Abstract
FlyBase (http://flybase.org/) is the primary database of genetic and genomic data for the insect family Drosophilidae. Historically, Drosophila melanogaster has been the most extensively studied species in this family, but recent determination of the genomic sequences of an additional 11 Drosophila species opens up new avenues of research for other Drosophila species. This extensive sequence resource, encompassing species with well-defined phylogenetic relationships, provides a model system for comparative genomic analyses. FlyBase has developed tools to facilitate access to and navigation through this invaluable new data collection.
A NEW LOOK TO FlyBase
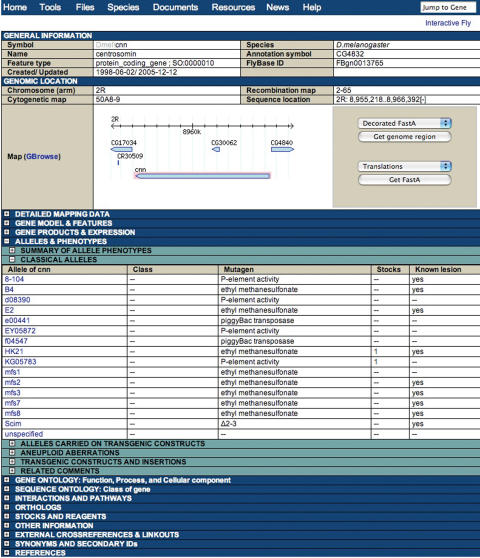
Over the past 2 years, FlyBase has effected a complete migration and integration of its underlying databases into a PostgreSQL chado genome database [(1), http://www.gmod.org/schema/]. This has enabled a reimplementation from the ground up of the FlyBase public interface, with a complete redesign of the Web pages, queries and reports (Figure 1). So, if you do not recognize us—take a second look! Detailed descriptions of the new FlyBase website will appear elsewhere.
Figure 1.
A FlyBase gene report. The new FlyBase design allows the user to choose which sections and subsections to view. In this example, the table showing classical alleles has been opened.
THE CHANGING CONTENT OF FlyBase
FlyBase is an integrated resource for a vast array of genetic and molecular data concerning the Drosophilidae, including interactive genomic maps, gene product descriptions, mutant allele phenotypes, genetic interactions, expression patterns, transgenic constructs and insertions of transgenic constructs, anatomy and images, and genetic stock collections (2). Data are captured from bulk data sources, by curation from the literature, and by annotation based on assessment of contributing evidence; data capture is organized around consistent attribution to primary sources. As far as possible, descriptive data are curated using controlled vocabularies (CV), including the Gene Ontology for molecular function, biological process and cellular component (3), the Sequence Ontology for sequence features (4) and an extensive CV for anatomical terms and developmental stages (available as part of the Open Biomedical Ontologies project, http://obo.sourceforge.net/).
Although FlyBase has since its inception curated genetic and genomic information on the family Drosophilidae, it is only with the recent whole-genome shotgun (WGS) sequencing and assembly of 11 additional species that substantial amounts of non-melanogaster data have appeared in FlyBase. Indeed, it will be interesting to see how the availability of these WGS sequence assemblies will affect Drosophila research through the ability to perform genome-wide comparative analyses at the sequence, phenotypic and biological process levels.
THE DROSOPHILA GENOMES (EMPHASIS ON THE PLURAL)
The genome sequences of 12 species of Drosophila are now available. The species and their phylogeny are shown in the left-hand side of Figure 2. For the genome of the primary biological research species, Drosophila melanogaster, the euchromatic arms have now been finished to high quality by the BDGP [(5), http://www.fruitfly.org/]. In the current release of the D.melanogaster genome assembly (Release 5; see Table 1 and http://www.fruitfly.org/sequence/release5genomic.shtml), the arms include several megabases of centric heterochromatin as well as the entirety of the euchromatin. The heterochromatin, sequenced by the BDGP and the Drosophila Heterochromatin Genome Project (DHGP) (6), also includes several major scaffolds that are currently unattached to the arms. The fully annotated arms are available from FlyBase and GenBank; the DHGP-annotated heterochromatic scaffolds should be contributed to FlyBase and GenBank in late 2006.
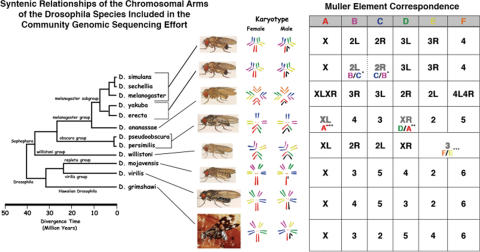
Figure 2.
