Abstract
Oligonucleotide-mediated gene targeting is emerging as a powerful tool for the introduction of subtle gene modifications in mouse embryonic stem (ES) cells and the generation of mutant mice. However, its efficacy is strongly suppressed by DNA mismatch repair (MMR). Here we report a simple and rapid procedure for the generation of mouse mutants using transient down regulation of the central MMR protein MSH2 by RNA interference. We demonstrate that under this condition, unmodified single-stranded DNA oligonucleotides can be used to substitute single or several nucleotides. In particular, simultaneous substitution of four adjacent nucleotides was highly efficient, providing the opportunity to substitute virtually any given codon. We have used this method to create a codon substitution (N750F) in the Rb gene of mouse ES cells and show that the oligonucleotide-modified Rb allele can be transmitted through the germ line of mice.
INTRODUCTION
Publication of the human genome sequence has opened a whole new area of genetic research. In the last decade, many locus-specific mutation databases have been constructed and made publicly available. For instance, the IARC p53 database contains information on 21 587 somatic mutations found in the human p53 gene (http://www-p53.iarc.fr). Site-specific modification of the mouse genome provides a powerful tool to functionally characterize the identified mutations and distinguish between polymorphisms and truly pathogenic mutations. However, current strategies based on homologous recombination require the design and construction of a targeting vector and multiple rounds of selection and clonal purification of modified cells, which are laborious and time-consuming (1). An alternative approach could be the introduction of subtle gene modifications into the mouse genome by single-stranded DNA oligonucleotides (2).
Numerous reports have shown that oligonucleotides that differ from the target locus by one or a few nucleotides can be used to introduce specific mutations into both episomally and chromosomally located genes (3–6). In most cases, chemically modified RNA–DNA chimeric oligonucleotides or single-stranded DNA oligonucleotides were used in which the chemical modifications served to protect the oligonucleotides from nucleolytic degradation. The mechanism of transfer of genetic information from the oligonucleotide to the target remains largely elusive. Many different cellular processes such as transcription (7,8), DNA replication (9), homologous recombination (10) and DNA mismatch repair (MMR) (11,12) seem to be involved. Several reports have noticed that cells targeted by chemically modified oligonucleotides underwent a G2 arrest that precluded their clonal expansion (13,14).
In mouse embryonic stem (ES) cells, oligonucleotide-mediated gene targeting frequencies appeared to be relatively low. Chemically modified oligonucleotides showed targeting frequencies that were ∼50-fold lower in ES cells than in CHO-K1 cells (3,15). Overexpression of the Redβ protein from bacteriophage lambda seemed to increase the targeting efficiency in ES cells, although the effect was not quantified (16). To date, none of these approaches have been followed up by experiments to show modification of an endogenous gene in ES cells and transmission of a modified allele through the mouse germ line.
Previously, we demonstrated that the efficiency of gene targeting by non-chemically modified single-stranded DNA oligonucleotides in mouse ES cells is strongly suppressed by the MMR system (11). In eukaryotic cells, DNA mismatches are recognized by MSH2 and its binding partners MSH3 and MSH6 (17). The MSH2/MSH6 heterodimer mainly recognizes single base substitutions and small loops of 1 or 2 nt, while the MSH2/MSH3 complex has more affinity for larger loops of unpaired bases (18,19). We showed that oligonucleotide-directed substitution, insertion or deletion of a few nucleotides was only effective in cells lacking the central MMR gene Msh2. However, the mutator phenotype associated with MMR deficiency (20) may lead to inadvertent genetic alterations on top of the oligonucleotide-mediated modification, hampering general application of oligonucleotide-mediated targeting.
Here we show that transient suppression of MSH2 by RNA interference allows effective oligonucleotide-mediated gene modification in wild-type mouse ES cells. Our method is generally applicable and provides the opportunity to modify in principle any given codon in the ES cell genome. The occurrence of frameshift mutations at simple sequence repeats was assessed to gain insight into the level of spontaneous mutagenesis resulting from transient MSH2 suppression. We have successfully applied the MSH2 knockdown strategy to substitute a codon in the Rb gene (N750F) of MMR-proficient ES cells and used these cells to generate the first mutant mouse line that was created via oligonucleotide-mediated gene targeting.
