Abstract
Beyond the well-known role of proteolytic machinery in protein degradation and turnover, many specialized proteases play a key role in various regulatory processes. Thousands of highly specific proteolytic events are associated with normal and pathological conditions, including bacterial and viral infections. However, the information about individual proteolytic events is dispersed over multiple publications and is not easily available for large-scale analysis. CutDB is one of the first systematic efforts to build an easily accessible collection of documented proteolytic events for natural proteins in vivo or in vitro. A CutDB entry is defined by a unique combination of these three attributes: protease, protein substrate and cleavage site. Currently, CutDB integrates 3070 proteolytic events for 470 different proteases captured from public archives (such as MEROPS and HPRD) and publications. CutDB supports various types of data searches and displays, including clickable network diagrams. Most importantly, CutDB is a community annotation resource based on a Wikipedia approach, providing a convenient user interface to input new data online. A recent contribution of 568 proteolytic events by several experts in the field of matrix metallopeptidases suggests that this approach will significantly accelerate the development of CutDB content. CutDB is publicly available at http://cutdb.burnham.org.
INTRODUCTION
Proteases degrade substrate proteins by cleaving peptide bonds. Many proteases are highly specific and cleave substrates only at specific sequence motifs. These proteases are responsible not only for degrading proteins but also for their activation/inactivation. Such proteolytic events (PEs) form highly organized and regulated networks. However, a comprehensive overview of proteolytic pathways has not yet been elucidated.
PEs are involved in multiple aspects of regulation in eukaryotic cells and play a major role in many natural processes, as well as in many diseases, including cancer, autoimmune diseases and bacterial and viral infections (1–3), and are subjects of intensive research. However, at present, the information about specific PEs is dispersed among original articles and is not organized in a systematic manner, such as information ‘metabolic pathways’ (4).
Several databases [e.g. MEROPS (5), HPRD (Human Protein Reference Database) (6) and UniProt (7)] contain some information about PEs; however, they are not a major focus of any of them. MEROPS is a database for classifying proteases and identifying proteases with tools, such as BLAST, synonymous names search, protease inhibitor information, comparative genome analysis tools and so on. It contains information on some PEs, usually in text form. MEROPS classification is a ‘gold standard’ in the protease world, but extracting information about specific PEs requires reading the text entries. HPRD and UniProt have a limited number of records of PEs in their entries. However, none of them is comprehensive enough to be used as a reference. Also, none of them is designed to easily accept annotations from a user community.
At present, most biological databases allow users to contribute to them only by using e-mail or feedback forms. Only recently some databases introduced a model of community annotation, with interfaces for users to directly edit their content [SEED (8) and VMD (9)]. This introduced a new paradigm in distributed annotations that matches the distribution of knowledge and expertise in the broad user community.
CutDB is a newly created community annotation database of PEs that ultimately aims to reconstruct all proteolytic pathways in their broad biological context. It was designed to store PEs reported in original, experimental articles and provide the PE data in a form accessible for large-scale bioinformatics analyses, but also for individual searches by experimental researchers. The content in CutDB is open to the public, and it can be edited both in structured fields and in the comments section, where users can express their opinions by using free text. Thus, this database has the potential to offer not only a comprehensive overview of proteolytic pathways but also the information that cannot be covered in the framework of the database, such as hypotheses, discussions of discrepancies in experimental results, negative results of experiments and so on.
The knowledge and information collected in CutDB will allow us to improve our understanding of proteases and the role of proteolysis in many important biological processes. CutDB is one of the first systematic efforts to collect information about PEs across all species, including humans. CutDB is a part of PMAP (http://pmap.burnham.org), an interactive in silico environment for interrogating data on cellular proteolytic pathways which is being developed at the NIH Roadmap Center for Proteolytic Pathways (see http://cpp.burnham.org).
