Abstract
Information on protein–protein interactions is still mostly limited to a small number of model organisms, and originates from a wide variety of experimental and computational techniques. The database and online resource STRING generalizes access to protein interaction data, by integrating known and predicted interactions from a variety of sources. The underlying infrastructure includes a consistent body of completely sequenced genomes and exhaustive orthology classifications, based on which interaction evidence is transferred between organisms. Although primarily developed for protein interaction analysis, the resource has also been successfully applied to comparative genomics, phylogenetics and network studies, which are all facilitated by programmatic access to the database backend and the availability of compact download files. As of release 7, STRING has almost doubled to 373 distinct organisms, and contains more than 1.5 million proteins for which associations have been pre-computed. Novel features include AJAX-based web-navigation, inclusion of additional resources such as BioGRID, and detailed protein domain annotation. STRING is available at http://string.embl.de/
INTRODUCTION
A fully comprehensive view of all functionally relevant protein interactions is still not available for any species, not even for relatively simple, single-celled model organisms. However, this information is essential for a systems-level understanding of cellular behavior, and it is needed in order to place the molecular functions of individual proteins into their cellular context.
For detecting direct physical binding between proteins, numerous small-scale and high-throughput experiments have been undertaken, and most of their reported interactions are available from dedicated interaction databases (1–4), as well as from multipurpose databases centered on specific model organisms (5–7). However, the growth of interaction data is severely lagging behind the pace of genome sequencing, so that for most genomes and proteins known to date no interaction data is available. Furthermore, proteins do not only interact physically: indirect associations such as genetic interactions or shared pathway memberships are equally important for a complete understanding of cellular function, but are for the most part not stored in interaction databases. Instead, they are available from a variety of pathway databases (8,9) and from the scientific literature.
The database STRING (‘Search Tool for the Retrieval of Interacting Genes/Proteins’) aims to collect, predict and unify most types of protein–protein associations, including direct and indirect associations. In order to cover organisms not yet addressed experimentally, STRING runs a set of prediction algorithms (10), and transfers known interactions from model organisms to other species based on predicted orthology of the respective proteins (11). STRING has grown from a purely predictive resource covering mainly prokaryotes (12) to a comprehensive tool integrating protein association information from all domains of life (Figure 1). Each interaction in the database is annotated with a benchmarked numerical confidence score, which can be used to filter the interaction network at any desired stringency. All data in STRING are stored in relational database tables. The interaction information is freely available for download, but download of the entire database content requires a license agreement to prevent redistribution (free for academic users who only access the previous version number).
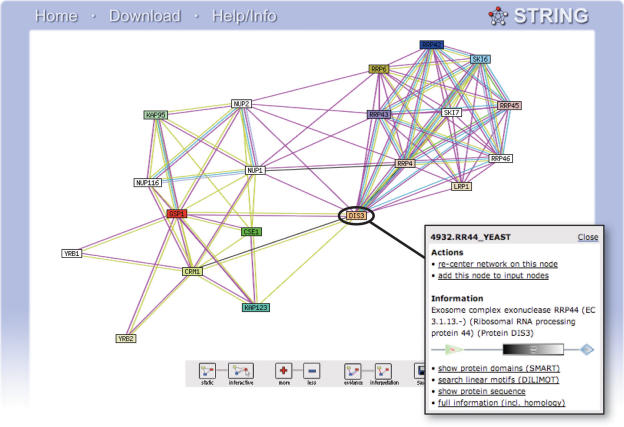
Figure 1.
Protein interaction network in STRING. Screenshot from STRING showing a network of Saccharomyces cerevisiae proteins [the exosome complex, upper right, is seen weakly associated with proteins from nuclear transport, lower left, see also Ref. (26)]. The inset shows the context menu available for all STRING proteins—in the context menu, annotation and domain architecture are shown directly, and links to other databases and tools are available (22,23). In the network, links between proteins signify the various interaction data supporting the network, colored by evidence type (see STRING website for color legend).