METHODS
Cell culture and transfection
We developed two selectable targeting reporter systems consisting of mutant neomycin genes (Figures 1a and 2a) (11). A single copy of these mutant reporter genes was stably integrated into the Rosa26 locus of Msh2−/−, Msh2+/−, Msh3−/−, Msh6−/− and wild-type ES cell lines as described previously (11,20,21). ES cells (E14) were originally derived from 129Ola and cultured in Glasgow minimal essential medium (GMEM) supplemented with 10% fetal calf serum, sodium pyruvate, non-essential amino acids, 1 mM 2-mercaptoethanol and 1000 U/ml of leukemia inhibitory factor.
Figure 1.
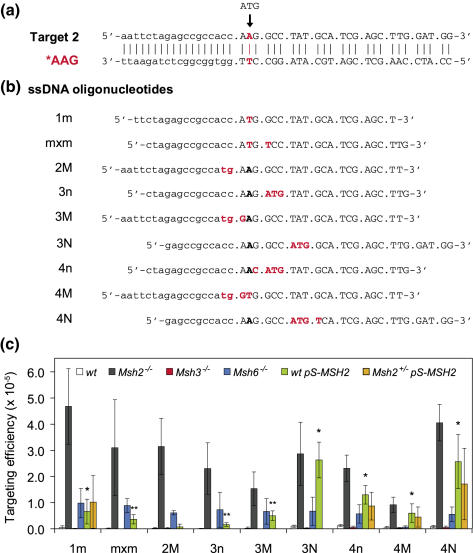
Oligonucleotide-mediated base substitution. (a) A single copy of a defective neomycin gene (neo) carrying an T to A point mutation in the start codon (Target 2) was inserted into the Rosa26 locus of Msh2−/−, Msh3−/−, Msh6−/−, Msh2+/− and wild-type ES cells. Upper case indicates codons. (b) Activity of the neo gene can be restored by single-stranded DNA oligonucleotides that substitute 1–4 nt to create a new ATG start codon (mismatching bases are indicated in red). Upper case indicates codons. (c) The efficiency of oligonucleotide-mediated base substitution in Msh3−/− (red bars), Msh6−/− (blue bars) and pS-MSH2 transfected wild-type (green bars) or Msh2+/− (orange bars) ES cells is the number of G418-resistant colonies per 105 cells that were plated after exposure to the oligonucleotides. Targeting data of Msh2−/− (black bars) and wild-type (white bars) ES cells are taken from Ref. (26) and shown as controls. For each oligonucleotide, the mean value is given of at least three independent experiments. Error bars show standard deviation. * P < 0.001, **P < 0.05 as compared with untreated wild-type cells (Student's t-test).
Figure 2.
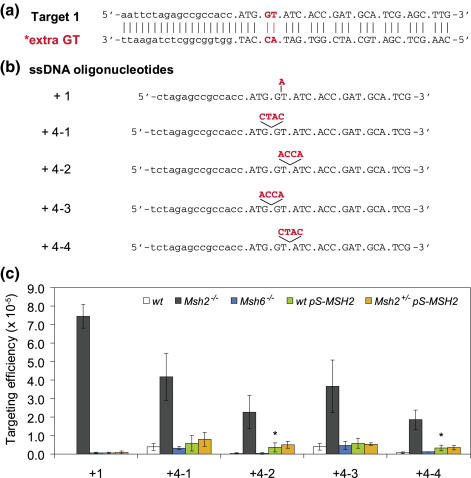
Oligonucleotide-mediated base insertion. (a) A single copy of a defective neomycin gene (neo) carrying a GT insertion behind the start codon (target 1) was inserted into the Rosa26 locus of Msh2−/−, Msh6−/−, Msh2+/− and wild-type ES cells. Upper case indicates codons. (b) Activity of the neo gene can be restored by single-stranded DNA oligonucleotides inserting either 1 or 4 nt to regenerate the open reading frame (inserted bases are shown in red). Upper case indicates codons. (c) The efficiency of oligonucleotide-mediated base insertion in Msh6−/− (blue bars) and pS-MSH2 transfected wild-type (green bars) or Msh2+/− (orange bars) ES cells is the number of G418-resistant colonies per 105 cells that were plated after exposure to the oligonucleotides. Targeting data of Msh2−/− (black bars) and wild-type (white bars) ES cells are taken from Ref. (26) and shown as controls. For each oligonucleotide, the mean value is given of at least three independent experiments. Error bars show standard deviation. * denotes P < 0.001 as compared with untreated wild-type cells (Student's t-test).
