Abstract
The recent explosion in protein data generated from both directed small-scale studies and large-scale proteomics efforts has greatly expanded the quantity of available protein information and has prompted the Saccharomyces Genome Database (SGD; http://www.yeastgenome.org/) to enhance the depth and accessibility of protein annotations. In particular, we have expanded ongoing efforts to improve the integration of experimental information and sequence-based predictions and have redesigned the protein information web pages. A key feature of this redesign is the development of a GBrowse-derived interactive Proteome Browser customized to improve the visualization of sequence-based protein information. This Proteome Browser has enabled SGD to unify the display of hidden Markov model (HMM) domains, protein family HMMs, motifs, transmembrane regions, signal peptides, hydropathy plots and profile hits using several popular prediction algorithms. In addition, a physico-chemical properties page has been introduced to provide easy access to basic protein information. Improvements to the layout of the Protein Information page and integration of the Proteome Browser will facilitate the ongoing expansion of sequence-specific experimental information captured in SGD, including post-translational modifications and other user-defined annotations. Finally, SGD continues to improve upon the availability of genetic and physical interaction data in an ongoing collaboration with BioGRID by providing direct access to more than 82 000 manually-curated interactions.
INTRODUCTION
The Saccharomyces Genome Database (SGD) collects, organizes and presents biological information about the genes and proteins of the budding yeast Saccharomyces cerevisiae. In 2003, in response to the community's needs for additional sequence-based predictive protein information, SGD introduced the Protein Information page, the PDB Homologs page, and the eMOTIF resource for the display of shared protein motifs (1). Since that time, there has been a marked increase in the number of studies focused on protein function, regulation and pathway/process involvement. These include a number of studies aimed at mapping the complete interactome by investigating the composition of protein complexes and specific protein–protein interactions, as well as other studies focused on proteome-wide post-translational modifications (2–6). Much of this research has become possible due to the increased use of proteome chip technologies, such as protein microarrays, as well as technological advances in mass spectrometry-based proteomics that result in increased sensitivity and higher throughput (7–9).
To meet the needs of both the traditional biochemist and the proteomics researcher, we have improved the integration and display of protein data at SGD by redesigning protein information pages, introducing a new sequence-based visualization tool and utilizing improved algorithms for the calculation of predictive information based on primary amino acid sequence.
ORGANIZATION OF THE PROTEIN INFORMATION PAGE(S)
The new Protein Information pages (http://www.yeastgenome.org/protein) are accessible via the ‘Protein’ tab located at the top of all Locus Summary pages for protein-encoding genes. Each Protein Information page presents basic locus-specific protein information and provides access to detailed information regarding HMM domains, protein family HMMs, motifs and physico-chemical properties by clicking on a sub-tab. The types of information displayed on these pages are listed in Table 1 and discussed in more detail below.
Table 1.
Summary of protein information available at SGD (http://www.yeastgenome.org/protein)
| Protein information page | Nomenclature fields |
| Description fields | |
| Predicted protein sequence and basic information | |
| Proteome Browser | |
| Summary links (physical interactions, HMM domains and homologs) | |
| Physico-chemical Properties page | Amino acid composition |
| Atomic composition | |
| Extinction coefficient | |
| Aliphatic index | |
| Estimated half-life | |
| Instability Index | |
| Coding region translation calculations | |
| Domains/Motifs page | InterPro-derived shared and unique domains |
| TMHMM-predicted transmembrane domains | |
| SignalP-predicted signal peptides |
The main Protein Information page has been redesigned to provide basic protein information clearly and concisely with a familiar and readily navigable layout similar to that of the SGD Locus Summary page. The top section of the page, devoted to nomenclature, provides standard and systematic protein names plus any associated aliases. The nomenclature section is followed by several descriptive information fields including: Description, which provides a brief synopsis of the function and/or role of the gene product within the cell; Name Description, which contains the expanded form of the standard gene name acronym; and gene product, which describes the specific function of the protein when it is known. These fields have been recently reviewed and rewritten using a standard, consistent format so that they accurately reflect the current state of knowledge for each gene product. The references for this information are found at the bottom of the page. This section also includes basic information including the predicted length, molecular weight and isoelectric point of the protein.