DATA CONTENTS
CutDB contains three primary sections (Figure 1c): PE, biological context and comments. A single PE record consists of three basic elements: a protease, a substrate (protein) and a unique cut site (cleavage site). The protease name and its classification follow the MEROPS rules. The protease data consist of a MEROPS protease definition, sequence code, peptidase code and organism name. The substrate data are mainly based on NCBI RefSeq data (10) and consist of the NCBI GI sequence number, RefSeq definition, amino acid sequence, organism name, cut site and size of products. The cut site data consist of the position number in the sequence and the eight amino acid residues around the cut site. The biological context consist of biological consequences, pathway, cellular localization, tissue specificity, cell line used in an experiment, disease, method of determination and PubMed ID (Table 1). CutDB allows redundant records in the case of multiple substrate names or substrate isoforms.
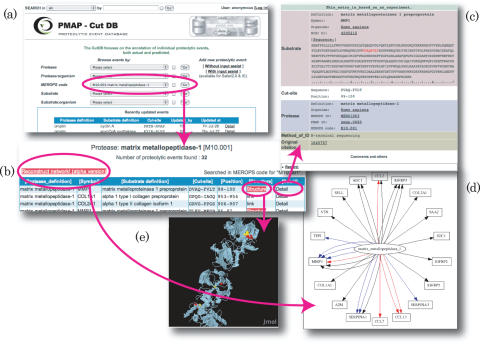
Figure 1.
(a) Front page of CutDB. (b) Search result using ‘M10.001 (matrix metallopeptidase-1)’ of MEROPS peptidase code. (c) Record describing ‘M10.001’ cleaves matrix metallopeptidase-1 preproprotein. (d) Network diagram centered around ‘M10.001’. (e) Structures of matrix metallopeptidase-1 in PDB with highlighted cut sites.
Table 1.
Objects and their attributes in the CutDB record
| Objects | Attributes |
|---|---|
| Protease | Name, organism, IDs |
| Substrate | Name, organism, IDs, sequence, cut site, size of products |
| Biological context | Consequences (e.g. activation/inactivation), pathway, tissue, cell line, cellular localization, disease |
| Metadata | Confirmed in experiments or predicted, methods of identification, original citation, curator, curated date, comments |
All other information that is not covered within designated structured fields can be put in the comment section as free text. The comment section is divided into several categories, such as ‘discussion’, ‘hypothesis’, ‘drugs in development’ and ‘other comment’. The user can edit all this information, including the PE and biological context sections. The user can also delete a specific entry.
CURRENT STATUS
Currently, the total number of PEs in CutDB is 3070 (Table 2). We specifically focused on collecting information about matrix metallopeptidase PEs from the articles reporting original experiments (Table 3).
Table 2.
Current number of proteolytic event records in CutDB
| Protease family | Number of records |
|---|---|
| Aspartic | 339 |
| Cysteine | 592 |
| Glutamic | 45 |
| Metallo | 1218 |
| Serine | 880 |
| Threonine | 6 |
Table 3.
Number of proteolytic events in major metallopeptidases
| Protease name | Peptidase code by MEROPS | Number of records |
|---|---|---|
| MMP-1 | M10.001 | 41 |
| MMP-8 | M10.002 | 16 |
| MMP-2 | M10.003 | 54 |
| MMP-9 | M10.004 | 177 |
| MMP-3 | M10.005 | 118 |
| MMP-7 | M10.008 | 75 |
| MMP-13 | M10.013 | 59 |
| MT1-MMP | M10.014 | 90 |
| MMP-20 | M10.019 | 18 |
| MMP-26 | M10.029 | 12 |
HYPERLINKS
All protease and MEROPS peptidase codes are linked to the MEROPS database. All substrates are linked to the NCBI site. Links to UniProt, SEED and PubMed data are also provided in the PE records.
