Abstract
Activation-induced cytidine deaminase (AID) likely initiates immunoglobulin gene-conversion (GC) by deaminating cytidines within the V-region of chicken B-cells. However, the intervening DNA lesion required to initiate GC remains elusive. GC could be initiated by a single strand break or a double strand break (DSB). To distinguish between these possibilities, we examined GC in the chicken DT40 B cell line deficient in non-homologous end joining (NHEJ). It is known that the NHEJ and homologous recombination DNA repair pathways compete for DSBs. In light of this, if a DSB is the major intermediate, deficiency in NHEJ should result in increased levels of GC. Here we show that DNA–PKcs−/−/− and Ku70−/− DT40 cells had 5- to 10-fold higher levels of GC relative to wildtype DT40 as measured by surface IgM reversion and sequencing of the V-region. These data suggest that a DSB is the major DNA lesion that initiates GC.
INTRODUCTION
Immunoglobulin (Ig) gene-conversion (GC) is a secondary antibody diversification mechanism used by B-cells of some animals (i.e. chickens, rabbits, horses, pigs and cows) (1,2). Unlike mice and humans who rely primarily upon VDJ recombination for the generation of antibody diversity, GC is the major contributor in these organisms (3). GC is a process through which templated changes are made into the V-region via homologous recombination (HR). The genetic information that stores these potential changes is contained in a series of pseudo-genes (ψV genes) upstream of the V-region. These ψV genes are characterized by their lack of a functional promoter and recombination signal sequences, and also frequently harbor 5′ and 3′ truncations. Through multiple rounds of GC, each derived from any of the possible donor ψV genes, considerable diversity is generated.
Since the discovery of activation-induced cytidine deaminase (AID) (4), tremendous progress has been made towards understanding the processes which underlie secondary antibody diversification. Class switch recombination (CSR), somatic hypermutation (SHM) and GC were previously thought to be unrelated (5,6). However, cells deficient in AID are unable to initiate all three processes (7–9), indicating that they are intimately linked. AID is thought to initiate these mechanisms through the deamination of cytidine in the DNA into uridine (10–12). The downstream processing of these deamination lesions is what leads to the distinct outcomes. In SHM, the mechanism has been broken down into separate stages whereby the uridine generated is subject to multiple fates. It can be replicated across, recognized by the mismatch repair proteins (MMRs) or removed by a uracil DNA glycosylase (UNG), leaving an abasic site (5,13), the point at which CSR and GC come into play. The abasic site is cleaved by APE1 (an apurinic/apyrimidinic endonuclease) to generate a DNA strand break. These strand breaks, when generated in switch regions can lead to CSR (14,15). If these strand breaks occur in the V-region of chicken B-cells, the DNA lesions could potentially be repaired by the HR machinery leading to GC events. This idea is supported by the recent findings of Saribasak et al. (16) indicating that DT40 cells deficient for UNG have severely reduced levels of GC, and instead accumulate transition mutations. What remains unclear however, is whether a single strand break (SSB) is enough to initiate GC events, or if a double strand break (DSB) is required (17). The difficulty in addressing this issue lies in the fact that both lesions can potentially be generated through the known mechanisms of AID action.
To address this question, we turned to the use of an indirect method for evidence of the DNA lesion being generated. It has been well established that repair of DSBs occurs through two primary pathways, non-homologous end joining (NHEJ) and HR (18). NHEJ is thought to be the predominant form of DNA repair in mammalian cells, and is responsible for the non-templated, imprecise joining of two DSB ends. This process is mediated by the proteins of the Ku complex, (Ku70 and Ku80) as well as DNA–PKcs and DNA ligase IV. In addition, while single-stranded DNA has been shown to bind to elements involved in NHEJ, this binding does not induce activation of the pathway (19). HR on the other hand, occurs through the use of homologous template DNA to repair lesions. HR is able to act upon DSBs, both one-ended and two-ended, as well as stalled and collapsed replication forks (20). It is now known that HR and NHEJ are both able to access DSB ends and are thus able to compete for them (21). Taking advantage of this phenomenon, we postulated that if GC is proceeding via a DSB, perturbing the NHEJ pathway, and thus the competition between HR and NHEJ, would result in an increase of GC. This effect would only be expected to occur if the initiating lesion is a DSB and not a SSB.
