Abstract
Pure mitochondria of the photosynthetic alga Chlamydomonas reinhardtii were analyzed using blue native-polyacrylamide gel electrophoresis (BN-PAGE). The major oxidative phosphorylation complexes were resolved: F1F0-ATP synthase, NADH-ubiquinone oxidoreductase, ubiquinol-cytochrome c reductase, and cytochrome c oxidase. The oligomeric states of these complexes were determined. The F1F0-ATP synthase runs exclusively as a dimer, in contrast to the C. reinhardtii chloroplast enzyme, which is present as a monomer and subcomplexes. The sequence of a 60-kD protein, associated with the mitochondrial ATP synthase and with no known counterpart in any other organism, is reported. This protein may be related to the strong dimeric character of the algal F1F0-ATP synthase. The oxidative phosphorylation complexes resolved by BN-PAGE were separated into their subunits by second dimension sodium dodecyl sulfate-PAGE. A number of polypeptides were identified mainly on the basis of their N-terminal sequence. Core I and II subunits of complex III were characterized, and their proteolytic activities were predicted. Also, the heterodimeric nature of COXIIA and COXIIB subunits in cytochrome c oxidase was demonstrated. Other mitochondrial proteins like the chaperone HSP60, the alternative oxidase, the aconitase, and the ADP/ATP carrier were identified. BN-PAGE was also used to approach the analysis of the major chloroplast protein complexes of C. reinhardtii.
The unicellular green alga Chlamydomonas reinhardtii is a model organism for the study of certain aspects of plant physiology, like chloroplast biogenesis (Harris, 2001). Nevertheless, C. reinhardtii mitochondria have not been well characterized because of difficulties in obtaining these organelles free of thylakoid contamination. The isolation of C. reinhardtii oxidative phosphorylation (OXPHOS) complexes, including the spectroscopical characterization of cytochrome bc1 complex (complex III) and cytochrome c oxidase (complex IV), was described earlier (Atteia et al., 1992; Atteia, 1994). However, the subunit composition of the OXPHOS complexes in the alga has not been studied in detail.
The mitochondrial genome of C. reinhardtii encodes five subunits of complex I, cytochrome b of complex III, and subunit I of complex IV (Michaelis et al., 1990). Until now, none of these subunits have been located on SDS-PAGE. Among the mitochondrial proteins of nuclear origin, few have been identified and their genes sequenced: subunits alpha, beta, and ATP6 of complex V (F1F0-ATP synthase; Franzén and Falk, 1992; Nurani and Franzén, 1996; Funes et al., 2002), and two subunits of complex III, the Rieske-type iron-sulfur protein (Atteia and Franzén, 1996) and cytochrome c1 (Atteia et al., 2002). The gene sequences of subunits COXIIA, COXIIB, and COXIII of the C. reinhardtii complex IV have been determined (Pérez-Martínez et al., 2000, 2001), but their protein products were not identified biochemically. Also, two genes encoding C. reinhardtii alternative oxidase (AOX), Aox1 and Aox2, have been sequenced (Dinant et al., 2001). Aox1, the more expressed of the two genes, encodes a protein similar to plant AOXs, but lacks a conserved Cys residue at its N terminus. This Cys is thought to participate in the regulatory dimerization of the plant enzymes (Umbach and Siedow, 1993, 2000). The biochemical characterization of C. reinhardtii AOX remains to be addressed. Until now, validation of the information of the gene sequences by the analysis on the protein level has been largely missing for the mitochondrial proteins of this photosynthetic alga.
Blue native (BN)-PAGE is a powerful tool for proteomics. This technique uses the charge shift induced by the binding of Coomassie Blue to solubilized proteins to separate and visualize membrane complexes under native conditions (Schägger, 1995). BN-PAGE was developed to study protein complexes of bovine mitochondria (Schägger and von Jagow, 1991) and later extended to study the mitochondrial complexes of yeast (Saccharomyces cerevisiae; Arnold et al., 1998), plants (Jänsch et al., 1996), and trypanosomatid kinetoplasts (Maslov et al., 1999). BN-PAGE has also been used to resolve chloroplast complexes of spinach (Spinacia oleracea; Kügler et al., 1997), mitochondrial complexes of Arabidopsis (Kruft et al., 2001), and simultaneously mitochondrial and chloroplast protein complexes of potato (Solanum tuberosum) leaves (Singh et al., 2000).
By applying pure mitochondria of C. reinhardtii (Eriksson et al., 1995) to BN-PAGE, we identified and characterized the OXPHOS complexes and their subunit composition. The oligomeric states of the complexes III to V and the AOX were analyzed. Finally, we used BN-PAGE to describe subcellular fractions containing both chloroplast and mitochondrial protein complexes from C. reinhardtii wild-type cells and from a photosynthetic mutant.
RESULTS
BN-PAGE of Mitochondrial Protein Complexes
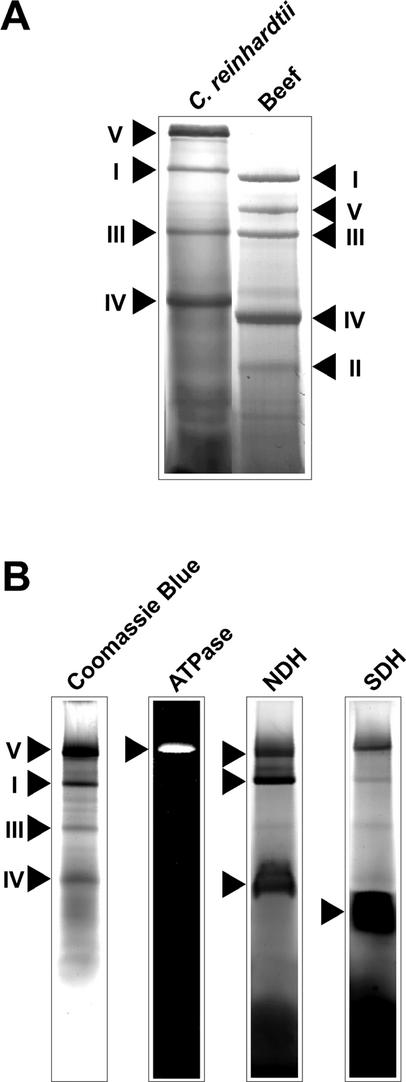
To separate the major OXPHOS complexes, pure C. reinhardtii mitochondria (Eriksson et al., 1995) from the 84CW15 strain were solubilized and applied to BN-PAGE. The protein profile exhibited four major bands and several weaker bands (Fig. 1A) that differed from that of bovine heart mitochondria in the position, amount, and intensity of the bands. The apparent molecular masses of C. reinhardtii OXPHOS complexes were estimated from the known molecular masses of the bovine complexes and are summarized in Table I. The BN-PAGE profile of C. reinhardtii mitochondria exhibited two main characteristics: a band with considerably lower electrophoretic mobility than bovine complex I, and the absence of bands that correspond to the bovine complex V and complex II (Fig. 1A). To establish the identities of the C. reinhardtii major complexes, specific activity stainings were performed.
Figure 1.
BN-PAGE of total mitochondrial proteins from C. reinhardtii and beef. A, Coomassie Blue-stained BN-PAGE gel lanes loaded with 800 (C. reinhardtii strain 84CW15) and 500 (beef) μg of total mitochondrial proteins. B, Gel lanes stained with Coomassie Blue and with specific activity stainings used for the detection of complexes V, I, and II (see “Materials and Methods”). Black arrows mark the major stained bands in each case. ATPase, ATPase activity; NDH, NADH dehydrogenase activity; SDH, succinate dehydrogenase activity.
Table I.
Estimated molecular masses of the respiratory complexes in C. reinhardtii and bovine mitochondria
| Complex No. | Estimated Molecular Mass
|
|
|---|---|---|
| C. reinhardtii | Beef | |
| kD | ||
| V | 1,600 | 600 |
| I | 800 | 750 |
| III | 500 | 500 |
| IV | 240 | 200 |
| II | 140a | 130 |
The molecular masses of the respiratory complexes of C. reinhardtii were estimated in comparison with the beef heart respiratory complexes reported earlier (Schägger and von Jagow, 1991).
Based on complex II specific staining, shown in Fig. 1B.
To localize the active C. reinhardtii complex V on BN-PAGE, a blue gel lane was incubated in the presence of ATP and CaCl2. Figure 1B shows that the uppermost band of 1,600 kD was able to hydrolyze ATP, as indicated by the formation of a calcium phosphate precipitate. The high apparent molecular mass of complex V on BN-PAGE suggests that this protein complex runs as a dimer.
NADH dehydrogenase activity was detected after incubation of a blue gel lane in the presence of NADH and nitroblue tetrazolium (NBT), which forms a purple precipitate upon reduction. With C. reinhardtii mitochondria, three bands of approximately 1,500, 800, and 200 kD were detected (Fig. 1B). The thin band of 1,500 kD detected by the NADH/NBT staining was identified as a dimer of complex I. The 800-kD band, exhibiting an electrophoretic mobility similar to that of bovine complex I (Fig. 1A), was identified as a complex I monomer. Previously, complex I of C. reinhardtii was estimated to be 850 kD on BN-PAGE (Duby et al., 2001). The diffuse band of 200 kD (Fig. 1B) was also observed in the bovine protein pattern (not shown) and considered to be a complex I subcomplex.
Succinate dehydrogenase activity in the gel was visualized by the precipitation of reduced NBT in the presence of succinate and phenazine methosulphate. Unlike bovine complex II, C. reinhardtii complex II did not appear as a defined band on the Coomassie Blue-stained gel, but as a diffuse band around 140 kD (Fig. 1B).