The middle of the Protein Information page includes a ‘thumbnail’ of the GBrowse graphical proteome viewer (see below), as well as links that summarize and provide access to other types of protein-related data such as physical interactions. These physical interactions are part of the extensive collection of genetic and physical interactions available as the result of an ongoing collaboration with BioGRID (http://www.thebiogrid.org/) and are mirrored by SGD. These data are manually curated from the primary literature using a standard format to describe the protein interactions and are updated monthly. As of September 2006, this collaboration documents 82 633 genetic and physical interactions (10,11).
Towards the bottom of the Protein Information page are the complete amino acid sequence and links to sequence records held at various external sequence databases. This is followed by a list of external databases that provide additional protein information and a list of references for all basic information presented on the page.
GBROWSE GRAPHICAL PROTEOME VIEWER
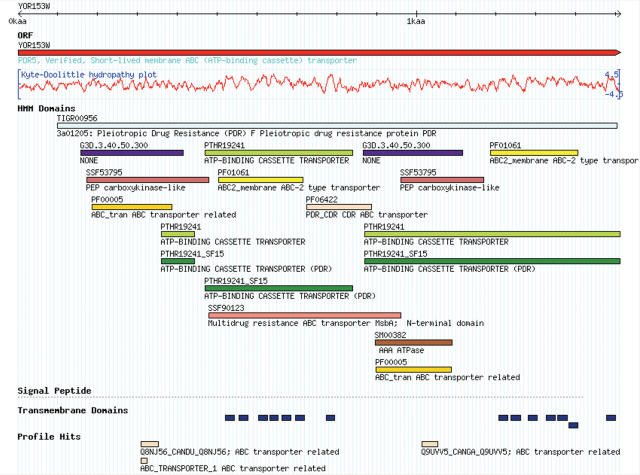
To improve the visualization and navigability of sequence-based protein information, SGD has introduced an interactive Proteome Browser (http://www.yeastgenome.org/scproteome) (Figure 1) based on GBrowse, a genome browser developed by the Generic Model Organism Database (GMOD) project [http://www.gmod.org/; (12)]. The Proteome Browser has been customized to view protein-centric information and consolidates the display of multiple types of information including HMM domains, protein family HMMs, motifs, signal peptides, profile hits, and hydropathy plots by populating individual tracks with distinct information types. Pop-up mouseover functionality provides easy access to InterPro-derived HMM domain details including source, name, description, database identifier and E-value or score of matches. This Proteome Browser also supports the interactive feature of GBrowse that allows the addition of user-provided annotations.
Figure 1.
The SGD Proteome Browser. The Pdr5p primary protein sequence is shown in the SGD Proteome Browser, a GBrowse-derived tool used to display predicted features, including: InterProScan-derived domains and motifs color-coded by HMM type, hidden Markov model-derived transmembrane domains (TMHMM), the Kyte-Doolittle hydropathy plot and profile hits. The Proteome Browser can also be used for the display of experimentally-derived features. HMM Domains are color-coded by originating resource: orange/yellows = Pfam, reds = Superfamily, purples = Gene3D, greens = Panther, blues = TIGRFAM and browns = SMART.