MATERIALS AND METHODS
Cell culture
DT40 cells deficient for DNA–PKcs and Ku70 were obtained from Dr Shunichi Takeda (22,23). The DT40 cell line is trisomic at the location of DNA–PKcs (chromosome 2) and thus the DT40 DNA–PKcs−/−/− cell line contains three disrupted alleles. Surface IgM (sIgM) negative variants of wild type DT40 (WT DT40) cells were obtained from Dr Michael Ratcliffe, while AID−/− DT40 cells were obtained from Dr Reuben Harris. Each cell line used was derived from the same parental CL18 DT40 cell line, which harbors a frameshift mutation in CDR1 of the Vλ region. WT, Ku70−/−, DNA–PKcs−/−/−, and AID−/− DT40 cells were cultured in RPMI 1640 supplemented with penicillin and streptomycin, 50 μm β-mercaptoethanol, 3% chicken serum (Gibco) and 7% bovine calf serum at 37°C in 6% CO2. Subcloning was performed by plating cells in Terasaki plates at ∼1 cell per well. Cells were allowed to settle for 1 h, and wells containing 1 cell were transferred to 96 well plates immediately.
Flow cytometry
Cells were stained for sIgM with polyclonal goat anti chicken IgM conjugated to flourescein isothiocyanate (Rockland, PA). Flow cytometry was performed on a FACSCalibur (Becton Dickson) flow cytometer and cell sorting of NHEJ mutants was performed on an EPICS elite (Beckman Coulter) while WT DT40 were sorted on a FACS Aria (Becton Dickson) cell sorter. Cells were considered positive for sIgM if their fluorescence intensity was at least 8-fold above the negative peak. Cells were sorted into RPMI media supplemented as above. Data shown in Figure 2B were analyzed by one-way ANOVA and the Tukey–Kramer Multiple comparisons test using GraphPad Istat Version 3.06.
PCR and cloning
PCR and cloning were done as described previously (24) with the following modifications. PCR program consisted of touchdown PCR [8 cycles at 95°C (15 s) 68–60°C (1°C per cycle) (15 s) 72°C (1 min); 32 cycles at 94°C (15 s) 60°C (15 s) 72°C (1 min); 1 cycle 72°C (5 min)]. Sequencing reactions were performed by Macrogen (Korea). Sequence changes were analyzed for GC events as described previously (24). Briefly, multiple nucleotide changes that could be contributed to a single pseudo gene with an identical string of 9 nt or more were considered a single continuous GC event. Single nucleotide changes that could also be contributed to a pseudo gene were classified as ambiguous. Single nucleotide changes that did not have compatible pseudo gene donors were considered point mutations.
Western blot
Cell lysates were run on 12% polyacrylamide gels and transferred onto nitro-cellulose membranes. Membranes were blocked overnight in 5% milk-TBS-T, washed 1× with TBS-T, then incubated with mouse anti-AID antibody (Cell Signalling) diluted 1:1000 in 5% milk-TBS-T overnight. Membranes were then washed 3× with TBS-T, and incubated for 1 h with goat anti-mouse IgG HRP (Southern Biotech) diluted 1:5000 in 5% milk TBS-T. This was followed by washing 7× with TBS-T and visualization with ECL plus (Amersham) and a Versadoc imaging system (Biorad).
RESULTS
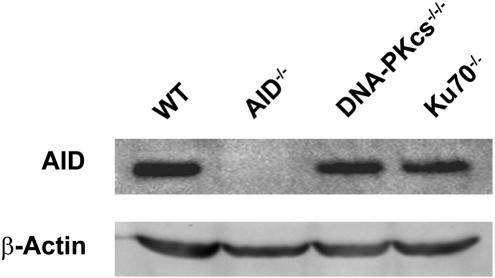
To determine whether or not DSBs are involved in the initiation of GC events, we utilized DT40 cells deficient in key factors involved in NHEJ (i.e. Ku70−/− and DNA–PKcs−/−/−). The DT40 cell line is derived from a chicken B-cell lymphoma and serves as a useful tool in that it constitutively undergoes rounds of GC in culture. Western blot analyses for AID in the WT, DNA–PKcs−/−/− and Ku70−/−,using an AID−/− cell line as a negative control, showed that the AID levels were similar in the three cell lines (Figure 1). This indicates that all DT40 clones should have similar levels of cytidine deamination, and thus similar levels of AID-induced DNA damage.
Figure 1.
Western blot analysis for AID. WT, AID−/−, DNA–PKcs−/−/− and Ku70−/− DT40 cells were analyzed by western blot for AID expression levels and β-Actin, as a loading control. Cell lysates were generated from founder populations of cells used for subsequent subcloning experiments.