For transfection, ES cells were seeded onto gelatine-coated six-well plates at a density of 7 × 105 per well in BRL-conditioned medium. The next day, cells were exposed to 3 μg of oligonucleotide per well using 27 μl of TransFast™ transfection agent (Promega) as described before (11). For transient down regulation of MSH2, wild-type or Msh2+/− ES cells were first transfected with 3 μg of pS-MSH2 using 27 μl TransFast™ in 1.4 ml of serum-free medium. After 75 min of exposure to the transfection mixture, 4 ml of (serum-containing) BRL-conditioned medium was added. The cells were incubated overnight and then refed with BRL-conditioned medium containing 20 μg/ml of puromycin (Sigma-Aldrich). After 2 days, cells were washed with PBS, trypsinized and seeded at a density of 7 × 105 cells per well. The next day (day 1, Figure 3), cells were transfected with 3 μg of oligonucleotide as described before (11). The next day, cells were counted and reseeded in selective medium containing 750 μg/ml (Target 2) or 600 μg/ml (Target 1) of G418 (Invitrogen-GIBCO). After 10 days, the number of G418-resistant colonies was counted. For the RbN750F mutation, targeted ES cells were expanded and plated in pools of 5000 cells per well onto four 96-well feeder plates containing irradiated mouse embryonic fibroblasts (MEFs). In three subsequent screening rounds, cells were seeded onto 96-well MEF feeder plates in pools of 100, 10 and 1 cells per well, respectively. All oligonucleotides, deprotected and desalted, were obtained from Sigma-Genosys Ltd.
Figure 3.
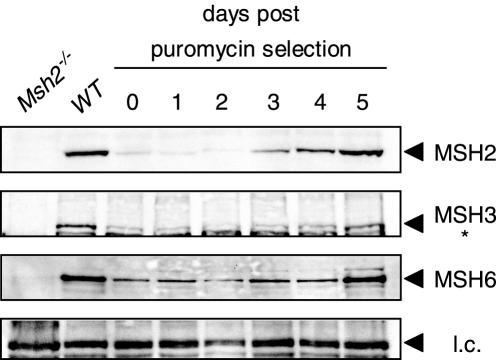
MMR protein levels after MSH2 suppression. Western blot analysis of wild-type ES cells transfected with pS-MSH2 followed by puromycin selection for 2 days. Whole-cell extracts were analyzed for MSH2, MSH3 and MSH6 for six consecutive days. Whole-cell extracts of Msh2−/− cells were used as negative control. WT, wild-type cells not treated with pS-MSH2; Asterisk indicates non-specific band; l.c., non-specific band as loading control.
RNA interference
We used the pSUPER vector containing a puromycin resistance gene for expression of shRNAs in murine ES cells (22). A 19 nt sequence corresponding to nucleotides 1231–1249 of the Msh2 gene (GenBank accession no. NM_008628), separated from the reverse complement of the same 19 nt sequence by a 9 nt non-complementary spacer (TTCAAGAGA), was inserted into the pSUPER vector (pS-MSH2). An empty vector with no gene-specific 19 nt sequence served as a non-silencing control (pS).