The Muller element arm synteny table. The phylogenetic relationships of the 12 sequenced species are shown to the left, color-coded diagrammatic karyotypes and a table of syntenic relationships are shown to the right. There is not a simple one-to-one correspondence of arms to Muller elements for all of the species, due to fusion and inversion events relative to D.melanogaster in five of the species.
Table 1.
Release 5 of the D.melanogaster genome assembly
| Current annotation status | Arm | GenBank accession no. | Annotated genes |
|---|---|---|---|
| Annotated Release 5.1 | X | AE014298.4 | 2332 |
| Annotated Release 5.1 | 2L | AE014134.5 | 2756 |
| Annotated Release 5.1 | 2R | AE013599.4 | 3028 |
| Annotated Release 5.1 | 3L | AE014296.4 | 2808 |
| Annotated Release 5.1 | 3R | AE014297.2 | 3553 |
| Annotated Release 5.1 | 4 | AE014135.3 | 91 |
| Annotation in progress | Heterochromatic sequences not integrated onto arms | ||
The other 11 species have all been sequenced in NHGRI-funded large-scale sequencing centers (Table 2), following the approval of three separate community-based white papers. The first white paper [(7), http://flybase.bio.indiana.edu/.data/docs/CommunityWhitePapers/DrosBoardWP2001.html] proposed the sequencing of a second species, Drosophila pseudoobscura, to support the annotation of D.melanogaster (8). The second white paper [(9), http://flybase.bio.indiana.edu/.data/docs/CommunityWhitePapers/] proposed the sequencing of several isolates of Drosophila simulans, a close relative of D.melanogaster, to understand the basis of variation within and between species, and the sequencing of a somewhat more distant member of the same species group, Drosophila yakuba, as an outgroup. The third white paper [(10), http://flybase.bio.indiana.edu/.data/docs/CommunityWhitePapers/GenomesWP2003.html] proposed the sequencing of eight additional species. Six of these species (Drosophila ananassae, Drosophila erecta, Drosophila grimshawi, Drosophila mojavensis, Drosophila virilis and Drosophila willistoni) were proposed principally to provide additional branch length for comparative genomic analysis in support of the annotation of D.melanogaster, as well as for the study of gene and chromosome evolution on a whole-genome scale. The other two species, D.persimilis and D.sechellia, are sibling species of D.pseudoobscura and D.simulans, respectively; these were chosen because the sibling species pairs can form fertile F1 hybrids and have been used to study genetic variation that underlies speciation.
Table 2.
Sequenced genomes of Drosophila species
| Species | GenBank WGS accession no. (CA Freeze 1) | Assembly size (bp) | Large-scale sequencing center |
|---|---|---|---|
| D.melanogaster | See Table 1 | 139 712 364 | BDGP/Celera |
| D.ananassae | AAPP01000000 | 230 993 012 | Agencourt Biosciences |
| D.erecta | AAPQ01000000 | 152 712 140 | Agencourt Biosciences |
| D.grimshawi | AAPT01000000 | 200 467 819 | Agencourt Biosciences |
| D.mojavensis | AAPU01000000 | 193 826 310 | Agencourt Biosciences |
| D.persimilis | AAIZ00000000 | 188 374 079 | Broad Institute |
| D.pseudoobscura | AADE01000000 | 148 799 920 | Baylor HGSC |
| D.sechellia | AAKO01000000 | 166 577 145 | Broad Institute |
| D.simulans | (Pending) | Washington University, GSC | |
| D.virilis | AANI01000000 | 206 026 697 | Agencourt Biosciences |
| D.willistoni | AAQB01000000 | 235 516 348 | J. Craig Venter Institute |
| D.yakuba | AAEU02000000 | 165 691 649 | Washington University, GSC |
REPRESENTATION OF THE DOZEN GENOMES IN FlyBase
A group called ‘Assembly, Annotation and Analysis’ (AAA) has been coordinating the community production and distribution of the relevant large datasets, the production of consensus annotation sets and the preparation of the initial reports of the results of these studies (http://rana.lbl.gov/drosophila/). By the end of 2006, it is expected that the major datasets will have been produced, publications submitted and data contributed to FlyBase and GenBank. For each species these data will include several independent homology-based and ab initio gene prediction sets, consensus mRNA and protein annotation sets, orthologies, gene family groupings, and syntenic relationships among the species, the latter extending the previously known large-scale syntenic conservations among the chromosome arms of the genus Drosophila (see Figure 2). The following discussion describes the major ways to interrogate and browse these genomes and their relationships to one another.