On the basis of their migrations and subunit composition (see below), which are comparable with the corresponding bovine complexes, the C. reinhardtii protein bands of 500 and 240 kD on BN-PAGE (Fig. 1A) were identified as complexes III and IV, respectively.
Resolution of C. reinhardtii OXPHOS Complexes into their Constitutive Subunits
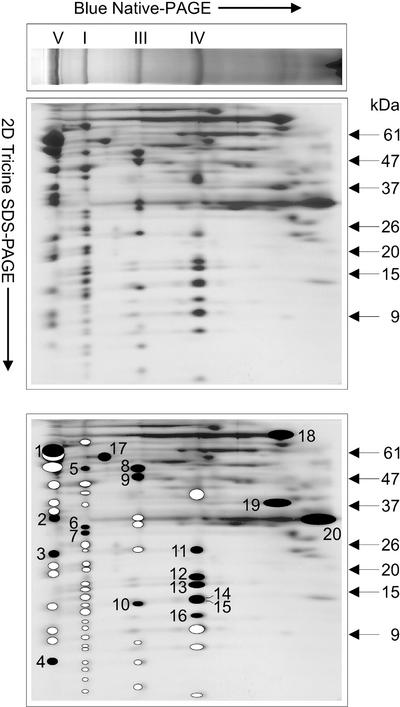
C. reinhardtii mitochondrial complexes V, I, III, and IV, separated by BN-PAGE, were resolved into their individual constituents on second dimension (2D)-SDS-PAGE (Fig. 2). The estimated molecular masses of the subunits are given in Table II.
Figure 2.
Two-dimensional resolution of the mitochondrial protein complexes from C. reinhardtii. The main OXPHOS complexes are indicated on the first dimension BN-PAGE. A BN gel lane was cut out and placed horizontally for subsequent resolution of the protein complexes into their respective components on 2D-Tricine-SDS-PAGE. In the schematic representation of the subunits (bottom), the numbered black spots depict those polypeptides that were subjected to Edman degradation. The corresponding sequences are shown in Table III. White spots represent the other putative subunits of each complex.
Table II.
Apparent number of subunits and the estimation of the molecular mass of the individual subunits of C. reinhardtii complexes V, I, III, and IV
| Complex No. | Apparent No. of Subunitsa | Estimated Molecular Mass of Subunits |
|---|---|---|
| kD | ||
| V | 14 | 60,60,52,45,38,35,31,24,21,19,13,9,8,7 |
| I | 25 | 75,52,45,41,37,29,28,26,25,22,20,16,15,14,13,12,11,10,9,8,8,7,7,6,5 |
| III | 9 | 53,48,32,30,25,13,8,7,6 |
| IV | 10 | 40,25,18,16,14,14,12,10,8,5 |
As detected by Coomassie Blue staining of Tricine SDS-polyacrylamide gel.
C. reinhardtii complex V was resolved into 13 polypeptides, three of which have been previously identified: the beta- (60 kD) and alpha- (52 kD) subunits of the F1 sector (Atteia et al., 1992; Franzén and Falk, 1992; Nurani and Franzén, 1996) and the ATP6 subunit (21 kD) of the F0 region (Funes et al., 2002). We determined the N-terminal sequence of the smallest polypeptide of 7 kD (Fig. 2; Table III, band 4). This N-terminal sequence was found to be encoded in the C. reinhardtii EST clone AW676361. The predicted protein corresponded to ATP9, a structural component of F0-ATP synthase. Similarly, the N-terminal sequence of the 32-kD polypeptide (Table III, band 2) was found in the deduced amino acid sequence of EST clones BE337293 and AV390953 and allowed its identification as the gamma subunit (predicted molecular mass of 30.8 kD). Also, the N-terminal sequence of the 24-kD polypeptide of complex V (Table III, band 3) was found in the deduced protein sequence of EST clones AW661069 and BG848206, identified as the delta subunit (predicted molecular mass of 22.6 kD). Finally, the EST clones BI532011 and BG860760 were found to encode the previously determined N terminus of the 45-kD subunit of complex V (Funes et al., 2002), but the deduced partial amino acid sequence (165 amino acids) did not show similarity to any ATP synthase subunit.
Table III.
Partial description of the mitochondrial proteome of C. reinhardtii
| Band No. | Amino Acid Sequencea | Protein Identity | Accession No. |
|---|---|---|---|
| Complex V | |||
| 1 | YVTALKVEFS/ELAARSAEFRAEQEA (int) | MASAP (60 kD) | AJ441255 |
| 2 | ASNQAVKQRI/(Funes et al., 2002) | Gamma subunit (31 kD) | BE337293EST |
| 3 | AKTAPKAEM(Funes et al., 2002) | Delta subunit (24 kD) | AW881069EST |
| 4 | SVLAASXMVGA | ATP9 subunit (7 kD) | AW678361EST |
| Complex I | |||
| 5 | STAAPAAGAPPPPPPPPAKT | 51-kD subunit family (53 kD) | AV386989EST |
| 6 | VSSQFFDAPNGPSVKQVLIED | 29-kD subunit (29 kD) | BM001979EST |
| 7 | ATNSTDIFNIHKDTPENNAA | 24-kD subunit family (28 kD) | BE212104EST |
| Complex III | |||
| 8 | QSAAKDVVATDANPFLRFSN | Core I (53 kD) | BG846882EST |
| 9 | More than one sequence | Probable core 2 + other protein(s) (48 kD) | |
| 10 | Blocked | Probable subunit IV (13 kD) | |
| Complex IV | |||
| 11 | GSHAAGHQTAKEFYM | COXIII subunit (25 kD) | AAG17279 |
| 12 | DAEVVEEEHAPPPPPPPPKK | COXVIb subunit (18 kD) | BE122218EST |
| 13 | MDAVPX(G/R)LNQ | COXIIB subunit (16 kD) | AAK32114 |
| 14 | GAPAEAKPSALSAEPGR | COXVb subunit (14 kD) | BG851120EST |
| 15 | DSPQPWQLLF | COXIIA subunit (14 kD) | AAK30367 |
| 16 | ASTTAGETIDKY | COXVIa subunit (12 kD) | BG857268EST |
| Other proteins | |||
| 17 | AAKDVRFGIEHRDLMLAGVNXLA | Mitochondrial HSP60 (60 kD) | NF |
| 18 | SXIAGAEKV(P/G)MSQFGP | Mitochondrial aconitate hydratase (90 kD) | AV397582EST |
| 19 | AAPSFGATRFXA | 38-kD protein | NF |
| 20 | GIGECFVR (Int) | Mitochondrial ATP/ADP carrier (31 kD) | S30259 |
Amino acid sequences of the protein bands subjected to Edman degradation (see Fig. 2). GenBank accession nos. are provided. Alternatively, the accession nos. of the ChlamyEST database clones that were used to identify the proteins (expressed sequence [EST] in superscript) are given. NF, Not found in the ChlamyEST database.
Sequences are amino terminal unless mentioned otherwise (Int, internal).
When performing 2D-SDS-PAGE in the presence of 8 m urea, an additional 60-kD protein was resolved in the complex V polypetide pattern. As shown in Figure 3, the 60-kD protein is not recognized by an anti-beta antibody. We determined the N-terminal sequence and an internal protein sequence of this polypeptide, here named MASAP (Table III, band 1). Subsequently, deoxyoligonucleotides were designed, a PCR product was obtained, and a corresponding cDNA was isolated from a λZAP cDNA library. From the deduced amino acid sequence, it was inferred that the MASAP is most likely soluble, exhibiting an apparent molecular mass of 60.5 kD and a pI of 5.66. No similarity to any mitochondrial protein in the databases was found. The protein presequence deduced from the cDNA was predicted to be mitochondrial using the TargetP V1.0 program (Emanuelsson et al., 2000). The function of this novel component remains to be established.
Figure 3.
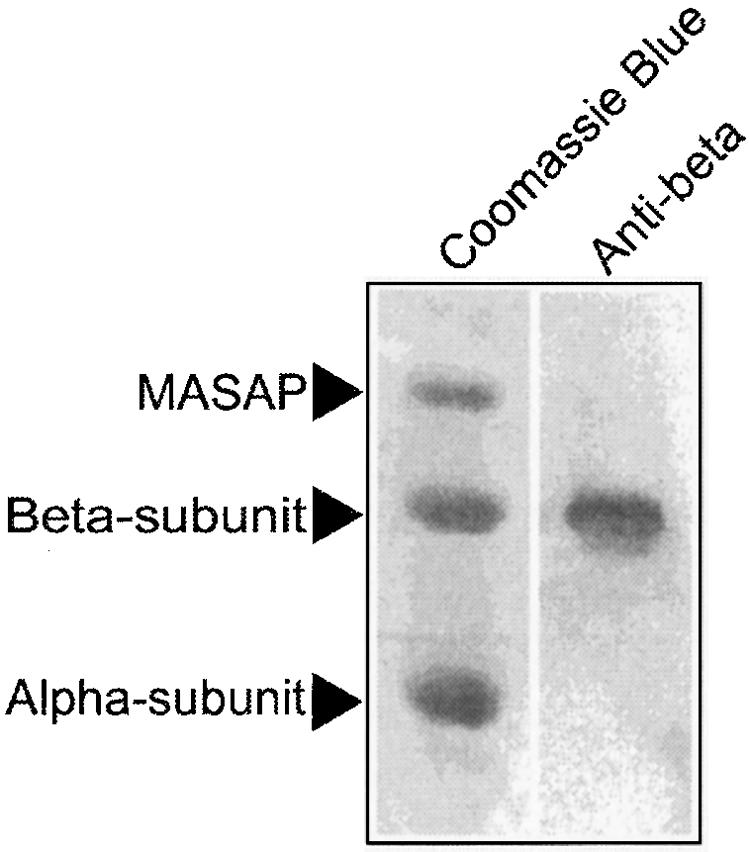
High-molecular mass subunits of C. reinhardtii complex V resolved on 2D-urea-SDS-PAGE. Complex V bands recovered from BN-PAGE were loaded onto a 2D-Tricine-SDS gel in the presence of 8 m urea. Only the largest subunits are shown. Left lane, Coomassie Blue staining; right lane, immunoblot analysis with an antibody against the beta-subunit. The immunoblot revealed that mitochondrial ATP synthase-associated protein (MASAP) is clearly distinct from the beta-subunit.