Western blot analysis
Cells were lysed in a buffer containing 60 mM Tris–HCl pH 6.8, 2% (w/v) SDS, 10% glycerol, 100 mM DTT and bromophenolblue. Protein extracts from 2 × 105 ES cells were separated by 7.5% SDS–PAGE and transferred to nitrocellulose membrane. Rabbit polyclonal antibodies against MSH2 (1:500) (23), MSH3 (kindly provided by G. Crouse; 1:500) and MSH6 (1:500) (21) were used as primary antibodies, and peroxidase-conjugated goat anti-rabbit IgG (BioSource International) was used as a secondary antibody. Signals were visualized with enhanced chemiluminescence.
Frameshift mutations at simple sequence repeats
We constructed two reporter cell lines consisting of a neo gene containing either a (G)10 or a (CA)15C repeat disrupting the open reading frame. The neo gene was derived from plasmid pMC1neo (24) in which the sequence between the first and the fifth codon was replaced by either a (G)10 or a (CA)15C repeat. The pMC1-(CA)15C-neo gene was placed immediately downstream of the promoterless histidinol-resistance gene in a Rosa26 targeting vector (11). The pMC1-(G)10-neo gene was inserted into the Rb targeting vector 129Rb-his which is based on 129Rb-hyg (25). These targeting vectors were subsequently introduced into Msh2−/−, Msh2+/− and wild-type ES cell lines by electroporation (25) to insert single copies of the (CA)15C and G10 reporters into Rosa26 and Rb locus, respectively.
Msh2−/−, Msh2+/− and wild-type cell lines containing the selectable slippage reporters were transfected with 3 μg of pS or pS-MSH2 followed by selection with puromycin for 2 days. After 7 days of cell culture, 105 Msh2−/− cells and 4 × 106 wild-type or Msh2+/− cells were plated onto two 100 mm dishes in selective medium containing 600 μg/ml G418. After 10 days, the number of resistant colonies was counted.
Spontaneous mutation frequency at Hprt locus
Msh2−/−, Msh2+/− and wild-type ES cell lines were transfected with 3 μg of pS or pS-MSH2 followed by selection with puromycin for 2 days. After 7 days of cell culture, 4 × 106 cells were plated onto two or seven 150 mm dishes in selective medium containing 10 μg/ml 6-thioguanine (Sigma-Aldrich). After 10 days, the number of resistant colonies was counted.
PCR-based detection of RbN750F mutation
A 738-bp product was amplified by PCR using primers 1 and 2 (Figure 4a) in a reaction mixture containing genomic DNA isolated from 2.5 × 104 ES cells, 1.25 U Taq polymerase, 1×PCR buffer containing 1.6 mM MgCl2, 12.5 pmol of each primer and 0.2 mM dNTPs in a total volume of 25 μl. After an initial denaturation step of 94°C for 5 min, amplification was carried out for 30 cycles of 94°C for 30 s, 60°C for 1 min, 72°C for 1 min 30 s and a final elongation step of 72°C for 10 min.
Figure 4.
