sequences. The user interface provides access to the actual sequences as well as the ability to switch easily to a browser displaying the other assemblies.
The Conservation track now has enhanced display options. Using the ‘Display chains between alignments’ configuration option, it is now easier to determine at a glance the exact status of the sequences, especially potential detection of the presence of retrotransposons. It is also now possible to view the codon translations using reading frames based on a variety of gene sets for any of the aligning species.
Simple Nucleotide Polymorphism (SNP) tracks
The UCSC Genome Browser now has an extensive revision of the data presentation for SNPs from NCBI's dbSNP (28). Data from dbSNP build 125 are mapped to the human, chimp and dog assemblies (hg17, panTro1 and canFam1, respectively) and from build 126 to human hg18 and mouse mm8. The redesigned details page, available by clicking on an individual SNP in the browser image, displays the results of extensive parsing of the dbSNP data and shows comparisons to the reference assembly.
The snp126 table in the hg18 database contains information about more than 12 million SNPs including the identity of both the SNP and the reference allele, the molecular type, class, validation status and heterozygosity information, where available. The user may choose to display or color subsets of SNPs based on these criteria. Another table records inconsistencies in the dbSNP record such as SNPs that align in more than one genomic location or where the reported flanking sequences and locations do not match. Individual exceptions are displayed on the SNP details page as ‘UCSC Annotations’. The full list of exceptions and their frequencies is available for download.
HapMap and other variation tracks
The HapMap LD track has been added to human assemblies in the Genome Browser. This track uses publicly available data from the HapMap Consortium (29) and shows linkage disequilibrium data for three populations, of European, African and Asian ancestry. These data are plotted graphically against the genomic coordinate scale, so that genes that are segregating non-randomly with respect to other genes within a 250 Mb distance are clearly distinguished. The track may be configured to display the differences as r2, D' or LOD values.
The HapMap LD and SNPs tracks augment existing Browser annotations that examine variation within the human population at a variety of scales (30). The SNPs track shows variation at the sub-kilobasepair scale, while the existing Structural Variation track shows copy-number polymorphism up to megabase scale. The seven subtracks of the Structural Variation track show results from a number of third-party contributors and span the range from the 5–10 kb scale [delHinds table (31)] to the near-megabase [cnpSebat table (32)].
A new set of tracks, Microsatellites, is now available on the browser for most assemblies. This track extends the display of genomic variation by showing stretches where two or three nucleotides are perfectly repeated 15 times or more. This is a subset of the Simple Repeats track and allows users to more easily locate these regions, which are often polymorphic in the population and therefore serve as valuable markers in linkage studies.
New track types: regulatory potential, gene trap
A new track on human and mouse assemblies, Regulatory Potential 7 Species (RP), is based on the principle that regions conserved across long evolutionary timescales are likely to be functionally significant. This track takes the concept further (33) by using known regulatory motifs from human and six other vertebrates as a training set to find short sequence patterns that cluster in regulatory sites, then locating recurrences of such clusters genome-wide. The quantitative RP score associated with each base in the assembly is displayed as continuously variable data in a format known as ‘wiggle’, which is optimized for rapid display in browser tracks at a wide range of viewing scales.
Data from the International Gene Trap Consortium (34) are presented in a new track for three mouse assemblies. Mapping information shows the location of gene inserts in mouse cell lines and link-outs to the Consortium database provide access to clone construction and purchasing details.