WEB INTERFACE
Users can access each PE record from the front page by using the pull-down menu or the text search (Figure 1a). The pull-down menu directly displays the content of the field in CutDB. The text search provides not only the fields prepared in the pull-down menu but also the following additional fields: cut site, creator name, last editor name and PubMed ID. Both searches from the pull-down menu and the text search return the set of PE records, which contain the query keywords in each record (Figure 1b). On the list page, some records have links to their structure that show the highlighted cut sites (Figure 1e). More detailed information can be displayed by clicking ‘Detail’ on the right-hand side of each record.
NETWORK DIAGRAM
On the list page, users can generate a clickable network diagram (Figure 1d) based on the listed PE records. At present, only human PEs can be converted into the network diagram. In the network diagram, the nodes correspond to proteases and substrates. The proteases are shown as ellipses and the substrates are shown as rectangles. The substrate names are converted into HUGO's gene symbols (11), which are stored in NCBI RefSeq records. By using the gene symbols, the network diagrams can be simplified by shortening the substrate definitions and converting the multiple substrate isoforms into one substrate symbol. The edges correspond to the relation between proteases and substrates. The colors of the edges show biological consequence: red for substrate activation and blue for substrate inactivation. Multiple edges are created in the case of multiple cut sites for the same substrate.
INPUT ASSISTANT
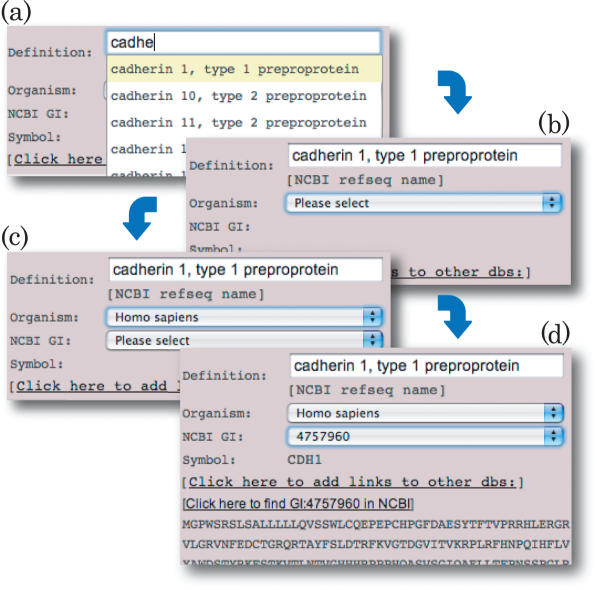
Input assistant is an option that allows the user to quickly add and edit the CutDB content. The user time for searching the substrate sequence and GI number is minimized (Figure 2). The information can be readily displayed without the need to search the NCBI site. The user can also alternatively enter the information manually into CutDB.
Figure 2.
(a) When the user inputs ‘cadhe’ into substrate definition, every human protein definition in RefSeq that contains ‘cadhe’ in the definition is immediately displayed in the pull-down menu. (b) When the user clicks one of the protein names, the server starts to search protein names and returns the chosen protein's organism name to the next item on the Web browser. (c) When the user selects ‘Homo sapiens’ from the organism names, the NCBI GI number that contains both the protein and organism names is displayed in the next pull-down menu. (d) The user selects one of the NCBI GI numbers; then, its amino acid sequence and gene symbol appear.
POSITION IDENTIFICATION TOOL
By using the position identification tool, the user can easily identify both the position numbers and the eight amino acid residues around cut sites. The user can access this interface by clicking ‘tool’ when adding or editing the record. The user has to copy and paste the sequence, put ‘∣’ in the cut site and click the ‘split’ button. As a result, the position numbers and amino acid residues of cut sites appear.
USER MANAGEMENT
Currently, the CutDB management system allows users to access the database without registration. User registration is necessary for adding and editing content. It is not necessary to register to see or retrieve content.
LITERATURE TRACK
Users can access Literature Track (LT) by clicking ‘Literature Track’ on the front page. LT is a literature database that stores two kinds of information: (i) information from the articles that are already curated and/or contain no cut site information or have only synthetic peptide information; and (ii) the set of candidate articles that will be read by curators in the near future. We intend to use LT to manage the articles and avoid re-reading identical articles by other curators. We will also use the data to build an automatic system to filter appropriate articles from PubMed. Only registered users can edit the content of LT.