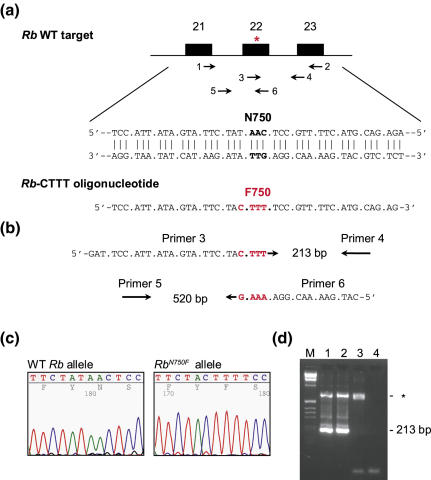
Oligonucleotide-mediated codon substitution in Rb. (a) Single-stranded DNA oligonucleotide Rb-CTTT was designed to replace an asparagine by a phenylalanine at position 750 in exon 22 of the Rb gene. Mismatching bases are indicated in red. Arrows indicate the location of PCR primers. (b) Primer pair 1/2 was used to amplify a 738 bp fragment from pools of cells. This fragment was used in a second PCR round using the nested primer pairs 3/4 or 5/6 of which primers 3 and 6 are specific for the CTTT mutation. (c) Sequence analysis of Rb mRNA in a purified mutant ES cell clone revealed the presence of the CTTT mutation, replacing the asparagine at position 750 by a phenylalanine. (d) PCR-based detection of RbN750F mutation in genomic DNA. PCRs were conducted with primer pairs 1/2 and 3/4, yielding a 213-bp product specific for the CTTT mutation. Lane M, molecular mass standards; lane 1, Rb+/N750F ES cell clone; lane 2, Rb+/N750F mouse; lane 3, wild-type littermate; lane 4, water control; * indicates non-specific band.
Similarly, a nested mutation-specific PCR was carried out with primers 3/4 or primers 5/6 (Figure 4b) using 1 μl of the first PCR and an annealing temperature of 58°C.
Sequence analysis of Rb mRNA
Total RNA was isolated from Rb+/N750F ES cells using RNA-Bee™ Total RNA Isolation Reagent (Campro Scientific). Rb cDNA was prepared by reverse transcription using a primer in exon 23 (primer 2). From this template, a 207 bp product spanning the oligonucleotide-mediated modification was amplified by PCR using primers in exon 21 (primer 1) and exon 23 (primer 4). This PCR product was cloned into the pGEM®-T Easy vector (Promega). Vector primers T7 and SP6 were used for sequencing.
RESULTS
Oligonucleotide-mediated targeting is suppressed by DNA MMR
To provide a simple in vivo readout for the efficacy of oligonucleotide-mediated gene modification, we have previously developed two selectable reporter gene systems consisting of mutant neomycin resistance genes (neo) (11). The neo cassette was mutated by either a single base substitution (ATG → AAG) in the start codon (Target 2, Figure 1a), or a 2 bp insertion (extra GT) immediately after the start codon, causing a frameshift mutation (Target 1, Figure 2a). A single copy of these mutant reporter genes was stably integrated into the Rosa26 locus of Msh2−/− and wild-type ES cells. Correction of the mutant sequence by oligonucleotides restores the open reading frame (Target 1) or generates an ATG start codon (Target 2) resulting in G418 resistance.
As we have previously shown, G418-resistant colonies were readily obtained in Msh2−/− cells, but not in wild-type cells, indicating that MMR activity imposes a strong barrier to oligonucleotide-mediated gene targeting (11,26).
Transient down regulation of MSH2
The necessity of MMR deficiency greatly restricts the application of oligonucleotide-mediated gene targeting for the generation of mutant mice. In order to establish a generally applicable protocol for oligonucleotide-mediated gene targeting in ES cells, we minimized the inadvertent effects of constitutive MMR deficiency by down regulating MMR activity in wild-type ES cells for only a brief period. For this purpose, we used the pSUPER vector containing a puromycin resistance gene to transiently express a short hairpin RNAi sequence against Msh2 mRNA (pS-MSH2) (22). After transfection of wild-type ES cells with pS-MSH2 and puromycin selection for 2 days, protein samples were taken for six consecutive days and analyzed by immunoblotting. MSH2 protein levels were 10-fold reduced for 3 days, and then gradually increased back to wild-type levels (Figure 3). This level of reduction was highly reproducible and also obtained in Msh2+/− ES cells (data not shown). We also analyzed MSH3 and MSH6 protein levels after pS-MSH2 transfection, since the stability of these proteins is known to be dependent on their interaction with MSH2 (20,21). Although both MSH3 and MSH6 protein levels were reduced upon down regulation of MSH2 (Figure 3), low levels of these proteins were present. This indicates that residual MSH2/MSH3 and MSH2/MSH6 complexes could still be formed in MSH2 knockdown cells.