VISIGENE
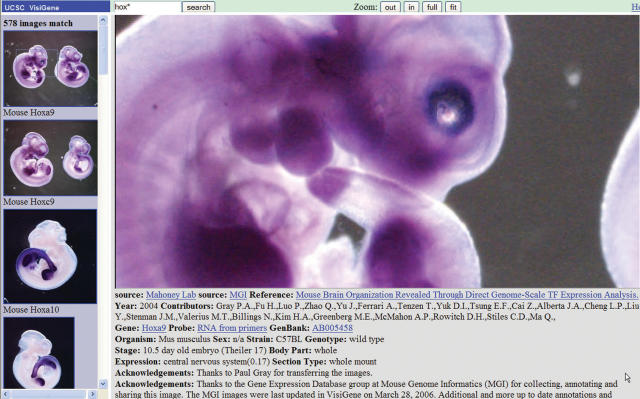
The new Genome Browser module, VisiGene, displays in situ hybridization images from several sources as a fully integrated feature of the human and mouse Known Genes tracks and of the Gene Sorter. Mouse images are presented from the Mahoney Center for Neurooncology, the Gene Expression Nervous System Atlas (GENSAT) (35), ABA and Mouse Genome Informatics (MGI) (21). Frog (Xenopus laevis) images from the Japanese National Institute of Basic Biology (NIBB) XDB project are also included. VisiGene images are also directly accessible from a link on the main index page of the web interface and are fully searchable by gene name or accession, including the use of the asterisk wildcard character.
VisiGene provides a series of thumbnail images in a panel to the left of the main viewing area and fully annotated full-resolution images in the main viewer area to the right (Figure 1). These are high-resolution images viewable in a virtual microscope interface that supports seven levels of zoom plus three levels of over-zoom.
Figure 1.
Screenshot of VisiGene display for mouse Hoxa9 gene.
Mirror sites
The UCSC Genome Browser is mirrored in many labs and institutions worldwide from the UCSC codebase (free for non-commercial users). Additionally, three locations in the US work closely with UCSC, syncing the data nightly, and keeping abreast of software updates. They are located at the Medical College of Wisconsin (http://genome.brc.mcw.edu/), Duke University (http://genome-mirror.duhs.duke.edu/) and now, Cornell University (partial mirror: http://genome-mirror.bscb.cornell.edu/). These mirrors may be used when the UCSC site is unavailable, but users are encouraged to use the UCSC site.
NEW FEATURES
Custom track enhancements
The custom track utility allows users to upload and maintain tracks of their own data, mapped to genomic coordinates. These tracks are displayed in the Browser along with the rich store of data resident at UCSC, allowing direct visual comparison with other annotation tracks. The interface for user-supplied custom tracks has been enhanced by the addition of the new module, hgCustom, providing enhanced user control over custom track features as well as the addition of new features.
User-supplied custom tracks may now be loaded from separate files at separate times without losing previously loaded custom tracks. Tracks may be added to an existing track set by any of three methods: file upload, copy/paste in a text box or via a URL to a file on a web-accessible server of the user's choice. Files may be uploaded in several compressed formats, including .gz, .Z or .bz2. All tracks now remain visible for at least 48 h and may be selectively modified or deleted via the Custom Track Management page.
The user may now provide a track description and access configuration options for a Custom track, features previously available only for UCSC-hosted tracks. A track description is useful for displaying information about the track's construction or experimental details closely associated with the data.
phyloGif
A new utility, phyloGif, enables users to show evolutionary relationships by drawing phylogenetic trees. Accessed via the ‘Utilities’ link on the main page, the tool accepts text input describing the relationships (and optionally, evolutionary distance) among the organisms. The user may choose to draw the evolutionary distances indicated by the branch-length values or to normalize the tree to equal branch lengths for all members of the tree.
hgConvert
As new assemblies are released for an organism, researchers are commonly faced with the task of converting coordinates for probes, clones, primers and annotations from a previous assembly to the newly released coordinates. The Browser provides an improved utility under the ‘Convert’ link in each assembly for conversion in either direction between two assemblies of the same organism. The conversion utility uses files created by the chaining and netting process (36) to find the best long-range orthology.
New display features
Several new display innovations and navigation aids have been added to the configuration page, which is accessible via the configuration button on the gateway page and directly below the browser image. Users are no longer constrained to display Browser tracks in the default order. Using the option ‘Enable track re-ordering’, tracks are easily rearranged within a track group, moved from one group to another, or collected together into a ‘user’ group along with custom tracks. Users may therefore customize a browser graphic display to suit their own needs, which is particularly useful for publications, in conjunction with the PDF and postscript image output options.