THE FlyBase BLAST TOOL: QUERIES ACROSS INSECT SPECIES
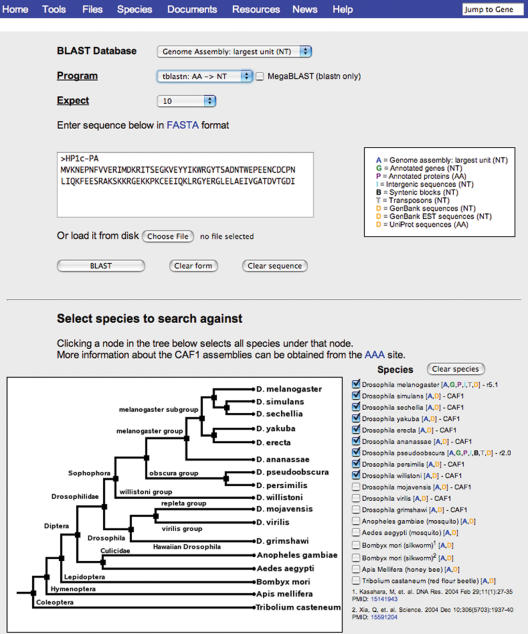
The FlyBase BLAST tool serves as a convenient entry point to data for the insect species for which genomic sequence data are available, including the 12 Drosophila species, mosquito (Anopheles and Aedes), silkworm, honey bee and Tribolium. The tool provides an array of options in an intuitive format (Figure 3). An extremely useful feature of the BLAST output presentation are links that go directly to a GBrowse view of the genomic region that corresponds to the BLAST hit.
Figure 3.
The FlyBase BLAST tool. ‘BLAST Database’ selection options are shown in the ‘BLAST database availability key’ and include the whole-genome assemblies, annotated transposons, protein sequences or intergenic sequences, as well as GenBank and UniProt datasets. The species for which data are available are shown in a hierarchical tree; clicking on any node in the tree selects all descendants of that node. Not all types of data are available for each of the featured species: an availability code (colored letters) is shown after each species listing. Advanced BLAST options (out of view at bottom of page) allow customization of BLAST parameters and output format; an ON/OFF toggle is provided for the ‘Low Complexity Filter’; the ‘Expect’ threshold parameter may be adjusted at the top of the page. The BLAST output includes a ‘BLAST HIT on Genome Map’ or ‘Feature on Genome Map’ link shown at the top of an aligned segment; this allows immediate access to a GBrowse view of the genomic region that corresponds to the BLAST hit, with the aligned region indicated by a gray panel.
THE GBrowse GENOME VIEWER: CUSTOMIZED VIEWS OF PREDICTIONS AND EVIDENCE
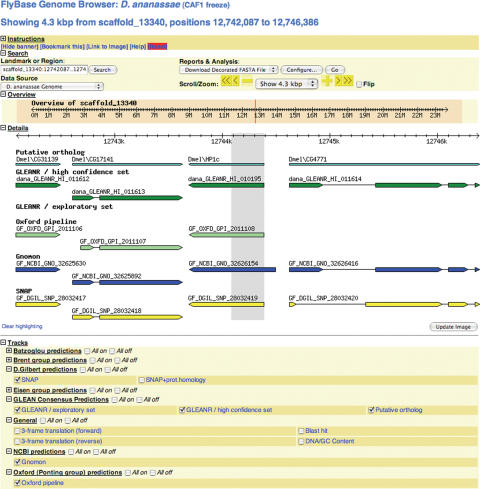
Interactive views of the data generated by the genomic sequencing projects are presented using a newly modified version of the GBrowse genome viewer [(11), http://www.gmod.org/?q=node/71]. Entry to a specific genomic region may be accomplished by running a BLAST search first, as described above. The tool may also be accessed from the FlyBase home page or from the ‘Tools’ menu found in the top bar on all FlyBase reports. Once the species to be viewed is chosen and the region of interest specified, the data to be viewed can also be specified and its presentation customized (Figure 4). For the newly sequenced genomes, the default view shows alignments to D.melanogaster putative orthologs and the GLEANR consensus predictions. GC content, translation stops and additional prediction sets may be selected for viewing, and the view may be modified by zooming or scrolling or flipping. As more data become available, they will be incorporated into the GBrowse presentation. The sequence and selected datasets for the genomic extent being viewed may be downloaded as a decorated FASTA file, a GFF file or a table.
Figure 4.