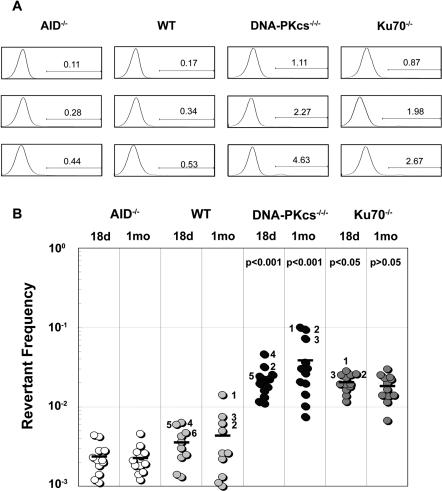
The four DT40 cell lines used in these experiments contain the same frameshift mutation in CDR1 of the V-region rendering them sIgM−. GC events can restore the reading frame of the V-region, making cells sIgM+. By subcloning a starting population that is sIgM−, the frequency of GC events can be estimated by assaying for the appearance of sIgM+ revertants. To overcome the stochastic nature of GC, at least 10 subclones were analyzed for each cell line to obtain average reversion frequencies. Cells were then analyzed by flow cytometry for sIgM expression.
AID−/−, WT, Ku70−/−, and DNA–PKcs−/−/− DT40 cells were subcloned, expanded in culture and analyzed at the 18-day and 1-month time points. At the 18-day time point, Ku70−/− and DNA–PKcs−/−/− DT40 subclones had ∼5-fold higher average reversion frequencies than that of WT DT40 cells (i.e. 6.2- and 5.3-fold for DNA–PKcs−/−/− and Ku70−/− cells, respectively) (Figure 2). Subclones were also analyzed at the 1 month time point to increase the sensitivity of the assay. At this time point, the average reversion frequency was ∼10 and ∼6 times higher in DNA–PKcs−/−/−and Ku70−/− DT40 cells, respectively, when compared to WT DT40 cells (Figure 2). Not surprisingly, the absence of AID resulted in the abrogation of reversion, as even though 18-day cultures appeared to have some sIgM+ cells, this proportion did not increase over time indicating the absence of ongoing GC events (Figure 2).
Figure 2.
IgM Reversion of sIgM− NHEJ deficient DT40 cells. (A) Representative histograms of IgM surface expression on subclones expanded for 18 days in culture. AID−/−, WT, DNA–PKcs−/−/− and Ku70−/− DT40 cell lines containing a frameshift in the Vλ-region were subcloned and expanded in culture. Cultures were stained with FITC-conjugated anti-chicken IgM and analyzed by flow cytometry. Gating for positive revertants was set at 8-times above the negative peak. Numbers indicate the percentage of IgM positive cells in each histogram. Three histograms are shown for each cell line that represent cultures that were low, medium and high for IgM+ cells. (B) Fluctuation analysis of IgM reversion for AID−/−, WT, DNA–PKcs−/−/− and Ku70−/− DT40 cell lines. Multiple subclones from each cell line were expanded and analyzed for IgM expression at 18 and 30 days. Average reversion frequency is denoted by a horizontal bar. Statistics are shown (see Materials and Methods) comparing the NHEJ-deficient cells to WT DT40 cells for the respective time points. Numbers beside symbols indicate clone number and correlate to sequence data shown in Figure 3, Table 1, and Supplementary Figures 1 and 2.
To validate the frequencies obtained via reversion analysis, unsorted WT and DNA–PKcs−/−/− DT40 subclones that were carried in culture for 1 month were sequenced at the Vλ light chain gene. To maximize the likelihood of finding GC events by sequencing, we used three subclones from each cell line that displayed the highest reversion frequencies (Figure 2). Sequences obtained are summarized in Figure 3A and raw sequence data is shown in Supplemental Figure 1. In the three wild type subclones analyzed, only one subclone carried a GC event (i.e. WT DT40 clone 2; Table 1, Figure 3). The three WT DT40 clones together gave an average GC frequency of ∼1.2%. In contrast, of the 3 DNA–PKcs−/−/− clones analyzed, 2 carried GC events. DNA–PKcs−/−/− clone 1 carried 2 unique GC events out of 29 sequences (∼6.9%) and clone 3 carried 1 unique GC out of 49 (∼2.0%) giving an average for the three clones of ∼3.0% by sequencing (Table 1, Figures 3A and Supplemental Figure 1). Interestingly, the frequency of point mutations was similar in WT and DNA–PKcs−/−/− DT40 cells (Table 1) suggesting that AID was accessing the V-region equally in each DT40 clone.