KNOWN AND PREDICTED INTERACTIONS
Known interactions in STRING are primarily imported from existing excellent interaction databases (1–5,8,9), and are complemented by automated text mining of PubMed abstracts and several other bodies of scientific text [such as from Ref. (6)]. As is the case for all interactions in STRING, imported interactions are mapped onto a consistent set of proteins and identifiers, thereby facilitating comparison between datasets. STRING does not store specific details regarding splicing isoforms or post-translational modifications, but instead reduces protein isoforms to a single protein per locus (usually as defined by the longest known protein-coding transcript). This level of resolution enables efficient storage and is compatible with most prediction/transfer algorithms, which usually operate only at the level of the gene locus.
Known interactions are further complemented by de novo interaction predictions derived from several comparative genomics prediction algorithms that are mainly applicable to prokaryotes (13–19). These algorithms systematically compare genomes, searching for frequently observed gene neighborhoods, gene fusion events and similarities in gene occurrence across genomes. For each prediction algorithm, dedicated viewers of the genomic evidence are available in STRING.
Interaction evidence from model organisms is often useful for other organisms as well, especially when orthologs of interacting proteins can be clearly identified in the second organism. STRING systematically executes such orthology transfers, using both precomputed orthologs from the COG database (20), as well as a homology-based orthology scheme computed de novo (11). STRING can thus immediately predict a large number of interactions for any newly sequenced genome, as soon as it is included into the system. The combination of known, predicted and transferred interactions is unique, making STRING the most comprehensive interaction resource available to date, especially for organisms not addressed experimentally.
The homology data stored in STRING form the basis for the interaction transfers, and are the result of more than 7 × 1011 pairwise protein comparisons using the sensitive Smith–Waterman dynamic programming algorithm. This dataset is a very useful asset in itself [see also (21)], and can be accessed independently of the protein interaction networks by locally installing the STRING database files. Users of the website can also browse all of the homologs detected for any protein of interest, and can inspect alignments with very fast response times (Figure 2).
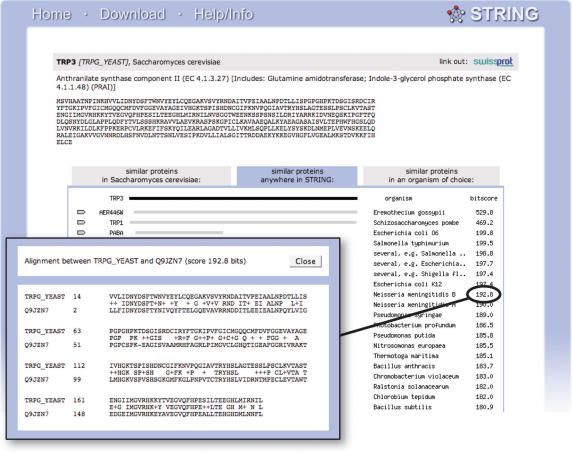
Figure 2.
Precomputed homology relations and alignments. For most genomes contained in STRING, sensitive all-against-all homology searches using the Smith–Waterman algorithm are included. These form the basis for assigning orthologs and transferring interaction information, but are also available directly to the user. Because they are stored in a relational database, access to homologs and alignments for any protein of interest is possible without the usual waiting time.
NEW FEATURES AND IMPROVEMENTS IN STRING 7
The network viewer in STRING (Figure 1) is the central information source and navigation hub for the user. It has been extended through a context-sensitive menu-box, which displays associated information for any protein in the network. This menu includes a graphical summary of protein domains and features, and allows the user to link out to other external resources such as the motif discovery tool DILIMOT (22). STRING is now also tightly integrated with the SMART protein architecture research tool (23). With the latter it shares a common set of genomes and proteins, for which consistent results are pre-computed and stored. This enables automatic interlinking between both resources (SMART includes interaction previews, and STRING includes domain architecture previews). The topology and evolution of interaction networks can thus be studied both at the level of proteins as well as at the level of individual domains.