IMPROVED PREDICTIVE INFORMATION
In response to community feedback, SGD has updated the algorithms used for the prediction of protein functional HMM domains, transmembrane domains and signal peptides. Beginning in 2005, HMM domains, protein family HMMs and motifs in S.cerevisiae proteins were predicted by software and datasets assembled by the InterPro database, using InterProScan (13). This included several HMM packages such as Pfam, SMART, TIGRFAM, Panther, Gene3D and Superfamily (14). Pfam is a comprehensive collection of protein families and HMM domains represented by multiple sequence alignments and profile HMMs. SMART contains a smaller library of HMM domains found in signaling, extracellular and chromatin-associated proteins. TIGRFAM is a collection of manually-curated protein families based on multiple sequence alignments, HMMs and additional annotations. Panther contains a large collection of protein families, subfamilies and domains, classified by experts based on function. Finally, Gene3D and Superfamily contain a collection of HMMs based on structural classification in the CATH and SCOP databases, respectively. The InterPro-derived results have been expanded to include two profile-based methods: BlastProDom and ProfileScan (ProSite) (15,16). InterProScan results are updated quarterly. Transmembrane domains are now predicted using TMHMM, which has been independently rated as superior for predicting transmembrane helices (17,18). Finally, the presence and location of signal peptides are predicted using SignalP (v. 3.0), a popular method that uses either a neural network or an HMM (19). SGD is using the HMM method of SignalP analysis. All predictions of functional domains, transmembrane domains, and signal peptides are included for display in the Proteome Browser (see above), providing an integrated view of multiple HMM and domain prediction packages.
On the Physico-chemical Properties page, we have included additional properties calculated by ExPASy's ProtParam tool, such as estimated protein half-life, instability index and extinction coefficient (20). A complete list of properties is available in Table 1.
SUMMARY
The increase in protein information generated using both traditional approaches and technology-driven large-scale studies has necessitated the expansion of protein annotation at SGD. The redesign of the main Protein Information page and Domains/Motifs page and the addition of a Physico-chemical Properties page have improved the organization of protein information. Furthermore, a Proteome Browser has been developed, enhancing the ability to visualize both predictive and experimentally-derived information. Finally, more robust algorithms have been employed to enhance the quality of sequence-based predictions at SGD. These changes will allow the integration of new data types in the future because the protein pages have been designed to be expandable. SGD is committed to increasing the ease of access to information about S.cerevisiae and welcomes all comments from the research community toward this end. Please send any suggestions about the Protein Information pages, the Proteome Browser or any other tool or resource at SGD to: yeast-curator@genome.stanford.edu.
Acknowledgments
The SGD project is supported by a P41 grant from the NHGRI HG001315 (J.M.C.). Funding to pay the Open Access publication charges for this article was provided by National Human Genome Research Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Weng S., Dong Q., Balakrishnan R., Christie K., Costanzo M., Dolinski K., Dwight S.S., Engel S., Fisk D.G., Hong E., et al. Saccharomyces Genome Database (SGD) provides biochemical and structural information for budding yeast proteins. Nucleic Acids Res. 2003;31:216–218. doi: 10.1093/nar/gkg054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krogan N.J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 3.Gavin A.C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L.J., Bastuck S., Dumpelfeld B., et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 4.Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 5.Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 6.Peng J., Schwartz D., Elias J.E., Thoreen C.C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S.P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 7.Tyers M., Mann M. From Genomics to Proteomics. Nature. 2003;422:193–197. doi: 10.1038/nature01510. [DOI] [PubMed] [Google Scholar]
- 8.Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 9.Aebersold R., Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 10.Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reguly T., Breitkreutz A., Boucher L., Breitkreutz B.J., Hon G.C., Myers C.L., Parsons A., Friesen H., Oughtred R., Tong A., et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J. Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gribskov M., McLachlan A.D., Eisenberg D. Profile analysis: detection of distantly related proteins. Proc. Natl Acad. Sci. USA. 1987;84:4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Servant F., Bru C., Carrere S., Courcelle E., Gouzy J., Peyruc D., Kahn D. ProDom: automated clustering of homologous domains. Brief. Bioinform. 2002;3:246–251. doi: 10.1093/bib/3.3.246. [DOI] [PubMed] [Google Scholar]
- 17.Moller S., Croning M.D.R., Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 18.Krogh A., Larsson G., von Heijne G., Sonnhammer E.L.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 19.Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 20.Gasteiger E., Hoogland C., Gattiker A., Duvand S., Wilkins M.R., Appel R.D., Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker J.M., editor. The Proteomics Protocols Handbook. Totowa, NJ: Humana Press; 2005. pp. 571–607. [Google Scholar]