Figure 3.
Summary of gene conversion sequence data. (A) Vλ regions from WT and DNA–PKcs−/−/− DT40 subclones that were maintained in cell culture for 1 month. The DT40 clones analyzed for sequencing correspond to those shown in Figure 2B. Each line represents identical sequences of the Vλ region. Numbers to the right of each line indicates the number of times that sequence was observed. Symbols on and above the line indicate deviations from the founder sequence. Black circles: point mutations; white circles: ambiguous mutations; white bars: deletions; black bars: minimum GC tracts, grey bars: maximum GC tracts. Numbers above each tract indicate pseudogene donor, slashes between numbers indicate multiple possible donors. At the bottom is the scale in nucleotides, with the CDRs shown as labeled bars above the scale. 0 = the first nucleotide of the second exon. (B) Same as A, except sequences were obtained from IgM+ sorted WT, DNA–PKcs−/−/− and Ku70−/− cells.
Table 1.
GC frequencies in DT40 cells determined by sequencing analysis.
| Cell type | Clonea | GC eventsb | Point mutationsb | Deletionsb | Ambiguousb | Clones sequenced | GC frequency | SHM frequencyc |
|---|---|---|---|---|---|---|---|---|
| WT | 1 | 0 | 1 | 0 | 2 | 33 | 0.0d | 6.7 |
| 2 | 1 | 2 | 0 | 1 | 26 | 3.9d | 17.1 | |
| 3 | 0 | 1 | 0 | 2 | 26 | 0.0d | 8.5 | |
| DNA–PKcs−/−/ − | 1 | 2 | 2 | 0 | 0 | 29 | 6.9d | 15.3 |
| 2 | 0 | 2 | 1 | 1 | 30 | 0.0d | 14.8 | |
| 3 | 1 | 0 | 0 | 1 | 49 | 2.0d | <4.5 | |
| WT sIgM+f | 4 | 4 | 1 | 0 | 2 | 15 | 93.3/97.4e | 14.8 |
| 5 | 5 | 0 | 0 | 3 | 14 | 92.9/96.4e | <15.9 | |
| 6 | 6 | 0 | 1 | 1 | 12 | 91.7/95e | <18.5 | |
| DNA–PKcs−/−/− sIgM+f | 2 | 1 | 1 | 1 | 0 | 10 | 20/79.1e | 22.2 |
| 4 | 1 | 0 | 1 | 1 | 18 | 72.2/79.8e | <12.3 | |
| 5 | 3 | 1 | 0 | 1 | 18 | 100/98.4e | 12.3 | |
| Ku70−/− sIgM+f | 1 | 2 | 1 | 0 | 1 | 19 | 88.2/98.3e | 11.7 |
| 2 | 2 | 0 | 0 | 1 | 19 | 100/99.1e | <13.1 | |
| 3 | 7 | 0 | 0 | 1 | 17 | 100/95.3e | <11.7 |
aDT40 clones analyzed for sequencing correspond to those shown in Figure 2B. Sequence data shown in Supplementary Figure 1 and illustration of this data in Figure 3, together with tract lengths and Pseudo gene usage.
bOnly unique GC events, point mutations, deletions, and ambiguous sequences are shown.
cSHM frequencies (× 10−5) were determined by dividing point mutations by nucleotides sequenced (i.e. clones sequenced × 450 nt/V region).
dPercent GC frequencies were determined by dividing unique GC events by all sequences analyzed in unsorted WT and DNA–PKcs−/−/− cells.
eFrequency of sIgM+ revertants as determined by sequencing (left side of column) or flow cytometry (right side of column).