C. reinhardtii complex I (800-kD band on BN-PAGE) was resolved into at least 25 subunits on 2D-SDS-PAGE (Fig. 2). The N-terminal sequences of three of its constituents are reported in Table III (bands 5–7). The 52-kD protein (band 5) of complex I exhibited an N-terminal sequence with an unusual high content of Pro. The EST clone AV386989 contained a sequence encoding the N terminus of this 52-kD protein, identified as a member of the 51-kD subunit family of complex I. Also, the EST clone BE212104 encoded the N-terminal sequence of the 28-kD subunit (band 7), a member of the 24-kD subunit family of complex I. Both the 51- and 24-kD subunit families are components of the flavoprotein fraction. The N-terminal sequence of the 29-kD subunit (band 6) was also found to be encoded in a clone of the ChlamyEST database (BM001979), but the deduced amino acid sequence did not allow its identification.
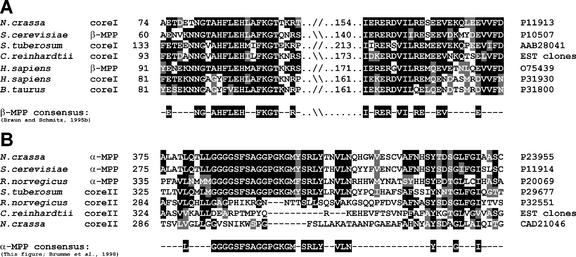
C. reinhardtii complex III was resolved on 2D-SDS-PAGE into nine subunits. The 53-kD subunit (Table III, band 8) was identified as the core I subunit by immunoblot analysis, using an antiserum against Neurospora crassa core I (see below). However, the N-terminal sequence of this band did not show any similarity with core I subunits from either plant or mammalian complex III. A clone from the EST database encoded the N-terminal sequence of this C. reinhardtii core I protein (BG846882). The whole sequence of core I was obtained from the overlapping EST clones BG846882, BI726156, AV633102, BG850841, and BG847806. The predicted mature core I protein (53.9 kD) contained 487 residues. The 48-kD protein of complex III (Fig. 2, band 9) is assumed to be the core II subunit, which probably comigrates with one or more proteins because a mixture of N-terminal sequences was obtained (not shown). In plants, cores I and II are known to represent the beta- and alpha-subunits of the mitochondrial processing peptidase (MPP), respectively. The alpha-MPP subunit does not possess MPP activity itself, but it is necessary for the beta-MPP activity. In most other organisms, the core proteins do not possess MPP activity, which is instead conferred by soluble, matrix-located alpha- and beta-MPP subunits (Braun and Schmitz, 1995a). The complete sequence of the core I of C. reinhardtii was analyzed for the presence of the consensus sequence for beta-MPP activity (Braun and Schmitz, 1995b). A multiple sequence alignment using core I and beta-MPP sequences (Fig. 4A) revealed that C. reinhardtii core I exhibits the consensus sequence, except for an Arg to Lys substitution at position 175. The ChlamyEST database also allowed us to construct the sequence of C. reinhardtii core II, based on EST clones BM000676, AV631099, BI727574, and BM001151. This sequence exhibits similarity to core II and alpha-MPP subunits from other organisms, but lacks the consensus sequence for alpha-MPP activity (Fig. 4B).
Figure 4.
Multiple sequence alignments of the core I and II proteins and the MPP subunits from various sources. The accession number for each sequence is shown on the right-hand side. The C. reinhardtii sequences were derived from the EST clones indicated in the text. A, Comparison of C. reinhardtii core I with other core I and beta-MPP amino acid sequences. C. reinhardtii core I exhibits the consensus sequence usually found for beta-MPP protease activity, including the zinc-binding motif (H-X-X-E-H) that is absent in the mammalian core I sequences. B, Alignment of core II and alpha-MPP amino acid sequences. The core II sequence of C. reinhardtii lacks the consensus sequence (G-G-G-G-S-F-S-A-G-G-P-G-K-G-M/S-R-L-Y) believed to be required for alpha-MPP activity.
In the 30-kD molecular mass range, C. reinhardtii complex III exhibits two subunits. Heme-specific 3,3′,5,5′-tetramethylbenzidine staining allowed the identification of the 30-kD protein as cytochrome c1 (not shown). Thus, the 32-kD protein above the cytochrome c1 is likely to be cytochrome b. The subunit of 25 kD was identified previously as the Rieske-type protein (Atteia and Franzén, 1996). The N terminus of the 13-kD subunit of complex III (Fig. 2, band 10) was not susceptible to Edman degradation.
Complex IV of the photosynthetic alga was resolved into 10 subunits. On the basis of its apparent molecular mass, the larger polypeptide (40 kD) was assigned as subunit I. The N-terminal sequences determined for the protein bands 11 (25 kD), 13 (16 kD), and 15 (14 kD) allowed their identification as subunits COXIII, COXIIB, and COXIIA, respectively (Pérez-Martínez et al., 2000, 2001). The N-terminal sequences of the proteins in bands 12 (18 kD), 14 (14 kD), and 16 (12 kD) were found to be encoded in the EST clones BE122218, BG851120, and BG857268, respectively. Homology searches led to the identification of band 12 as COXVIb (18 kD), band 13 as COXVb (14 kD), and band 14 as COXVIa (12 kD). In Figure 2, bands 14 and 15 were not resolved; however, the SDS-polyacrylamide gels used for N-terminal sequencing did allow the complete separation of these subunits.
Identification of Other Mitochondrial Proteins
The N-terminal sequences of other dominant proteins in C. reinhardtii mitochondria were also determined. The 38-kD protein (Fig. 2, band 19) could not be identified by its N-terminal sequence (Table III). The 31-kD protein (Fig. 2, band 20) was blocked at its N terminus. Nevertheless, the sequence of a tryptic fragment (Table III, band 20) matched a region from residues 54 to 61 of the C. reinhardtii ADP/ATP carrier (Sharpe and Day, 1993). The ADP/ATP translocator—as detected by Coomassie Blue staining—appeared to smear on BN-PAGE (Fig. 2).
The identity of the 60-kD protein (Table III, band 17) was established based on the similarity of its N-terminal sequence with that of mitochondrial chaperonin HSP60 (heat shock protein 60) of plants. On BN-PAGE, C. reinhardtii HSP60 was found to run as a faint band of approximately 650 kD (Fig. 2), indicating its multimeric nature. The HSP60 particle in the photosynthetic alga is probably a 14 mer, as in potato (Jänsch et al., 1996).
The N-terminal sequence of the 90-kD protein (Table III, band 18) was found to be encoded by an EST clone (AV397582) and corresponds to mitochondrial aconitate hydratase (aconitase). This soluble Krebs cycle enzyme that catalyzes the formation of isocitrate from citrate in the mitochondrial matrix appears to be a major constituent of the C. reinhardtii mitochondrial proteome. The entire amino acid sequence of the mature protein (776 residues, 83.2 kD) could be constructed on the basis of EST clones AV397582, AV631772, BI873612, BI873370, BF859712, and BF863471.
Oligomeric States of the OXPHOS Complexes
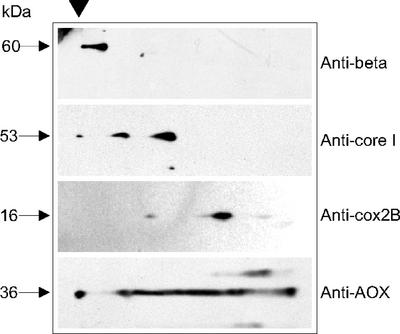
The oligomeric states of C. reinhardtii OXPHOS complexes were analyzed by immunoblot analysis of 2D-SDS-polyacrylamide gels subsequent to the application of pure mitochondria to BN-PAGE (Fig. 5). An antiserum against the beta-subunit of Polytomella sp. complex V recognized only the most upper band of BN-PAGE, previously identified as complex V (see above).
Figure 5.
Oligomeric states of mitochondrial protein complexes as detected by immunoblotting. Proteins resolved by 2D-Tricine-SDS-PAGE gels were transferred onto nitrocellulose membranes and immunoblotted with the indicated antibodies (from top to bottom, anti-beta subunit of Polytomella sp. ATP synthase, anticore I of N. crassa, anti-COXIIB of Polytomella sp., and anti-AOX of C. reinhardtii). The arrow indicates the position of the well on the first dimension BN-PAGE, where a small portion of total proteins precipitates before entering the stacking gel.
As revealed by immunoblot analysis with an antibody against N. crassa core I subunit, the major form of C. reinhardtii complex III was a dimer of 500 kD (Fig. 5). The antibody also recognized a minor form of 1,000 kD.
C. reinhardtii complex IV was detected immunochemically with an antibody against the COXIIB subunit of Polytomella sp. and appeared to be present in BN-PAGE in several oligomeric states, with apparent molecular masses of 530, 240, and 160 kD (Fig. 5). The 240-kD form was the most abundant.