Since the last update (11), STRING has grown substantially both in terms of data sources and number of organisms covered. Five new databases are included [MINT, HPRD, BioGRID, DIP and Reactome (2–5,8)], as well as 194 new organisms. Especially due to this latter increase in completely sequenced organisms, the architecture of STRING had to be substantially upgraded so that it can accommodate present and future growth. With respect to the user interface, this required changes in the viewers for the genomic context data, which could no longer show all of the genomes simultaneously by default. Instead, STRING uses a phylogenetic tree of species to collapse redundant genomes; this tree has been derived from concatenated alignments of a small number of universal protein families (24). Users can navigate the tree by expanding or collapsing its sub-branches, thus choosing which organisms to focus on. AJAX technology (‘Asynchronous JavaScript and XML’) is then used to fetch the requested information into the existing, pre-loaded browser page, thus increasing useability and speed.
With respect to the underlying database structure, changes were necessary in the way homology data and interaction transfers are stored. Both can no longer be computed and stored in an ‘all-against-all’ fashion, because of their quadratic scaling with the number of genomes. Beginning with version 7, STRING therefore adopts a two-layered approach when accommodating fully sequenced genomes (Figure 3): important model organisms and those for which experimental data are available form the ‘core genomes’, all other genomes form the periphery. Within the core, homology searches and interaction transfers are still executed in an all-against-all fashion, whereas for peripheral genomes only searches against the core are included. These and other changes in STRING dramatically improve the scalability of the resource, leading to faster update cycles even when the number of sequenced genomes is to increase as fast as currently projected. Together with future plans to increase the scope and specificity of the stored interaction information, STRING should thus continue to facilitate not only network research but also wider projects that range from phylogenetics to metagenomics (24,25).
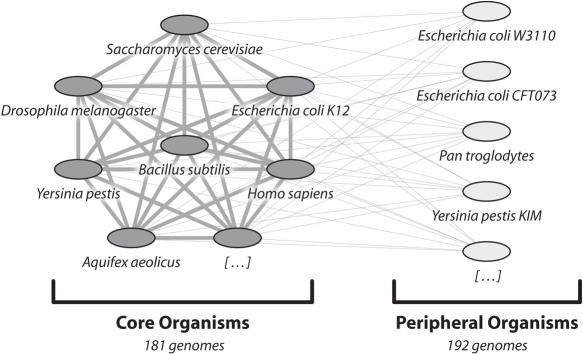
Figure 3.
Organisms covered by STRING. STRING currently contains 373 fully sequenced organisms. These are divided into ‘Core Organisms’ and ‘Peripheral Organisms’. The former include all important model organisms for which experimental data are available, as well as selected representatives for cases of redundant genome sequencing (e.g. when several closely related strains of a bacterial species have been sequenced, only one strain is included). The ‘Peripheral Organisms’ form the remainder; they tend to be somewhat redundant, and usually have little more than genomic sequence information annotated. For the core organisms, homology relations and interaction transfers are fully computed, whereas the peripheral organisms are only connected to the core but not among themselves (the graphic shows only a small selection of organisms; lines indicate homology searches and interaction transfers). This architecture allows STRING to encompass all sequenced genomes, while still keeping database size and computation time within reasonable limits.
Acknowledgments
The authors wish to thank Dianna Fisk from the Saccharomyces Genome Database for access to the Gene Summary Paragraphs, and Toby Gibson, Martijn Huynen, Victor Neduva, Rune Linding and members of the Bork group for continued feedback and discussions. This work was supported in part by grants from the Bundesministerium für Forschung und Bildung, Germany, as well as through the ADIT Integrated Project, contract number LSHB-CT-2005-511065, and through the BioSapiens Network of Excellence, contract number LSHG-CT-2003-503265, both funded by the European Commission FP6 Programme. Funding to pay the Open Access publication charges for this article was provided by the University of Zurich, through its Research Priority Program ‘Systems Biology and Functional Genomics’.