Although the sequencing analysis showed an increase in GC frequencies in NHEJ-deficient versus WT DT40 cells, the relatively few GC-positive sequences observed in the two cell lines were insufficient to draw any conclusions. This prompted us to further validate the fluctuation analysis by verifying that the frequency data we had obtained from the sIgM reversion experiments was indeed representative of GC events. Although Ig reversion in WT DT40 cells is known to proceed predominantly through GC (25), it is conceivable that sIgM reversion in NHEJ-deficient DT40 cells could proceed via non-GC events, such as compensatory insertions/deletions. By sorting for these positive events, and sequencing the Vλ region, we were able to directly observe the mechanism by which the frameshift had been repaired in these sIgM+ cells. To this end, we sorted 18-day cultures of WT, DNA–PKcs−/−/− and Ku70−/− DT40 subclones to enrich for sIgM+ cells. Cultures that were at least ∼80% sIgM+ by post-sort flow cytometry were sequenced at the Vλ gene. Sequencing analysis of sorted WT, DNA–PKcs−/−/−and Ku70−/− DT40 clones revealed that nearly every sequence obtained had undergone a GC event that resulted in restoration of the reading frame (Figure 3B, raw sequence data shown in Supplemental Figure 2). For WT clones, post sort analysis revealed >94% purity, and practically all sequences obtained revealed restorative GC events (Table 1, Figure 3B). Similarly, Ku70−/− DT40 clones 1–3, post-sort analysis showed sIgM reversion was >95% and again, similar results were obtained by sequencing (Table 1, Figures 3B and Supplememtary Figure S2). For DNA–PKcs−/−/− clones 4 and 5, 80 and 98% of the cells were sIgM+, respectively, and had restorative GC events at a frequency of 72 and 100% (Table 1, Figures 3B and Supplementary Figure S2). Although DNA–PKcs−/−/− clone 2 was ∼80% sIgM+ by flow cytometry, only 20% of the sequences had restorative GC events, while 40% showed an untemplated 4 nt deletion in the CDR1 (Figures 3B and Supplemental Figure 2). The unsorted founder DT40 clone 2 also contained the same deletion (Figures 3A and Supplemental Figure 1). Thus, with the exception of one clone, sequencing analysis showed that the majority of sIgM reversion in WT, DNA–PKcs−/−/− and Ku70−/− DT40 clones occurred through restorative GC events. These findings validate the frequency data obtained via sIgM reversion, and indicate that NHEJ deficient cell lines repair frameshift mutations in the Vλ region primarily through GC, doing so at a higher frequency than WT cells.
DISCUSSION
In the whirlwind of activity that has ensued since the discovery of AID, the mechanistic details behind secondary antibody diversification have been quickly coming to light. It has been proposed that the action of AID can lead to the generation of a number of DNA lesions. Among them are abasic sites, SSBs and DSBs. DSBs have been observed in hypermutating V-regions (26,27), although the frequency of these types of DNA breaks appears to be significantly lower than SSBs (28). Work by Maria Jasin and co-workers has shown that GC of non-Ig genes is initiated by DSBs (29). However, it is possible that GC at Ig genes could be induced by any of the DNA lesions produced by AID so long as a free 3′ end is generated that can be used to initiate homology search and prime template strand synthesis.
In this study we took advantage of the competition between HR and NHEJ for DSBs and examined the impact of NHEJ-deficiency on GC. Our results indicate that GC is increased in the absence of NHEJ providing evidence for the stimulation of GC by AID-induced DSBs. While this finding does not rule out the possibility that other DNA lesions can initiate GC, the significant increase of GC events seen in NHEJ-deficient cells (5- to 10-fold) indicates that NHEJ-accessible DSBs likely initiate the majority of events. Based on this, one would expect in WT DT40 cells that these AID-induced DSBs are being resolved either by HR or NHEJ. In this case, resolution by HR could lead either to normal repair or GC events, while resolution by NHEJ would result predominantly in precise rejoining via the complementary ends of a staggered break or, less frequently, rejoining with associated insertion/deletion.
Interestingly, the number of unique GC events observed via sequencing was higher in sorted IgM+ WT DT40 than most of the sorted IgM+ NHEJ mutants (Figure 3B), a result that seemingly contradicts the data obtained from the reversion analysis. It is important to note that the sorting step introduces a selective event thus making frequency determination unreliable. Nevertheless, this phenomenon can be explained by the random nature of GC initiation. The mutant subclones we sequenced likely underwent a reversion event early in their outgrowth. This allowed that event to dominate the culture during expansion of the subclone, effectively masking other unique GCs. The differences in the number of unique GCs between IgM+ WT and NHEJ-deficient DT40 clones is likely due to chance and if the remaining subclones analyzed in Figure 2 were sorted and sequenced, some WT and NHEJ-deficient DT40 clones would likely contain sequences with large numbers of unique GCs, while other subclones would likely contain sequences dominated by 1 or 2 revertants. This argument is supported by the fact that sequencing of unsorted populations revealed more GC events in DNA–PKcs−/−/− than WT DT40. In addition, since the founder sequence shares the most homology with ψV8, following initial repair, silent GC events could be occurring at a higher frequency in the NHEJ mutants than the WT but would be undetectable via sequencing since the donor and template are identical.