In the absence of a specific antibody for complex I, it could nonetheless be inferred from Figure 1B that a minor fraction of complex I runs as a dimer. A BN-PAGE band of 1,500 kD was reproducibly detected by the specific staining for NADH dehydrogenase activity. Furthermore, on 2D-SDS-PAGE, the polypeptide pattern of this high-molecular mass complex seemed to be identical to that of complex I, although this cannot be clearly discerned in Figure 2 due to its low abundance and proximity to complex V.
C. reinhardtii AOX
AOX is a mitochondrial key enzyme in photosynthetic organisms (Vanlerberghe and McIntosh, 1997). In the BN-PAGE analyses of plant mitochondria reported so far, no mention of the AOX has been made. In this study, antibodies were raised against the overexpressed C terminus of C. reinhardtii AOX1 and used to localize the corresponding protein. Immunoblots of 2D-SDS-polyacrylamide gels revealed the presence of the 36-kD AOX all over the width of the gel (Fig. 5). In contrast to the other respiratory complexes, C. reinhardtii AOX was not resolved as a discrete band under the conditions used (2 mg n-dodecyl maltoside mg−1 mitochondrial protein). The behavior of the AOX on BN-PAGE is likely due to its propensity to form aggregates (Berthold and Siedow, 1993). At this stage, it is not known whether BN-PAGE is suitable to obtain a good resolution of the AOX protein or of other membrane-bound proteins such as the ADP/ATP carrier.
A Proteomic Approach to the Analysis of Subcellular Fractions, Different Growth Conditions, and Mutants
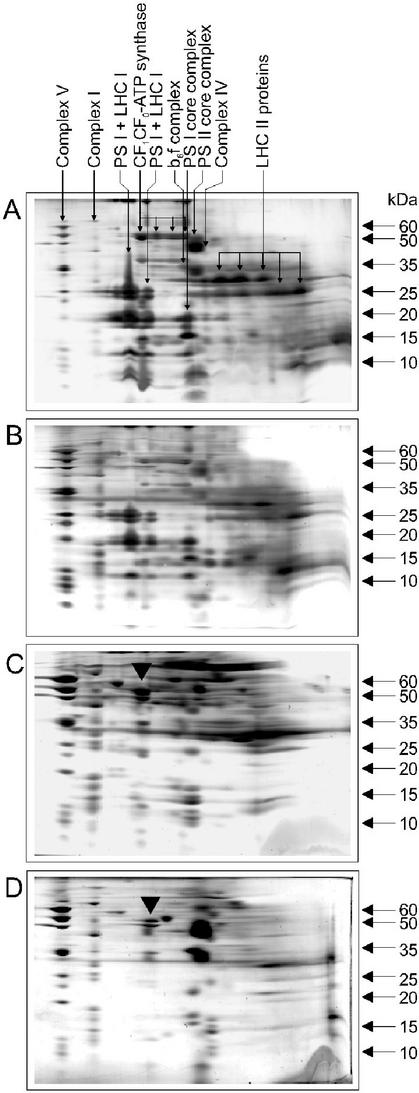
We have explored different uses of BN-PAGE for the comprehensive characterization of C. reinhardtii mitochondrial protein components. The purification procedure of C. reinhardtii mitochondria consists of cell rupture, two differential centrifugations, and a Percoll gradient centrifugation step that removes remnant chloroplast proteins (Eriksson et al., 1995). To follow the enrichment of mitochondria during this procedure, the pellets of the two differential centrifugations (P1 and P2) were analyzed on BN-PAGE (Fig. 6). In pellet P1, resulting from the centrifugation of the cell homogenate at 2,000g, the photosynthetic complexes were dominantly present (Fig. 6A). The distribution of these complexes on BN-PAGE is roughly comparable with that of spinach (Kügler et al., 1997) and potato chloroplast complexes (Singh et al., 2000). PSII (300 kD) was identified by immunoblotting with an antibody against the D1 protein (not shown). The chloroplast ATP synthase (CF0CF1-ATP synthase) was identified by its typical subunit composition. Apart from the monomer of 500 kD (Fiedler et al., 1995), at least three subcomplexes of CF0CF1-ATP synthase could be separated on BN-PAGE (Fig. 6), including the CF1 entity of approximately 350 kD. In pellet P1, mitochondrial complexes V, I, and IV could be detected by Coomassie Blue staining. Figure 6A reveals the great contrast in the electrophoretic behavior between chloroplast and mitochondrial ATP synthases in the green alga. Although several chloroplast ATP synthase oligomeric forms and subcomplexes were visible, only a single, high molecular form of the mitochondrial enzyme was observed. Pellet P2 represents the crude mitochondrial fraction that results from the second centrifugation step at 5,000g and shows a pronounced enrichment in mitochondrial protein complexes. Complexes V, I, and IV were clearly visible, whereas complex III was obscured by the chloroplast ATP synthase and PSI (Fig. 6B). Pure mitochondria were obtained after Percoll density gradient centrifugation and are typified by the virtual absence of chloroplast protein complexes (Fig. 6C).
Figure 6.
2D-Gly SDS-polyacrylamide gels comparing different fractions of the isolation procedure for mitochondria from the C. reinhardtii 84CW15 strain and the photosynthetic mutant BF4.F54.F14. The indicated sample (350 μg total protein) was subjected to BN-PAGE and to subsequent denaturing 2D. The main mitochondrial and photosynthetic complexes are indicated by arrows. LHC I and II, Light-harvesting complex I and II; CF1CF0 ATP synthase, chloroplast ATP synthase. The first three panels correspond to fractions of the C. reinhardtii 84CW15 strain. A, P1, the first pellet after cell disruption and centrifugation at 2,000g. B, P2, Pellet obtained after centrifugation at 5,000g of the supernatant resulting from the first centrifugation that constitutes the crude mitochondrial fraction. C, Mitochondria, purified by Percoll density gradient centrifugation. D, Pellet P2 from the triple photosynthetic mutant BF4.F54.F14. This mutant was treated with N-cetyltrimethylammonium bromide (CTAB) to enable cell rupture by glass beads, as indicated in “Materials and Methods.” The black bold arrow indicates the position of complex III.
We also analyzed the crude mitochondria of the photosynthetic mutant strain BF4.F54.F14 (Fig. 6D, comparable with fraction P2 in Fig. 6B). This mutant is devoid of PSI, CF0CF1-ATP synthase (Chua et al., 1975; Piccioni et al., 1981), and most of the light-harvesting complexes (Olive et al., 1981). To obtain mitochondria from this cell wall-containing strain, the cells were pretreated with CTAB. As expected, the only photosynthetic complexes found in the crude mitochondria were the b6f complex and PS II. No differences in the mitochondrial protein patterns were observed between the mutant strain and the wild-type strain. Nevertheless, in the mutant, the mitochondrial complex III subunits appeared clearly on the 2D gels (Fig. 6C, arrow).
DISCUSSION
The Electron Transfer Complexes and Their Oligomeric States
Previous works have analyzed the mitochondrial proteome of the model plant Arabidopsis (Kruft et al., 2001; Millar et al., 2001). Besides land plants (Streptophyta), green algae (Chlorophyta) are the other main constituent of Chlorobionta. In this work, we addressed the study of mitochondria from C. reinhardtii, a unicellular model system for photosynthetic cells. To characterize the mitochondria of C. reinhardtii, we used BN-PAGE, a powerful analytical technique for both membrane and soluble proteins. A critical parameter to study the mitochondrial proteome is the purity of the sample to be analyzed. C. reinhardtii intact mitochondria were prepared according to Eriksson et al. (1995). These mitochondria were assessed to be basically free of chloroplast contamination by comparing the 2D-SDS-PAGE polypeptide pattern of the different fractions obtained during the purification procedure (Fig. 6).
The estimation of the molecular mass of proteins from their migration on BN-PAGE is approximate because this technique separates according to size but also according to charge (Schägger and von Jagow, 1991). It was inferred that the behavior of OXPHOS complexes on BN-PAGE resembles their physiological state in the mitochondrial inner membrane at the time of solubilization. For yeast and mammalian mitochondria, when low detergent to protein ratios were used for solubilization, the association of different protein complexes in supercomplexes was revealed (Schägger and Pfeiffer, 2000). These complex-complex interactions seem to reflect functional associations that exist in vivo, the so-called respirosome.
The resolution of the mitochondrial protein complexes of C. reinhardtii in BN-PAGE was clearly distinct from the pattern obtained with Arabidopsis mitochondria (Kruft et al., 2001). In all BN-PAGE experiments, C. reinhardtii complex I was found to run mainly as a monomer. Two other forms could be detected by activity staining: a minor form of high molecular mass (1,500 kD) that probably corresponds to a dimer and a subcomplex of 200 kD. In agreement with the results of Cardol et al. (2002), the 200-kD band represents a soluble fraction that contains the hydrophilic 49- and 76-kD subunits of the complex I peripheral arm. It is likely that the complex I monomer represents the physiological state of this protein in mitochondria because even in the most mild solubilization conditions, it was always found as a monomer (Schägger and Pfeiffer, 2000). In addition, complex I has been shown to associate with complexes III and IV (Schägger and Pfeiffer, 2000). Nevertheless, the 1,500-kD band in C. reinhardtii is thought to represent only dimeric complex I because immunoblot analysis of 2D-SDS-PAGE with antibodies against subunits of complexes III and IV (Fig. 5) never indicated the presence of supercomplexes. The possible physiological role of dimeric complex I remains to be established.