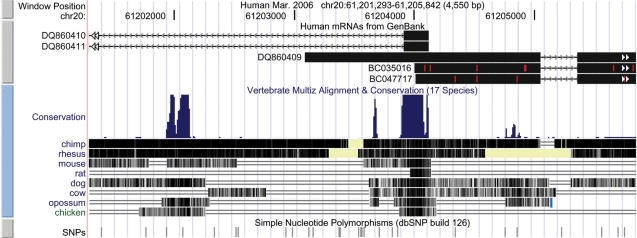
The new ‘Next/previous item navigation’ option, off by default, draws small double-headed arrows on either end of a gene annotation in the browser display (Figure 2) if the annotation extends offscreen. A click on the arrows enables a quick jump to the next exon.
Figure 2.
New display features. Region of HAR1 gene on chromosome 20 (37). Blue or gray mini-button on the left affords access to track configuration. mRNA track: red ticks, bases differ from reference; double arrows navigate to next exon. Conservation track: yellow, aligning sequence has missing data; double lines, unaligned DNA exists on both reference and aligning species; blue tick in opossum track, end of synteny block.
The configuration page for an individual track is accessible from several locations, including a mini-button in the browser image, a link in the track controls and via the configuration buttons. A new configurable display feature in mRNA tracks for all assemblies allows the user to visualize differences in codon translation relative to the reference assembly even when zoomed out to low resolution. The ‘Color track by codons: different mRNA bases’ option allows the user to view the entire GenBank collection of mRNAs at any scale and visually detect the occurrence of SNPs relative to the reference assembly.
FUTURE DIRECTIONS
In the coming years, the browser will be expanded into the area of disease-related genes, including medical resequencing. A whole-genome viewer to show results of genetic association studies is under development. We will be releasing browsers for numerous low-coverage (2x) assemblies using a new automated process. We intend to implement a ‘personalized cart’ function to allow the user to save, revisit and share browser configurations. Our custom track feature will be expanded to include modifications that will greatly increase the time a track will remain available to users.
Acknowledgments
The UCSC Genome Browser project is funded by grants from the National Human Genome Research Institute (NHGRI), the Howard Hughes Medical Institute (HHMI) and the National Cancer Institute (NCI). We would like to thank the many collaborators who have contributed annotation data to our project, as well as our users for their feedback and support. We would also like to thank the dedicated system administrators who have provided an excellent computing environment: Jorge Garcia, Patrick Gavin, Chester Manuel, Victoria Lin and Paul Tatarsky. Funding to pay the Open Access publication charges for this article was provided by HHMI.
Conflict of interest statement. R.M.K., D.K., A.S.Z., H.T., D.J.T., A.T., C.W.S., K.E.S., A.S., K.R.R., B.R., B.J.R., A.P., F.H., A.S.H., R.A.H., M.D., H.C., G.P.B., R.B., D.H. and W.J.K. receive royalties from the sale of UCSC Genome Browser source code licenses to commercial entities.