The initiating DSB can be further classified as either one-ended or two-ended. One-ended DSBs could be generated by replication across AID induced SSBs while two-ended DSBs could be generated through the proximal formation of two SSBs on opposite DNA strands. We believe that two-ended DSBs are the more likely of the two as an initiator of GC events. While it is clear that both NHEJ and HR can resolve two-ended DSBs, one-ended DSBs can only be resolved correctly through HR. Association of NHEJ factors with one-ended DSBs would presumably result in an unresolvable complex which would persist until dissociation of the NHEJ factors followed by normal HR. HR would therefore not be expected to increase when NHEJ is removed. In fact, recent findings by Hochegger et al. (30) indicate that NHEJ is suppressed by Parp1, thus favoring HR whenever possible. In support of this idea, the V-region of DT40 cells is rich with WRCY/RGYW hotspot motifs clustered on both strands near the CDRs, many of them in close proximity possibly serving as substrates for AID induced two-ended DSB formation (E. Tang and A. Martin, unpublished observation). Complicating this interpretation however, is the fact that cells deficient for NHEJ are resistant to SSB inducing agents such as camptothecin, a topoisomerase-1 inhibitor, indicating that NHEJ is inhibiting HR repair of single ended DSBs (31). It is possible that this effect is most relevant when numerous one-ended DSBs are present as with the use of DNA-damaging agents, while the lower relative frequency of breaks generated by AID activity might reduce the impact of NHEJ on one-ended DSB metabolism. Nevertheless, as the exact role of NHEJ at one-ended DSBs is unclear, our results cannot rule out the possibility that they contribute to the initiation of GC.
NHEJ-deficient DT40 cells were previously analyzed for GC at the V-region and were not found to be significantly different from WT cells (24). This discrepancy might be attributed to the method of analysis. While we measured sIgM gain, a previous group measured for sIgM loss (24). Measuring for sIgM gain might be a more appropriate readout for GC for two reasons. First, sIgM loss could potentially occur through multiple mechanisms while sIgM gain can happen only through repair of the frame-shift itself. Precedent for this idea exists in a previous study (9) where sorting for a population of sIgM-loss variants from sIgM+ AID−/− DT40 cells did not yield enrichment of sIgM− cells, indicating that a small fraction of cells had transiently undergone loss of sIgM expression. This phenomenon could obscure GC frequency differences when measuring by sIgM-loss. Second, Ig GC in DT40 cells results almost exclusively in productive conversions, indicating that GC events rarely generate sIgM loss. In light of this, it seems reasonable to conclude that assaying via sIgM gain allowed us to observe differences that were undetectable by sIgM loss. It should also be noted that the role of NHEJ on GC was not the primary focus of this paper (24).
While NHEJ is known to function throughout the cell cycle, HR is preferentially used in late S and G2 (32). This suggests three possibilities. One is that NHEJ is directly competing with HR for DSBs being formed during late S and G2. The second possibility is that DSBs generated in G0 and G1 are persisting into S and G2, where the GC machinery can now act to repair the lesion. However, the cell cycle requirement of HR is thought to reflect the necessity for the presence of a sister chromatid for templated repair. In the case of pseudo-genes, which exist throughout cell cycle, the need for a second DNA copy may be abrogated. Thus, a third possibility is that GC and NHEJ are competing for DSBs at all stages of the cell cycle. It will be interesting to see what the requirements are in terms of genome copy number, replication and cell cycle for the induction of GC events and HR in general.