Immunoblot analysis allowed the identification of oligomeric forms of the respiratory complexes III and IV (Fig. 5). The major form of complex III is a dimer of 500 kD coexisting with a minor form of 1,000 kD. In other organisms, complex III is mainly present as a dimer as well. It was found that the beef complex III dimer is more active than the monomer (Nalecz and Azzi, 1985). In addition, cytochrome c binds to only one recognition site of the dimeric yeast bc1 complex (Lange and Hunte, 2002), and the dimeric yeast bc1 complex oxidizes ubiquinol by an alternating, half-of-the-sites mechanism (Gutiérrez-Cirlos and Trumpower, 2002).
Antibodies against the COXIIB subunit of the colorless C. reinhardtii relative Polytomella sp. showed that C. reinhardtii complex IV is present mainly in a 240-kD form. In potato, the 160-kD monomeric form was predominant, but a portion of 230 kD was also present (Jänsch et al., 1996). The 240-kD form in C. reinhardtii is smaller than the theoretical dimer (300 kD) and may represent a dimeric cytochrome c oxidase exhibiting anomalous migration in BN-PAGE. The crystal structure of beef complex IV is clearly dimeric (Tsukihara et al., 1996), although solubilized dimers are difficult to maintain and easily dissociate into monomers (Musatov et al., 2000). Also, monomers have been reported to be more active than dimers (Nalecz et al., 1983).
Although BN-PAGE allowed the high resolution of several C. reinhardtii OXPHOS complexes, other proteins, such as complex II, the AOX, and the ADP/ATP carrier, ran as diffuse bands or smeared along the gel. The pattern on 2D-SDS-PAGE for the ADP/ATP carrier suggested that it was present on the first dimension either in multiple oligomeric forms, as partial aggregates, or both (Fig. 2). The AOX, a membrane-bound protein, also aggregates under the electrophoretic conditions applied. Surprisingly, the same is true for the aconitase, which is clearly a soluble protein. The high resolution of some complexes, along with the aggregation of some other proteins under the same conditions, might be an inherent property of BN-PAGE. With this technique, it was claimed that several mitochondrial dehydrogenases in yeast form supramolecular complexes (Grandier-Vazeille et al., 2001). However, care must be taken to distinguish supercomplexes from contamination that originates from smeared proteins. The comigration of proteins in discrete regions of BN-PAGE may reflect a contribution of aggregates and not necessarily indicate in vivo associations. The associations should exhibit a certain stoichiometry, and the conclusions should be corroborated using an independent method, i.e. cross-linking, gradient centrifugation, or gel filtration experiments.
C. reinhardtii Mitochondrial Complex V Is Atypical
C. reinhardtii complex V is resolved on 2D-SDS-PAGE into at least 13 distinct subunits (Funes et al., 2002; this work), comparable with 13 subunits in beef (Schägger and von Jagow, 1991), in potato (Jänsch et al., 1996), in Arabidopsis (Kruft et al., 2001), and in Polytomella sp. (Atteia et al., 1997; A. Atteia and R. van Lis, unpublished data). This work allowed the identification of subunits gamma (31 kD), delta (24 kD), and ATP9 (7 kD). These subunits do not exhibit amino acid extensions as do the alpha- and beta-subunits (Atteia et al., 1992; Franzén and Falk, 1992; Nurani and Franzén, 1996). In contrast to mitochondrial ATP synthases from plant or mammalian sources, the gamma-subunit in C. reinhardtii (31 kD) is not the third largest protein of the complex because three unidentified proteins of 45, 38, and 35 kD were present in the polypeptide pattern of complex V.
When using 2D-SDS-PAGE supplemented with 8 m urea, an additional 60-kD polypeptide was resolved from C. reinhardtii complex V separated on BN-PAGE. This polypeptide, named MASAP, was previously found to be associated with C. reinhardtii complex V isolated by Suc density gradients (Atteia, 1994). Because solubilization was performed with 5% (w/v) Triton X-100 and the gradients contained 0.5 m potassium phosphate and 0.2% (w/v) Triton X-100, it can be concluded the MASAP tightly interacts with complex V. The previously reported N-terminal amino acid sequence of MASAP (Atteia, 1994) was confirmed in this work, and the complete sequence of the corresponding cDNA was obtained. The deduced amino acid sequence did not show similarity to other mitochondrial proteins in the databases. Yet, its presequence has all the characteristics of a mitochondrial targeting sequence. A 66-kD protein, identified as the HSP66 chaperonin, has been found associated to yeast ATP synthase (Gray et al., 1990). However, MASAP does not show any similarity to heat shock proteins, making it unlikely to be a chaperonin.
Assuming that the 14 proteins in C. reinhardtii are genuine constituents of complex V, the expected monomer of this complex would be 740 kD. Nevertheless, this complex exhibited the lowest electrophoretic mobility on BN-PAGE with an estimated molecular mass of 1,600 kD. In contrast, monomeric complex V from yeast, plants, and mammals has a molecular mass of 550 to 580 kD on BN-PAGE (Schägger, 1995; Jänsch et al., 1996; Arnold et al., 1998; Kruft et al., 2001). Also, the C. reinhardtii chloroplast ATP synthase exhibited a molecular mass of 500 kD (Fig. 6). On the same gels, the mitochondrial and chloroplast ATP synthases of the green alga clearly behaved differently. In addition, both specific staining and immunolabeling could not reveal the presence of a mitochondrial F1-ATP synthase moiety. This also contrasts with BN-PAGE analysis of plant, trypanosomatid, and mammalian mitochondria, which invariably revealed the presence of dissociated F1-ATP synthase particles (Schägger and von Jagow, 1991; Jänsch et al., 1996; Kügler et al., 1997; Maslov et al., 1999; Singh et al., 2000; Kruft et al., 2001). Clearly, the behavior of C. reinhardtii complex V on BN-PAGE differs from the ones observed in other organisms.
Complex V dimers have been observed on BN-PAGE with mammalian and yeast mitochondria but only as a small fraction of the total amount. In the case of yeast complex V, dimeric forms were observed when mitochondrial membranes were solubilized with low detergent to protein ratios. Three additional small subunits—g, h, and Tim 11—are believed to be involved in the dimerization of the yeast complex (Arnold et al., 1998). Altogether, our data strongly suggest an unprecedented strong dimerization of C. reinhardtii mitochondrial complex V and an uncommon resistance to dissociation of the F1 sector. We hypothesize that MASAP, by itself or in conjunction with the three unidentified proteins of 45, 38, and 35 kD, participate in the formation of highly stable complex V dimers in C. reinhardtii. Also, the unique amino acid extensions identified in the alpha- and beta-subunits (Franzén and Falk, 1992; Nurani and Franzén, 1996) could play a role in the dimerization of complex V.
The Core Proteins in C. reinhardtii Complex III
In eukaryotes, complex III has core I and core II subunits, two mitochondrial matrix-exposed proteins not involved in electron transfer. In plants, these proteins function as a MPP, and may have originated from a protease that was integrated into the bc1 complex during early stages of the endosymbiotic event that gave rise to mitochondria (Braun and Schmitz, 1995b). In contrast to plants, the MPP activity in the photosynthetic alga C. reinhardtii was shown to be soluble (Nurani et al., 1997). Also, complex III of Polytomella sp., a non-photosynthetic relative of C. reinhardtii, is proteolytically inactive (Brumme et al., 1998). In this work, we identified C. reinhardtii core I subunit and determined its complete sequence using the ChlamyEST database. The deduced protein exhibits similarity to beta-MPP and core I subunits from different organisms. Core I exhibits the complete inverse zinc-binding motif (HXXEH), which was shown to be essential for the proteolytic activity of MPP in rat mitochondria (Kitada et al., 1995). The core I of C. reinhardtii has the beta-MPP consensus sequence (Braun and Schmitz, 1995b), except for a single Arg to Lys substitution at position 175. However, this substitution is unlikely to be responsible of the lack a beta-MPP activity. In addition, the proposed core II sequence derived from the ChlamyEST database did not exhibit the consensus sequences for alpha-MPP. This raises the possibility that the MPP activity in C. reinhardtii could be organized as in N. crassa (Hawlitschek et al., 1988), with the core I protein exhibiting beta-MPP activity and the alpha-MPP being a soluble protein in the mitochondrial matrix. In the study of Nurani et al. (1997), the soluble fraction of C. reinhardtii was shown to exhibit proteolytic activity. It is likely that the preparation of this soluble fraction by sonication might have caused a certain level of dissociation of the core I subunit from complex III, giving rise to the observed soluble MPP activity.
C. reinhardtii Complex IV
This work provides new insights into the subunit composition of complex IV of the photosynthetic alga. The identification of COXIIA and COXIIB as distinct subunits of 14 and 16 kD indicates that the C. reinhardtii subunit COXII is a heterodimer, as previously shown for Polytomella sp. (Pérez-Martínez et al., 2001). In contrast to Polytomella sp., C. reinhardtii COXIIA and COXIIB subunits are well separated on 15% (w/v) Tricine-SDS polyacrylamide gels. The N-terminal sequence of C. reinhardtii COXIII and COXIIA determined in this study confirmed the prediction of the cleavage site in the preproteins. However, the sequence determined for COXIIB does not coincide with the N terminus predicted from the gene (Pérez-Martínez et al., 2001). This sequence was found to correspond to an internal sequence starting at residue 96 of the deduced mature protein. The same internal sequence was determined for COXIIB from Polytomella sp. (Pérez-Martínez et al., 2001). It was suggested that the COXIIB N terminus is blocked and that the observed sequence represents a region of the protein that is cleaved during Edman degradation. Three additional subunits of C. reinhardtii complex IV (COXVIb, COXVIa, and COXVb) were also identified. COXVb sequence is atypical because its first 40 residues and the last 40 residues show very poor similarity with is mammalian counterparts. Also, the first 60 residues of C. reinhardtii mature COXVIb did not show any similarity to other COXVIb subunits; this extension accounts for the fact that the green algal COXVIb has a molecular mass at least twice that of typical COXVIb subunits. The N-terminal sequence of COXVIb is characterized by a high content of Pro and charged residues, with a highly acidic theoretical pI of 4.39. The atypical sequences of some constituents of C. reinhardtii complex IV raise questions on the assembly and interactions of the complex IV subunits in the inner mitochondrial membrane.