Conflict of interest statement. None declared.
REFERENCES
- 1.Alfarano C., Andrade C.E., Anthony K., Bahroos N., Bajec M., Bantoft K., Betel D., Bobechko B., Boutilier K., Burgess E., et al. The biomolecular interaction network database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zanzoni A., Montecchi-Palazzi L., Quondam M., Ausiello G., Helmer-Citterich M., Cesareni G. MINT: a Molecular INTeraction database. FEBS Lett. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 4.Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mishra G.R., Suresh M., Kumaran K., Kannabiran N., Suresh S., Bala P., Shivakumar K., Anuradha N., Reddy R., Raghavan T.M., et al. Human protein reference database—2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirschman J.E., Balakrishnan R., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., Fisk D.G., Hong E.L., Livstone M.S., Nash R., et al. Genome Snapshot: a new resource at the Saccharomyces Genome Database (SGD) presenting an overview of the Saccharomyces cerevisiae genome. Nucleic Acids Res. 2006;34:D442–D445. doi: 10.1093/nar/gkj117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarz E.M., Antoshechkin I., Bastiani C., Bieri T., Blasiar D., Canaran P., Chan J., Chen N., Chen W.J., Davis P., et al. WormBase: better software, richer content. Nucleic Acids Res. 2006;34:D475–D478. doi: 10.1093/nar/gkj061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joshi-Tope G., Gillespie M., Vastrik I., D'Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K.F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Mering C., Huynen M., Jaeggi D., Schmidt S., Bork P., Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Mering C., Jensen L.J., Snel B., Hooper S.D., Krupp M., Foglierini M., Jouffre N., Huynen M.A., Bork P. STRING: known and predicted protein–protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snel B., Lehmann G., Bork P., Huynen M.A. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valencia A., Pazos F. Computational methods for the prediction of protein interactions. Curr. Opin. Struct. Biol. 2002;12:368–373. doi: 10.1016/s0959-440x(02)00333-0. [DOI] [PubMed] [Google Scholar]
- 14.Huynen M.A., Bork P. Measuring genome evolution. Proc. Natl Acad. Sci. USA. 1998;95:5849–5856. doi: 10.1073/pnas.95.11.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pellegrini M., Marcotte E.M., Thompson M.J., Eisenberg D., Yeates T.O. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enright A.J., Iliopoulos I., Kyrpides N.C., Ouzounis C.A. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 17.Marcotte E.M., Pellegrini M., Ng H.L., Rice D.W., Yeates T.O., Eisenberg D. Detecting protein function and protein–protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 18.Dandekar T., Snel B., Huynen M., Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 19.Overbeek R., Fonstein M., D'Souza M., Pusch G.D., Maltsev N. The use of gene clusters to infer functional coupling. Proc. Natl Acad. Sci. USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rattei T., Arnold R., Tischler P., Lindner D., Stumpflen V., Mewes H.W. SIMAP: the similarity matrix of proteins. Nucleic Acids Res. 2006;34:D252–D256. doi: 10.1093/nar/gkj106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neduva V., Russell R.B. DILIMOT: discovery of linear motifs in proteins. Nucleic Acids Res. 2006;34:W350–W355. doi: 10.1093/nar/gkl159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letunic I., Copley R.R., Pils B., Pinkert S., Schultz J., Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciccarelli F.D., Doerks T., von Mering C., Creevey C.J., Snel B., Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 25.Tringe S.G., von Mering C., Kobayashi A., Salamov A.A., Chen K., Chang H.W., Podar M., Short J.M., Mathur E.J., Detter J.C., et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 26.Dimaano C., Ullman K.S. Nucleocytoplasmic transport: integrating mRNA production and turnover with export through the nuclear pore. Mol. Cell. Biol. 2004;24:3069–3076. doi: 10.1128/MCB.24.8.3069-3076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]