Our work has revealed the involvement of the NHEJ pathway in the processing of AID-induced lesions within the V-region. The imprecise nature of NHEJ could potentially contribute to the process of SHM, or alternately be a hindrance to it through the introduction of frameshift mutations. Consistent with this idea is the involvement of the NHEJ pathway in resolving the DSBs that are induced in CSR. CSR is thought to proceed via a two-ended DSB intermediate, initiated by AID deaminating cytidines on opposite strands (33–35). Though there is some controversy in the field (36), a mounting body of evidence suggests that in CSR, NHEJ plays a key role. Removal of NHEJ results in severely attenuated levels of CSR (37) and an altered spectrum of switching events. This may represent an insight into an alternate pathway of AID induced mutation. In addition to the current model of AID-induced point mutation, AID induced DSBs in conjunction with NHEJ could result in the formation of frameshift mutations. This alternate pathway may however be more of a hindrance to SHM than a boon, as frameshift mutations would result in truncated gene-products. This idea is corroborated by evidence of low levels of frameshift mutation in actively hypermutating cell lines (38). Thus, the generation of frameshift mutations through the induction of the NHEJ pathway may represent a novel aspect of SHM. Of further interest is the implicit dependence on DNA–PKcs for the processing of the AID-induced DSBs. What this means though is currently unclear as although it is becoming apparent that NHEJ has at least two pathways, one that requires DNA–PKcs and one that does not (39), the differences between these two pathways remain nebulous at best.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We are grateful to Drs Marc Shulman, Mark Baker, and Michael Ratcliffe for helpful discussions and for reading the manuscript. We are also thankful to Dr Michael Ratcliffe for the WT DT40, Dr Shunichi Takeda for the DNA–PKcs−/−/−and Ku70−/− DT40 cells and Dr Reuben Harris for the AID−/− DT40 cells. This research is supported by a grant from the Canadian Institute of Health Research (MOP66965) to A.M. who is supported by a Canada Research Chair award. E.T. is supported by an Ontario Graduate Scholarship award. Funding to pay the Open Access publication charges for this article was provided by a grant from the Canadian Institute of Health Research (MOP66965).
Conflict of interest statement. None declared.
REFERENCES
- 1.Ratcliffe M.J. Antibodies, immunoglobulin genes and the bursa of Fabricius in chicken B cell development. Dev. Comp. Immunol. 2006;30:101–118. doi: 10.1016/j.dci.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 2.Knight K.L., Winstead C.R. Generation of antibody diversity in rabbits. Curr. Opin. Immunol. 1997;9:228–232. doi: 10.1016/s0952-7915(97)80140-9. [DOI] [PubMed] [Google Scholar]
- 3.Reynaud C.A., Anquez V., Grimal H., Weill J.C. A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell. 1987;48:379–388. doi: 10.1016/0092-8674(87)90189-9. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu M., Sankaranand V.S., Anant S., Sugai M., Kinoshita K., Davidson N.O., Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 5.Martin A., Scharff M.D. AID and mismatch repair in antibody diversification. Nature Rev. Immunol. 2002;2:605–614. doi: 10.1038/nri858. [DOI] [PubMed] [Google Scholar]
- 6.Nussenzweig M.C., Alt F.W. Antibody diversity: one enzyme to rule them all. Nature Med. 2004;10:1304–1305. doi: 10.1038/nm1204-1304. [DOI] [PubMed] [Google Scholar]
- 7.Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 8.Arakawa H., Hauschild J., Buerstedde J.M. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 2002;295:1301–1306. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 9.Harris R.S., Sale J.E., Petersen-Mahrt S.K., Neuberger M.S. AID Is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr. Biol. 2002;12:435–438. doi: 10.1016/s0960-9822(02)00717-0. [DOI] [PubMed] [Google Scholar]
- 10.Martin A., Bardwell P.D., Woo C.J., Fan M., Shulman M.J., Scharff M.D. Activation-induced cytidine deaminase turns on somatic hypermutation in hybridomas. Nature. 2002;415:802–806. doi: 10.1038/nature714. [DOI] [PubMed] [Google Scholar]
- 11.Petersen-Mahrt S.K., Harris R.S., Neuberger M.S. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–104. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 12.Bransteitter R., Pham P., Scharff M.D., Goodman M.F. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl Acad. Sci. USA. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rada C., Williams G.T., Nilsen H., Barnes D.E., Lindahl T., Neuberger M.S. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 14.Kenter A.L. Class-switch recombination: after the dawn of AID. Curr. Opin. Immunol. 2003;15:190–198. doi: 10.1016/s0952-7915(03)00018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaudhuri J., Alt F.W. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nature Rev. Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 16.Saribasak H., Saribasak N.N., Ipek F.M., Ellwart J.W., Arakawa H., Buerstedde J.M. Uracil DNA glycosylase disruption blocks Ig gene conversion and induces transition mutations. J. Immunol. 2006;176:365–371. doi: 10.4049/jimmunol.176.1.365. [DOI] [PubMed] [Google Scholar]