Toward Functional Proteomics
The application of different subcellular fractions to BN-PAGE, either membranous, soluble, or whole organelles, enables a comprehensive study of the effect of growth conditions, mutations, and other factors that can influence biogenesis and metabolism. This is exemplified by the resolution of the chloroplast complexes together with their mitochondrial counterparts and by the analysis of the BF4.F54.F14 mutant strain. Among the many mutant strains available in C. reinhardtii, only few have been characterized at the biochemical level (de Vitry and Vallon, 1999; Duby et al., 2001). The impact of mutations in nuclear and organellar genes is likely to be better understood using a proteomic approach. The method developed in this work to isolate intact mitochondria from strains that have cell walls using CTAB makes BN-PAGE studies amenable for any C. reinhardtii mutant or wild-type strain.
We presented a partial catalog of the C. reinhardtii mitochondrial proteome based on BN-PAGE. With this approach, the behavior and composition of protein complexes was revealed, novel proteins were described (MASAP), some unusual structural features of proteins encoded by previously characterized genes were demonstrated (COXIIA and COXIIB), and novel predictions were made based on newly obtained sequences (cores I and II). With the genome project of C. reinhardtii approaching finalization, a more complete picture of the mitochondrial proteome may be obtained.
MATERIALS AND METHODS
Cell Growth and Isolation of Mitochondria
All Chlamydomonas reinhardtii strains were grown at 25°C to 26°C in Tris-acetate phosphate medium (Harris, 1989) in continuous light and agitation. For the cell wall-less strain 84CW15, the medium was supplemented with 1% (w/v) sorbitol. Mitochondria from 84CW15 cells were isolated in their late exponential growth phase as described by Eriksson et al. (1995). To isolate mitochondria from strains containing cell walls, the cells were resuspended in washing buffer (20 mm HEPES [pH 7.2]) to a concentration of 50 mg wet weight mL−1. Subsequently, 50 μm CTAB was added from a 10 mm stock solution, and the cells were incubated at room temperature with agitation for 5 min. Before cell disruption with glass beads, the cells were diluted 5-fold and washed twice in washing buffer. The major portion of the orange precipitate that formed on top of the pellet of the second centrifugation of the mitochondrial purification procedure (Eriksson et al., 1995) was removed by pipetting and discarded; this enabled the application of the sample to BN-PAGE.
BN-PAGE
Sample preparation and BN-PAGE were carried out as described by Schägger and von Jagow (1991) with the following modifications: Isolated mitochondria or other cell fractions were first washed with 0.25 m sorbitol and 15 mm Bis-Tris (pH 7.0) and then resuspended in sample buffer (50 mm Bis-Tris and 0.75 m amino caproic acid [pH 7.0]). Pure mitochondria (final protein concentration of 5 mg mL−1) was solubilized in the presence of 1% (w/v) n-dodecyl maltoside. Other fractions were solubilized in the presence of 2% (w/v) n-dodecyl maltoside at the same protein concentration. From mitochondrial fractions of cell wall-containing strains that were pretreated with CTAB, any residual orange precipitates were removed during the washing steps and also after solubilization. The solubilization was carried out with samples prepared the same day. Once solubilized, the proteins could be stored on ice at 4°C up to a week. Linear polyacrylamide gradients varied from 5% to 10% to 5% to 15% (w/v). To minimize protein aggregation in the sample wells or in the gel, the stacking gel was poured immediately onto the resolving gel before it polymerized. For electrophoresis, either the Vertical Gel Electrophoresis System VI6 (4–5-h run at 20–25 mA; Gibco-BRL, Cleveland) or the Bio-Rad Mini Protean II system (1 h run at 15 mA; Bio-Rad Laboratories, Hercules, CA) was used. No prerun was performed.
Specific Staining of the OXPHOS Complexes
Covalently linked hemes were detected on SDS-polyacrylamide gels by their peroxidase activity in the presence of 3,3′,5,5′-tetramethylbenzidine (Thomas et al., 1976). Other specific stainings were carried out directly on the blue gel lanes. NADH dehydrogenase activity was detected in 100 mm Tris-HCl (pH 7.4) containing 1 mg mL−1 NBT and 100 mm NADH (Kuonen et al., 1986). Succinate dehydrogenase activity was assayed in a buffer containing 50 mm phosphate buffer (pH 7.4), 100 mm sodium succinate, 200 μm phenazine methosulphate, and 2 mg mL−1 NBT (Jung et al., 2000). ATPase activity was located in situ by the method of Horak and Hill (1972), incubating the lane of BN-PAGE overnight in 10 mm ATP and 30 mm CaCl2 in 50 mm HEPES (pH 8.0).
2D-Tricine-SDS-PAGE
Entire lanes from BN-PAGE were used to resolve the subunits in the 2D-Tricine-SDS-PAGE (15% [w/v] acrylamide) as described by Schägger and von Jagow (1991). Alternatively, Gly-SDS-PAGE (15% [w/v] acrylamide) was used (Laemmli, 1970). Where indicated, 2D-Tricine-SDS-PAGE was run in the presence of 8 m urea. Apparent molecular masses were estimated using BenchMark protein standards (Invitrogen, Carlsbad, CA).
Protein Analysis
Protein concentrations were determined as described by Markwell et al. (1978). Samples containing chlorophyll were precipitated using methanol and chloroform (Wessel and Flugge, 1984) before protein determination. After electrophoresis, proteins were electrotransferred onto nitrocellulose (Bio-Rad) or ProBlot membranes (Amersham-Pharmacia Biotech, Uppsala) using 50 mm H3BO3 and 50 mm Tris (no pH adjustment) as transfer buffer (tank transfer system). Immunodetection was carried out using the ECL kit (Amersham-Pharmacia Biotech) or the Pico kit (Pierce Chemical, Rockford, IL). The antisera used were raised against C. reinhardtii AOX (see below), Neurospora crassa core I subunit, and the COXIIB and beta-ATP synthase subunits of Polytomella sp. For N-terminal sequencing, the bands of the protein complexes resolved by BN-PAGE were excised from preparative gels. The slices were incubated in cathode buffer containing 1% (v/v) β-mercaptoethanol for 20 min, rinsed with cathode buffer, and loaded as a stack on top of a Tricine-SDS-PAGE. N-terminal analysis of electroblotted proteins onto polyvinylidene difluoride membranes was performed by automated Edman degradation at the Faculty of Medicine, Universidad Nacional Autónoma de México (LF 3000 Beckman sequencer, Beckman Instruments, Fullerton, CA) or at the Institut Pasteur, Paris (Procise 494 or 473A sequencers, PE-Applied Biosystems, Foster City, CA), all equipped with on-line HPLC apparatus. Internal sequencing after trypsinolysis was carried out as previously described (Atteia et al., 1997).
Cloning of the cDNA Encoding the MASAP
Using the degenerate oligodeoxynucleotides 5′-TAC GT(G/C) AC(G/C) GC(G/C) CT(G/C) AAG G-3′ and 5′-CTC CTG CTC (G/C) GC (G/C) C GGA AC-3′, designed on the N-terminal and internal amino acid sequences of MASAP, a PCR product of 1,173 bp was obtained using as template a mass excision plasmid preparation from a λZAP II cDNA library of C. reinhardtii. Samples were denatured for 5 min at 95°C and subjected to three cycles of 30-s denaturation at 95°C, 40 s of annealing at 62°C, and 2-min extension at 72°C, followed by 32 cycles of 30-s denaturation at 95°C, 40 s of annealing at 64°C, 2-min extension at 72°C, and a last 10-min extension at 72°C. The fragment was cloned into the pGEM-T easy vector (Promega, Madison, WI) and sequenced using the T7 and SP6 primers. The amplified DNA fragment was used to screen the cDNA library of C. reinhardtii. A cDNA of 2.4 kb was obtained and sequenced. The complete sequence is available at the GenBank/EBI Data Bank (accession no. AJ441255).
AOX Carboxy Terminus Overexpression and Antibody Production
Primers were designed based on the sequence of the C. reinhardtii Aox1 gene (accession no. AF352435): 5′-GAC GAG CTC CTG CTG TCG CCG CGC AC-3′ and 5′-CTG AAG CTT GGG CAG CTG GCT GGC GC-3′. Underlined are the added SacI and HindIII restriction sites. PCR amplification with Taq polymerase was done using as a template a plasmid preparation obtained by mass excision from a λ ZAPII C. reinhardtii cDNA library. Samples were denatured for 5 min at 95°C and subjected to three cycles of 30-s denaturation at 95°C, 40 s of annealing at 62°C, and 1-min extension at 72°C, followed by 32 cycles of 30-s denaturation at 95°C, 40 s of annealing at 64°C, and 1-min extension at 72°C and a last 10-min extension at 72°C. The 360-bp product was cloned into the restriction sites SacI and HindIII of the pQE30 vector (Qiagen USA, Valencia, CA), and the C-terminal region of the AOX protein of 11 kD containing a six-residue His tag was overexpressed and purified using a nickel-nitrilotriacetic acid agarose resin according to the manufacturer's instructions. The purified overexpressed C-terminal AOX fragment was used to raise antibodies in a rabbit.