Nucleotide substitutions in MSH2 knockdown ES cells
We next investigated whether transient down regulation of MSH2 rendered cells permissive for nucleotide substitutions by oligonucleotides. Wild-type ES cells were transfected with pS-MSH2 and cultured in the presence of puromycin for 2 days. pS-MSH2 containing cells were transfected with different oligonucleotides to substitute 1, 2, 3 or 4 nt in the mutated neo reporter gene (Figure 1b).
For the majority of substitution oligonucleotides, transient down regulation of MSH2 significantly increased the targeting efficiency [Figure 1c, compare green and white bars; efficiencies in wild-type cells were taken from Ref. (26)]. In particular, 4 nt could now efficiently be substituted reaching levels of 60% of the levels found in Msh2−/− cells [Figure 1c, oligonucleotides 4n, 4M and 4N; efficiencies in Msh2−/− cells were taken from Ref. (26)]. The performance of oligonucleotides substituting 3 nt was more variable ranging from 7% (oligonucleotide 3n) to 90% (oligonucleotide 3N) of the efficiencies found in Msh2−/− cells. Substitution of a single (oligonucleotide 1m) or 2 nt (oligonucleotides mxm and 2M) modestly improved (Figure 1c, compare green and white bars), reaching levels of only 2–14% of the frequencies found in Msh2−/− cells. We also investigated whether MSH2 knockdown had a stronger effect in Msh2+/− ES cells than in wild-type ES cells. This was not the case: the targeting efficiencies of oligonucleotides 1m, 4n, 4M and 4N did not significantly differ in both cell types (Figure 1c, compare orange and green bars).
In general, larger stretches of nucleotides could efficiently be substituted in pS-MSH2 transfected wild-type ES cells, while 1 or 2 nt substitutions were still significantly suppressed. This indicates that transient knockdown of MSH2 has not fully abrogated MMR activity. Residual MMR complexes exist that still detect simple mismatches (1 and 2 nt), while complex mismatches of 4 nt tend to escape this detection. Recognition of 3 nt mismatches appears to be dependent on the sequence context as some (3n and 3M) are well recognized, while another (3N) escaped detection.
Nucleotide insertions in MSH2 knockdown ES cells
Surprisingly, the efficiency of 1 or 4 nt insertions was only marginally improved after transient down regulation of MSH2 in wild-type and Msh2+/− ES cells (Figure 2c, green and orange bars). Oligonucleotides +4–2 and +4–4 performed significantly better after pS-MSH2 transfection compared to untreated wild-type cells, while the efficiency of oligonucleotides +4–1 and +4–3 did not increase. The efficiencies of these 4 nt insertions in pS-MSH2 transfected wild-type ES cells remained at only 14–18% of the levels found in Msh2−/− cells. Insertion of a single base (+1) was hardly efficient with a frequency of 1% compared to Msh2−/− cells.
To rule out that the pS-MSH2 vector was stably integrated after puromycin selection, protein samples were taken of 16 G418-resistant colonies (eight wild-type and eight Msh2+/− ES cell colonies) and immunoblotted for MSH2. All colonies showed wild-type levels of MSH2 (data not shown). Furthermore, none of these colonies survived in medium supplemented with puromycin, indicating that the effects of pS-MSH2 transfection were truly transient.
Taken together, transient down regulation of MSH2 rendered cells permissive for 4 nt substitutions, but not for simple nucleotide substitutions or 4 nt insertions, although these were all highly efficient in Msh2−/− cells.
Oligonucleotide-mediated targeting in Msh3−/− and Msh6−/− cells
The MSH2/MSH6 heterodimer has been suggested to recognize single base pair mismatches and one or two unpaired nucleotides, while the MSH2/MSH3 complex is predominantly involved in the recognition of loops of two to five unpaired bases (18,19). In order to study which of these complexes is responsible for suppression of oligonucleotide-mediated gene targeting, we have introduced the neo reporter genes into Msh3−/− and Msh6−/− ES cells (21) and studied the performance of insertion and substitution oligonucleotides in these cells.