- 17.Sale J.E. Immunoglobulin diversification in DT40: a model for vertebrate DNA damage tolerance. DNA. Repair. (Amst). 2004;3:693–702. doi: 10.1016/j.dnarep.2004.03.042. [DOI] [PubMed] [Google Scholar]
- 18.Richardson C., Jasin M. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol. Cell. Biol. 2000;20:9068–9075. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammarsten O., Chu G. DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc. Natl Acad. Sci. USA. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishino Y., Nishino T., Morikawa K. Mechanisms of maintaining genetic stability by homologous recombination. Chem. Rev. 2006;106:324–339. doi: 10.1021/cr0404803. [DOI] [PubMed] [Google Scholar]
- 21.Pierce A.J., Hu P., Han M., Ellis N., Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes. Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takata M., Sasaki M.S., Sonoda E., Morrison C., Hashimoto M., Utsumi H., Yamaguchi-Iwai Y., Shinohara A., Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takata M., Sasaki M.S., Tachiiri S., Fukushima T., Sonoda E., Schild D., Thompson L.H., Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sale J.E., Calandrini D.M., Takata M., Takeda S., Neuberger M.S. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 2001;412:921–926. doi: 10.1038/35091100. [DOI] [PubMed] [Google Scholar]
- 25.Buerstedde J.M., Reynaud C.A., Humphries E.H., Olson W., Ewert D.L., Weill J.C. Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J. 1990;9:921–927. doi: 10.1002/j.1460-2075.1990.tb08190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papavasiliou F.N., Schatz D.G. The activation-induced deaminase functions in a postcleavage step of the somatic hypermutation process. J. Exp. Med. 2002;195:1193–1198. doi: 10.1084/jem.20011858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zan H., Wu X., Komori A., Holloman W.K., Casali P. AID-dependent generation of resected double-strand DNA breaks and recruitment of Rad52/Rad51 in somatic hypermutation. Immunity. 2003;18:727–738. doi: 10.1016/s1074-7613(03)00151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong Q., Maizels N. DNA breaks in hypermutating immunoglobulin genes: evidence for a break and repair pathway of somatic mutation. Genetics. 2001;158:369–378. doi: 10.1093/genetics/158.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouet P., Smih F., Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl Acad. Sci. USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hochegger H., Dejsuphong D., Fukushima T., Morrison C., Sonoda E., Schreiber V., Zhao G.Y., Saberi A., Masutani M., Adachi N., et al. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 2006;25:1305–1314. doi: 10.1038/sj.emboj.7601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonoda E., Hochegger H., Saberi A., Taniguchi Y., Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 32.Rothkamm K., Kruger I., Thompson L.H., Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X., Kinoshita K., Honjo T. Variable deletion and duplication at recombination junction ends: implication for staggered double-strand cleavage in class-switch recombination. Proc. Natl Acad. Sci. USA. 2001;98:13860–13865. doi: 10.1073/pnas.241524898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rush J.S., Fugmann S.D., Schatz D.G. Staggered AID-dependent DNA double strand breaks are the predominant DNA lesions targeted to S mu in Ig class switch recombination. Int. Immunol. 2004;16:549–557. doi: 10.1093/intimm/dxh057. [DOI] [PubMed] [Google Scholar]
- 35.Petersen S., Casellas R., Reina-San-Martin B., Chen H.T., Difilippantonio M.J., Wilson P.C., Hanitsch L., Celeste A., Muramatsu M., Pilch D.R., et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bosma G.C., Jiyoon K., Urich T., Fath D.M., Cotticelli M.G., Ruetsch N.R., Radic M.Z., Bosma M.J. DNA-dependent protein kinase activity is not required for immunoglobulin class switching. J. Exp. Med. 2002;196:1483–1495. doi: 10.1084/jem.20001871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manis J.P., Gu Y., Lansford R., Sonoda E., Ferrini R., Davidson L., Rajewsky K., Alt F.W. Ku70 is required for late B cell development and immunoglobulin heavy chain class switching. J. Exp. Med. 1998;187:2081–2089. doi: 10.1084/jem.187.12.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W., Bardwell P.D., Woo C.J., Poltoratsky V., Scharff M.D., Martin A. Clonal instability of V region hypermutation in the Ramos Burkitt's lymphoma cell line. Int. Immunol. 2001;13:1175–1184. doi: 10.1093/intimm/13.9.1175. [DOI] [PubMed] [Google Scholar]
- 39.Adachi N., Iiizumi S., So S., Koyama H. Genetic evidence for involvement of two distinct nonhomologous end-joining pathways in repair of topoisomerase II-mediated DNA damage. Biochem. Biophys. Res. Commun. 2004;318:856–861. doi: 10.1016/j.bbrc.2004.04.099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.