Sequence Analysis in Silico
Protein sequences were obtained from ENTREZ at the NCBI server (www.ncbi.nlm.nih.gov), and alignments were made with the FASTA program (vega.igh.cnrs.fr/bin/fasta-guess.cgi). EST clones of C. reinhardtii were obtained from the ChlamyEST database (http://www.biology.duke.edu/chalmy) using the WU-TBLASTN program. Multiple alignments were done with ClustalW (Thompson et al., 1994; searchlauncher.bcm.tmc.edu). Molecular masses and pI calculations were done with the compute pI/molecular mass tool (Bjellqvist et al., 1993), and the prediction of intracellular sorting was done with the TargetP V1.0 program (Emanuelsson et al., 2000), both from the ExPASy Molecular Biology Server (www.expasy.ch).
Note added in proofs
Recent data on the bovine heart complex I indicate a molecular mass of over 900 kDa instead of 750 kDa, as mentioned in this work (Carroll et al., 2002). This would modify the estimated molecular masses of the bands on BN-PAGE, corresponding to the C. reinhardtii complexes I and V, to around 1000 kDa and 2000 kDa, respectively.
ACKNOWLEDGMENTS
We thank Drs. David W. Krogmann (Purdue University, West Lafayette, IN), Samuel I. Beale (Brown University, Providence, RI), and Dominique Drapier (Institute de Biologic Physico-Chimqique, Paris) for critical comments to the manuscript. Our gratitude goes to Dr. Jacques d'Alayer (Institut Pasteur, Paris) for the determination of N-terminal and internal sequences. We also thank Dr. Dominique Drapier (Institute de Biologic Physico-Chimqique) for the kind gifts of C. reinhardtii strains, Dr. John P. Davies (Iowa State University, Ames) for providing the cDNA library of C. reinhardtii, Dr. Hans-Peter Braun (Hannover University, Germany) for providing the anticore I antibody of N. crassa, and Dr. Robert Bassi (University of Verona, Italy) for the antibody against the D1 protein of C. reinhardtii. We thank Miriam Vázquez-Acevedo (Universidad Nacional Autónoma de México, Mexico City) for the supply of the antibody against the Polytomella sp. COXIIB subunit, Hector Malagón Rivero (Universidad Nacional Autónoma de México, Mexico City) for his help with the production of the anti-AOX antibody, and Drs. Antonio Peña (Universidad Nacional Autónoma de México, Mexico City), Jorge Ramírez (Universidad Nacional Autónoma de México, Mexico City), and George Dreyfus (Universidad Nacional Autónoma de México, Mexico City) for the use of material and equipment at their labs at the Instituto de Fisiología Celular (Universidad Nacional Autónoma de México, Mexico City).
Footnotes
This work was supported by Consejo Nacional de Ciencia y Tecnología (grant no. 27754N), by Dirección General de Asuntos para el Personal Académico (Mexico; grant no. IN204595), by Dirección General de Estudios de Posgrado-Universidad Nacional Autónoma de México (PhD student fellowship to R.v.L.), and by the National Science Foundation (grant no. MCB–9975765 to the Chlamydomonas genome project).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.018325.
LITERATURE CITED
- Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schägger H. Yeast mitochondrial F1F0-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 1998;17:7170–7178. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atteia A. Identification of mitochondrial respiratory proteins from the green alga Chlamydomonas reinhardtii. C R Acad Sci III. 1994;317:11–19. [PubMed] [Google Scholar]
- Atteia A, de Vitry C, Pierre Y, Popot JL. Identification of mitochondrial proteins in membrane preparations from Chlamydomonas reinhardtii. J Biol Chem. 1992;267:226–234. [PubMed] [Google Scholar]
- Atteia A, Dreyfus G, González-Halphen D. Characterization of the alpha and beta-subunits of the F0F1-ATPase from the alga Polytomella spp., a colorless relative of Chlamydomonas reinhardtii. Biochim Biophys Acta. 1997;1320:275–284. doi: 10.1016/s0005-2728(97)00031-5. [DOI] [PubMed] [Google Scholar]
- Atteia A, Franzén L-G. Identification, cDNA sequence and deduced amino acid sequence of the mitochondrial Rieske iron-sulfur protein from the green alga Chlamydomonas reinhardtii: implications for protein targeting and subunit interaction. Eur J Biochem. 1996;237:792–799. doi: 10.1111/j.1432-1033.1996.0792p.x. [DOI] [PubMed] [Google Scholar]
- Atteia A, van Lis R, Wetterskog D, Gutiérrez-Cirlos E-B, Ongay-Larios L, Franzén L-G, González-Halphen D. Structure, organization and expression of the genes encoding mitochondrial cytochrome c1 and the Rieske iron-sulfur protein in Chlamydomonas reinhardtii. Mol Genet Genomics. 2002;1320:275–284. doi: 10.1007/s00438-002-0779-x. [DOI] [PubMed] [Google Scholar]
- Berthold DA, Siedow JN. Partial purification of the cyanide-resistant alternative oxidase of skunk cabbage (Symplocarpus foetidus) mitochondria. Plant Physiol. 1993;101:113–119. doi: 10.1104/pp.101.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjellqvist B, Hughes GJ, Pasquali Ch, Paquet N, Ravier F, Sanchez J-Ch, Frutiger S, Hochstrasser DF. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14:1023–1031. doi: 10.1002/elps.11501401163. [DOI] [PubMed] [Google Scholar]
- Braun HP, Schmitz UK. The bifunctional cytochrome c reductase/processing peptidase complex from plant mitochondria. J Bioenerg Biomembr. 1995a;27:423–436. doi: 10.1007/BF02110005. [DOI] [PubMed] [Google Scholar]
- Braun HP, Schmitz UK. Are the “core” proteins of the mitochondrial bc1 complex evolutionary relics of a processing protease? Trends Biochem Sci. 1995b;20:171–175. doi: 10.1016/s0968-0004(00)88999-9. [DOI] [PubMed] [Google Scholar]
- Brumme S, Kruft V, Schmitz UK, Braun HP. New insights into the co-evolution of cytochrome c reductase and the mitochondrial processing peptidase. J Biol Chem. 1998;273:13143–13149. doi: 10.1074/jbc.273.21.13143. [DOI] [PubMed] [Google Scholar]
- Cardol P, Matagne RF, Remacle C. Impact of mutations affecting ND mitochondria-encoded subunits on the activity and assembly of complex I in Chlamydomonas: implication for the structural organization of the enzyme. J Mol Biol. 2002;319:1211–1221. doi: 10.1016/S0022-2836(02)00407-2. [DOI] [PubMed] [Google Scholar]
- Carroll J, Shannon RJ, Fearnley IM, Walker JE, Hirst J. Definition of the nuclear encoded protein composition of bovine heart mitochondrial complex I. Indentification of two new subunits. J Biol Chem. 2002;277:50311–50317. doi: 10.1074/jbc.M209166200. [DOI] [PubMed] [Google Scholar]
- Chua N-H, Matlin K, Bennoun P. A chlorophyll-protein complex lacking in photosystem I mutants of Chlamydomonas reinhardtii. J Cell Biol. 1975;67:361–377. doi: 10.1083/jcb.67.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vitry C, Vallon O. Mutants of Chlamydomonas: tools to study thylakoid membrane structure, function and biogenesis. Biochimie. 1999;81:631–643. doi: 10.1016/s0300-9084(99)80120-5. [DOI] [PubMed] [Google Scholar]
- Dinant M, Baurain D, Coosemans N, Joris B, Matagne RF. Characterization of two genes encoding the mitochondrial alternative oxidase in Chlamydomonas reinhardtii. Curr Genet. 2001;39:101–108. doi: 10.1007/s002940000183. [DOI] [PubMed] [Google Scholar]
- Duby F, Cardol P, Matagne RF, Remacle C. Structure of the telomeric ends of mt DNA, transcriptional analysis and complex I assembly in the dum-24 mitochondrial mutant of Chlamydomonas reinhardtii. Mol Genet Genomics. 2001;266:109–114. doi: 10.1007/s004380100529. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Eriksson M, Gardeström P, Samuelsson G. Isolation, purification and characterization of mitochondria from Chlamydomonas reinhardtii. Plant Physiol. 1995;107:479–483. doi: 10.1104/pp.107.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler HR, Schmid R, Leu S, Shavit N, Strotmann H. Isolation of CF0CF1 from Chlamydomonas reinhardtii cw15 and the N-terminal amino acid sequences of the CF0CF1subunits. FEBS Lett. 1995;377:163–166. doi: 10.1016/0014-5793(95)01332-6. [DOI] [PubMed] [Google Scholar]
- Franzén L-G, Falk G. Nucleotide sequence of cDNA clones encoding the beta subunit of the mitochondrial ATP synthase from the green alga Chlamydomonas reinhardtii: the precursor protein encoded by the cDNA contains both an N-terminal presequence and a C-terminal extension. Plant Mol Biol. 1992;19:771–780. doi: 10.1007/BF00027073. [DOI] [PubMed] [Google Scholar]
- Funes S, Davidson E, Claros MG, van Lis R, Pérez-Martínez X, Vázquez-Acevedo M, King MP, González-Halphen D. The typically mitochondrial DNA-encoded ATP6 subunit of the F1F0-ATPase is encoded by a nuclear gene in Chlamydomonas reinhardtii. J Biol Chem. 2002;277:6051–6058. doi: 10.1074/jbc.M109993200. [DOI] [PubMed] [Google Scholar]