DISCUSSION AND FUTURE DIRECTIONS
CutDB is a unique database that focuses on the relations between proteases and their substrates. Our ultimate goal is to collect a complete dataset on PEs in the cell and to reconstruct all the proteolytic pathways in a broad biological context. Currently, we have accumulated mainly PEs involving metallopeptidases in human cells.
The CutDB information can be expanded by the implementation of additional networks of molecular interactions, such as transcriptional regulation and protein–protein interactions. We plan to import and implement these data, most of which are publicly available, into CutDB in order to provide integrated proteolytic pathways.
In addition, much research is being undertaken to implement automatic annotation approaches. Although complete automatic annotation from literature is impossible as yet, it is helpful for the curated database. The automatic annotation system for CutDB will be based on coordination with the Literature Track. We are currently engaging in novel algorithmic/semiautomatic approaches to add content to CutDB based on literature text-mining methods. Still, most of the information is added by hand and current CutDB contributors are members of several laboratories actively working in the field of proteolysis and focusing on a limited set of human proteases. The content of CutDB is expanding rapidly; however, a much more extensive community effort will be required to achieve more comprehensive coverage of existing and rapidly growing knowledge about PEs in a variety of species.
IMPLEMENTATION
All frameworks for the Web interface are implemented using ‘Ruby on Rails’. The database in the background is MySQL. The Web server is Lighttpd. The network diagram is generated by Graphviz. The protein structure is displayed using Jmol. The information processing to transfer data into MySQL was performed using BioRuby.
Acknowledgments
We extend our thanks to Dr Nobuya Tanaka for introducing Ruby on Rails and his help in the early stages. We would also like to thank Prof. Iris Lindberg, Prof. Alex Bateman and Ms Olivia Haggis for their suggestions and sending us their proteolytic event data. Finally, we thank Dr Boris Ratnikov and many others at the Burnham Institute for Medical Research for curating the data. This research is funded by NIH grant number 5 U54 RR020843-03, ‘Center on Proteolytic Pathways’. Funding to pay the Open Access publication charges for this article was provided by NIH/Burnham Institute for Medical Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nagase H., Woessner J.F., Jr Matrix metalloproteinases. J. Biol. Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 2.Fuentes-Prior P., Salvesen G.S. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem. J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strongin A.Y. Mislocalization and unconventional functions of cellular MMPs in cancer. Cancer Metastasis Rev. 2006;25:87–98. doi: 10.1007/s10555-006-7892-y. [DOI] [PubMed] [Google Scholar]
- 4.Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K.F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawlings N.D., Tolle D.P., Barrett A.J. MEROPS: the peptidase database. Nucleic Acids Res. 2004;32:D160–D164. doi: 10.1093/nar/gkh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peri S., Navarro J.D., Amanchy R., Kristiansen T.Z., Jonnalagadda C.K., Surendranath V., Niranjan V., Muthusamy B., Gandhi T.K., Gronborg M., et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Apweiler R., Bairoch A., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Overbeek R., Begley T., Butler R.M., Choudhuri J.V., Chuang H.Y., Cohoon M., de Crecy-Lagard V., Diaz N., Disz T., Edwards R., et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tripathy S., Pandey V.N., Fang B., Salas F., Tyler B.M. VMD: a community annotation database for oomycetes and microbial genomes. Nucleic Acids Res. 2006;34:D379–D381. doi: 10.1093/nar/gkj042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eyre T.A., Ducluzeau F., Sneddon T.P., Povey S., Bruford E.A., Lush M.J. The HUGO Gene Nomenclature Database, 2006 updates. Nucleic Acids Res. 2006;34:D319–D321. doi: 10.1093/nar/gkj147. [DOI] [PMC free article] [PubMed] [Google Scholar]