REFERENCES
- 1.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kent W.J., Hsu F., Karolchik D., Kuhn R.M., Clawson H., Trumbower H., Haussler D. Exploring relationships and mining data with the UCSC Gene Sorter. Genome Res. 2005;15:737–741. doi: 10.1101/gr.3694705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu F., Pringle T.H., Kuhn R.M., Karolchik D., Diekhans M., Haussler D., Kent W.J. The UCSC Proteome Browser. Nucleic Acids Res. 2005;33:D454–D458. doi: 10.1093/nar/gki100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bejerano G., Siepel A.C., Kent W.J., Haussler D. Computational screening of conserved genomic DNA in search of functional noncoding elements. Nature Methods. 2005;2:535–545. doi: 10.1038/nmeth0705-535. [DOI] [PubMed] [Google Scholar]
- 8.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2006;34:D16–D20. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 10.Hsu F., Kent W., Clawson H., Kuhn R., Diekhans M., Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 11.The Gene Ontology Consortium The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu C.H., Apweiler R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., et al. The Universal Protein resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamosh A., Scott A.F., Amberger J., Bocchini C., Valle D., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30:52–55. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lenhard B., Hayes W.S., Wasserman W.W. GeneLynx: a gene-centric portal to the human genome. Genome Res. 2001;11:2151–2157. doi: 10.1101/gr.199801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Safran M., Chalifa-Caspi V., Shmueli O., Olender T., Lapidot M., Rosen N., Shmoish M., Peter Y., Glusman G., Feldmesser E., et al. Human Gene-Centric Databases at the Weizmann Institute of Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res. 2003;31:142–146. doi: 10.1093/nar/gkg050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eyre T.A., Ducluzeau F., Sneddon T.P., Povey S., Bruford E.A., Lush M.J. The HUGO Gene Nomenclature Database, 2006 updates. Nucleic Acids Res. 2006;34:D319–D321. doi: 10.1093/nar/gkj147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strausberg R.L., Buetow K.H., Emmert-Buck M.R., Klausner R.D. The Cancer Genome Anatomy Project: building an annotated gene index. Trends Genet. 2000;16:103–106. doi: 10.1016/s0168-9525(99)01937-x. [DOI] [PubMed] [Google Scholar]
- 18.Mishra G.R., Suresh M., Kumaran K., Kannabiran N., Suresh S., Bala P., Shivakumar K., Anuradha N., Reddy R., Raghavan T.M., et al. Human protein reference database—2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diehn M., Sherlock G., Binkley G., Jin H., Matese J.C., Hernandez-Boussard T., Rees C.A., Cherry J.M., Botstein D., Brown P.O., et al. SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res. 2003;31:219–223. doi: 10.1093/nar/gkg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birney E., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blake J.A., Eppig J.T., Bult C.J., Kadin J.A., Richardson J.E. Mouse Genome Database Group. The Mouse Genome Database (MGD): updates and enhancements. Nucleic Acids Res. 2006;34:D562–D567. doi: 10.1093/nar/gkj085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imanishi T., Itoh T, Suzuki Y, O'Donovan C, Fukuchi S, Koyanagi K.O., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., et al. Integrative annotation of 21037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:856–875. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K.F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sunkin S. Towards the integration of spatially and temporally resolved murine gene expression databases. Trends Genet. 2006;22:211–217. doi: 10.1016/j.tig.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 25.Gerhard D.S., Wagner L., Feingold E.A., Shenmen C.M., Grouse L.H., Schuler G., Klein S.L., Old S., Rasooly R., Good P., et al. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smigielski E., Sirotkin K., Ward M., Sherry S. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 2000;28:352–355. doi: 10.1093/nar/28.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The International HapMap Consortium. The international HapMap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 30.Thomas D.J., Trumbower H., Kern A.D., Rhead B., Kulin R.M., Haussler D., Kent W.J. Variation Resources at UCSC. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkl953. (Database Issue), in press (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hinds D., Stuve L., Nilsen G., Halperin E., Eskin E., Ballinger D., Frazer K., Cox D. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 32.Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 33.King D.C., Taylor J., Elnitski L., Chiaromonte F., Miller W., Hardison R.C. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15:1051–1060. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nord A.S., Chang P.J., Conklin B.R., Cox A.V., Harper C.A., Hicks G.G., Huang C.C., Johns S.J., Kawamoto M., Liu S., et al. The International Gene Trap Consortium Website: a portal to all publicly available gene trap cell lines in mouse. Nucleic Acids Res. 2006;34:D642–D648. doi: 10.1093/nar/gkj097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heintz N. Gene Expression Nervous System Atlas (GENSAT) Nature Neurosci. 2004;7:483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- 36.Kent W.J., Baertsch R., Hinrichs A., Miller W., Haussler D. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl Acad. Sci. USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollard K.S., Salama S.R., Lambert N., Lambot M., Coppens S., Pedersen J.S., Katzman S., King B., Onodera C., Siepel A., et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]