A GBrowse view. The region of the D.ananassae genome homologous to the D.melanogaster HP1c gene has been accessed via a link from the FlyBase BLAST output. The region of BLAST alignment is indicated by a gray panel. The default selection of data includes alignments of D.melanogaster putative orthologs and the GLEANR consensus predictions; in this view, additional gene prediction tracks have been selected. The presentation of the data options may be customized further by using a ‘Configure tracks’ option (out of view at bottom of page). The species being viewed may be changed at any time by choosing from the ‘Data Source’ menu. Once viewing the species of choice, a particular region can be specified using sequence coordinates or a ‘landmark’. For the newly sequenced species, the initial set of landmarks consists of the gene predictions, identified by their alphanumeric designations, and the D.melanogaster putative orthologs. The options for downloading the data shown are listed in the menu ‘Reports and Analysis’; the FASTA output may be customized by using the ‘Configure’ option.
BULK DATA DOWNLOADS
Data files for all classes of data in FlyBase are available for download by FTP in several formats, including GFF3 for sequence data. Links to the bulk data repositories may be accessed from the ‘Files’ menu, ‘Precomputed files’ option, at the top of all FlyBase pages; from there, the ‘Genomes: Annotation and Sequence’ section provides access to genome data for each (or all) of the sequenced species. In addition, bulk queries can be performed and downloaded via the ‘QueryBuilder’ tool, accessed from the top page or the ‘Tools’ menu.
MORE ON THE SPECIES OF FAMILY DROSOPHILIDAE
From the ‘Species’ menu on the top bar of the FlyBase home page and all report pages, additional information on the Drosophilidae may be accessed. At present there are four items to choose from: ‘Phylogeny’ links to an index of species, each linked to its position in the Drosophilidae phylogenetic tree; ‘Synteny table’ goes to the presentation of syntenic relationships of the chromosomal arms of the 12 sequenced species shown in Figure 2; ‘Drosophilidae’ links to a compilation of color images of species within this family, originally published by the University of Texas at Austin School of Biological Sciences; and ‘Abbreviations’ accesses a list of the four-letter genus–species codes for all species found in FlyBase. The ‘Species’ resources will be updated periodically, as appropriate community resources and data become available.
FlyBase continues to curate and present traditional genetic data for all the Drosophilid species. Now, availability and integration of genomic data for 12 well-characterized species provide a powerful resource that will allow the research community to take full advantage of the family Drosophilidae as a model for comparative genomic and phylogenetic analyses.
Acknowledgments
FlyBase is supported by grant P41 HG00739 from the National Human Genome Research Institute, National Institutes of Health (USA), with additional support from the Medical Research Council (UK) grant G05000293. Funding to pay the Open Access publication charges for this article was provided by the NHGRI FlyBase grant award.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zhou P., Emmert D., Zhang P. Using chado to store genome annotation data. In: Baxevanis A.D., Davison D.B., editors. Current Protocols in Bioinformatics. Vol. 2. Hoboken, NJ: John Wiley & Sons, Inc.; 2005. pp. 9.6.1–9.6.28. [DOI] [PubMed] [Google Scholar]
- 2.Grumbling G., Strelets V., and FlyBase Consortium FlyBase: anatomical data, images and queries. Nucleic Acids Res. 2006;34:D484–D488. doi: 10.1093/nar/gkj068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris M., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eilbeck K., Lewis S., Mungall C., Yandell M., Stein L., Durbin R., Ashburner M. The sequence ontology: a tool for unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Celniker S.E., Wheeler D.A., Kronmiller B., Carlson J.W., Halpern A., Patel S., Adams M., Champe M., Dugan S.P., Frise E., et al. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 2002;3:RESEARCH0079. doi: 10.1186/gb-2002-3-12-research0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoskins R.A., Smith C.D., Carlson J.W., de Carvalho A.B., Halpern A., Kaminker J.S., Kennedy C., Mungall C.J., Sullivan B.A., Sutton G.G., et al. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 2002;3:RESEARCH0085. doi: 10.1186/gb-2002-3-12-research0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooley L., Desplan C., Gaul U., Geyer P., Kaufman T., Krasnow M., Rubin G., Gelbart W. Drosophila White Paper 2001. 2001. [Google Scholar]
- 8.Richards S., Liu Y., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., et al. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Begun D.J., Langley C.H. 2003. Proposal for sequencing of Drosophila yakuba and Drosophila simulans, revised. [Google Scholar]
- 10.Clark A., Gibson G., Kaufman T., McAllister B., Myers E., O'Grady P. 2003. Proposal for Drosophila as a model system for comparative genomics. [Google Scholar]
- 11.Stein L., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J., Harris T., Arva A., et al. The generic genome browser: a building block for a model organism database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]