- GrandierVazeille X, Bathany K, Chaignepain S, Camougrand N, Manon S, Schmitter J-M. Yeast mitochondrial dehydrogenases are associated in a supramolecular complex. Biochemistry. 2001;40:9758–9769. doi: 10.1021/bi010277r. [DOI] [PubMed] [Google Scholar]
- Gray RE, Grasso DG, Maxwell RJ, Finnegan PM, Nagley P, Devenish RJ. Identification of a 66 KDa protein associated with yeast mitochondrial ATP synthase as heat shock protein hsp60. FEBS Lett. 1990;268:265–268. doi: 10.1016/0014-5793(90)81024-i. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Cirlos EB, Trumpower BL. Inhibitory analogs of ubiquinol act anti-cooperatively on the yeast cytochrome bc1 complex: evidence for an alternating, half-of-the-sites mechanism of ubiquinol oxidation. J Biol Chem. 2002;277:1195–1202. doi: 10.1074/jbc.M109097200. [DOI] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. San Diego: Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- Harris EH. Chlamydomonasas a model organism. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:363–406. doi: 10.1146/annurev.arplant.52.1.363. [DOI] [PubMed] [Google Scholar]
- Hawlitschek G, Schneider H, Schmidt B, Tropschug M, Hartl FU, Neupert W. Mitochondrial protein import: identification of processing peptidase and of PEP, a processing enhancing protein. Cell. 1988;53:795–806. doi: 10.1016/0092-8674(88)90096-7. [DOI] [PubMed] [Google Scholar]
- Horak A, Hill RD. Adenosine triphosphate of bean plastids. Its properties and site of formation. Plant Physiol. 1972;49:365–370. doi: 10.1104/pp.49.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jänsch L, Kruft V, Schmitz UK, Braun HP. New insights into the composition, molecular mass and stoichiometry of the protein complexes of plant mitochondria. Plant J. 1996;9:357–368. doi: 10.1046/j.1365-313x.1996.09030357.x. [DOI] [PubMed] [Google Scholar]
- Jung C, Higgins CMJ, Xu Z. Measuring the quantity and activity of mitochondrial electron transport chain complexes in tissues of central nervous system using blue native polyacrylamide gel electrophoresis. Anal Biochem. 2000;286:214–223. doi: 10.1006/abio.2000.4813. [DOI] [PubMed] [Google Scholar]
- Kitada S, Shimokata K, Niidome T, Ogishima T, Ito A. A putative metal-binding site in the beta subunit of rat mitochondrial processing peptidase is essential for its catalytic activity. J Biochem. 1995;117:1148–1150. doi: 10.1093/oxfordjournals.jbchem.a124836. [DOI] [PubMed] [Google Scholar]
- Kruft V, Eubel H, Jansch L, Werhahn W, Braun H-P. Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 2001;127:1694–1710. [PMC free article] [PubMed] [Google Scholar]
- Kügler M, Jänsch L, Kruft V, Schmitz UK, Braun H-P. Analysis of the chloroplast protein complexes by blue-native polyacrylamide gel electrophoresis (BN-PAGE) Photosynth Res. 1997;53:35–44. [Google Scholar]
- Kuonen DR, Roberts PJ, Cottingham IR. Purification and analysis of mitochondrial membrane proteins on nondenaturing gradient polyacrylamide gels. Anal Biochem. 1986;153:221–226. doi: 10.1016/0003-2697(86)90084-9. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lange C, Hunte C. Crystal structure of the yeast cytochrome bc1 complex with its bound substrate cytochrome c. Proc Natl Acad Sci USA. 2002;99:2800–2805. doi: 10.1073/pnas.052704699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwell MAK, Hass SM, Biber LL, Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- Maslov DA, Nawathean P, Scheel J. Partial kinetoplast-mitochondrial gene organization and expression in the respiratory deficient plant trypanosomatid Phytomonas serpens. Mol Biochem Parasitol. 1999;99:207–221. doi: 10.1016/s0166-6851(99)00028-6. [DOI] [PubMed] [Google Scholar]
- Michaelis G, Vahrenholz C, Pratje E. Mitochondrial DNA of Chlamydomonas reinhardtii: the gene for apocytochrome band the complete functional map of the 15.8 kb DNA. Mol Gen Genet. 1990;223:211–216. doi: 10.1007/BF00265056. [DOI] [PubMed] [Google Scholar]
- Millar AH, Sweetlove LJ, Giegé P, Leaver CJ. Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol. 2001;127:1711–1727. [PMC free article] [PubMed] [Google Scholar]
- Musatov A, Ortega-Lopez J, Robinson NC. Detergent-solubilized bovine cytochrome c oxidase: dimerization depends on the amphiphilic environment. Biochemistry. 2000;39:12996–13004. doi: 10.1021/bi000884z. [DOI] [PubMed] [Google Scholar]
- Nalecz KA, Bolli R, Azzi A. Preparation of monomeric cytochrome coxidase: its kinetics differ from those of the dimeric enzyme. Biochem Biophys Res Commun. 1983;114:822–828. doi: 10.1016/0006-291x(83)90855-0. [DOI] [PubMed] [Google Scholar]
- Nalecz MJ, Azzi A. Functional characterization of the mitochondrial cytochrome b-c1complex: steady-state kinetics of the monomeric and dimeric forms. Arch Biochem Biophys. 1985;240:921–993. doi: 10.1016/0003-9861(85)90101-8. [DOI] [PubMed] [Google Scholar]
- Nurani G, Eriksson M, Knorpp C, Glaser E, Franzen L-G. Homologous and heterologous protein import into mitochondrial isolated from the green alga Chlamydomonas reinhardtii. Plant Mol Biol. 1997;35:973–980. doi: 10.1023/a:1005878614878. [DOI] [PubMed] [Google Scholar]
- Nurani G, Franzén L-G. Isolation and characterization of the mitochondrial ATP synthase from Chlamydomonas reinhardtii: cDNA sequence and deduced protein sequence of the alpha subunit. Plant Mol Biol. 1996;31:1105–1116. doi: 10.1007/BF00040828. [DOI] [PubMed] [Google Scholar]
- Olive J, Wollman F-A, Bennoun P, Recouvreur M. Ultrastructure of thylakoid membranes in C. reinhardtii: evidence for variations in the partition coefficient of the light-harvesting complex containing particles upon membrane fracture. Arch Biochem Biophys. 1981;208:456–467. doi: 10.1016/0003-9861(81)90532-4. [DOI] [PubMed] [Google Scholar]
- Piccioni RG, Bennoun P, Chua NH. A nuclear mutant of Chlamydomonas reinhardtiidefective in photosynthetic photophsophorylation. Eur J Biochem. 1981;117:93–102. doi: 10.1111/j.1432-1033.1981.tb06307.x. [DOI] [PubMed] [Google Scholar]
- Pérez-Martínez X, Antaramian A, Vázquez-Acevedo M, Funes S, Tolkunova E, d'Alayer J, Claros MG, Davidson E, King MP, González-Halphen D. Subunit II of cytochrome c oxidase in Chlamydomonasalgae is a heterodimer encoded by two independent nuclear genes. J Biol Chem. 2001;276:11302–11309. doi: 10.1074/jbc.M010244200. [DOI] [PubMed] [Google Scholar]
- Pérez-Martínez X, Vázquez-Acevedo M, Tolkunova E, Funes S, Claros MG, Davidson E, King MP, González-Halphen D. Unusual location of a mitochondrial gene: subunit III of cytochrome c oxidase is encoded in the nucleus of Chlamydomonasalgae. J Biol Chem. 2000;275:30144–30152. doi: 10.1074/jbc.M003940200. [DOI] [PubMed] [Google Scholar]
- Schägger H. Native electrophoresis for isolation of mitochondrial oxidative phosphorylation protein complexes. Methods Enzymol. 1995;260:190–203. doi: 10.1016/0076-6879(95)60137-6. [DOI] [PubMed] [Google Scholar]
- Schägger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Sharpe JA, Day A. Structure, evolution and expression of the mitochondrial ADP/ATP translocator gene from Chlamydomonas reinhardtii. Mol Gen Genet. 1993;237:134–144. doi: 10.1007/BF00282794. [DOI] [PubMed] [Google Scholar]
- Singh P, Jansch L, Braun HP, Schmitz UK. Resolution of mitochondrial and chloroplast membrane protein complexes from green leaves of potato on blue-native polyacrylamide gels. Indian J Biochem Biophys. 2000;37:59–66. [PubMed] [Google Scholar]
- Thomas PE, Ryan D, Levin W. An improved staining procedure for the detection of the peroxidase activity of P450 on sodium dodecyl sulfate polyacrylamide gels. Anal Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome coxidase at 2.8Å. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- Umbach AL, Siedow JN. The cyanide-resistant alternative oxidases from the fungi Pichia stipitis and Neurospora crassaare monomeric and lack regulatory features of the plant enzyme. Arch Biochem Biophys. 2000;378:234–245. doi: 10.1006/abbi.2000.1834. [DOI] [PubMed] [Google Scholar]
- Umbach L, Siedow JN. Covalent and noncovalent dimers of the cyanide-resistant alternative oxidase protein in higher plant mitochondria and their relationship to enzyme activity. Plant Physiol. 1993;103:845–854. doi: 10.1104/pp.103.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L. Alternative oxidase: from gene to function. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:703–734. doi: 10.1146/annurev.arplant.48.1.703. [DOI] [PubMed] [Google Scholar]
- Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]