We found that most 4 nt insertions could efficiently be introduced in Msh3−/− cells (26), but not in Msh6−/− cells (Figure 2c, blue bars). This indicates that 4 nt insertions are mainly suppressed by MSH2/MSH3 complexes. In MSH2 knockdown cells, most 4 nt insertions were less efficient than in Msh3−/− cells, indicating that after MSH2 knockdown, residual MSH2/MSH3 activity was still sufficiently high to suppress 4 nt insertions.
In contrast, nucleotide substitutions were supported by MSH6 deficiency (Figure 1c, blue bars) rather than MSH3 deficiency (Figure 1c, red bars). In Msh6−/− cells, the efficiency of nucleotide substitutions was significantly increased, except for oligonucleotide 4M. This indicates that nucleotide substitutions are primarily suppressed by MSH2/MSH6 complexes. Strikingly, simple nucleotide substitutions (1m, mxm and 2M) and the 3 nt substitutions 3n and 3M were less efficient in MSH2 knockdown cells than in Msh6−/− cells, while complex substitutions (4n, 4M and 4N) and the 3 nt substitution 3N, were more efficient in MSH2 knockdown cells than in Msh6−/− cells. These findings suggest that suppression of nucleotide substitutions requires different levels of MSH2/MSH6 activity: low levels are sufficient for suppression of simple substitutions, while suppression of 4 nt substitutions requires high levels of MSH2/MSH6.
Thus, in pS-MSH2 transfected wild-type cells, residual levels of both MSH2/MSH3 and MSH2/MSH6 complexes were still able to suppress nucleotide insertions and simple substitutions.
Effects of transient MSH2 suppression
To obtain an indication of the level of accumulation of spontaneous mutations as a result of transient suppression of MSH2 activity, we assessed the occurrence of frameshift mutations at simple sequence repeats. For this purpose, we have used two frameshift reporter constructs consisting of a neo gene in which the open reading frame was disrupted by either a (G)10 or a (CA)15C repeat. Polymerase slippage errors that remain unrepaired in the absence of MMR activity may lead to e.g. deletion of a single G or addition of a CA restoring the open reading frame and resulting in G418 resistance. At the (G)10 repeat, pS-MSH2 transfected wild-type cells showed a slippage frequency that was ∼400-fold higher than in control wild-type cells but 7-fold lower than in Msh2−/− cells (Table 1). At the (CA)15C repeat, MSH2 suppression resulted in only a 2-fold increase in slippage frequency compared to untreated wild-type cells which was ∼30-fold lower than in Msh2−/− cells (Table 1). Similar results were obtained in Msh2+/− ES cells, where the slippage frequency increased 5-fold upon pS-MSH2 transfection, but was still 25-fold lower compared to Msh2−/ − cells.
Table 1.
Frameshift mutations at simple sequence repeats
| Genotype | Number of G418-resistant colonies/106 cells | |
|---|---|---|
| (G)10 repeata | (CA)15C repeatb | |
| WT pS | 0.75 (±0.0) | 83 (±22) |
| WT pS-MSH2 | 322 (±36) | 159 (±51) |
| Msh2+/− pS | ND | 44 (±13) |
| Msh2+/− pS-MSH2 | ND | 214 (±19) |
| Msh2−/− pS | 2285 (±1039) | 5085 (±1239) |
aResults from two independent experiments.
bResults from four independent experiments.
In addition, we have determined the mutation frequency at the Hprt locus, assayed by the appearance of 6-thioguanine resistant cells. We initially plated cells at a density of 106 per 85 cm2. The number of 6-thioguanine (6TG)-resistant colonies per 106 pS-MSH2 transfected wild-type cells varied between 0 and 2, whereas one Hprt mutant was found in untreated wild-type cells (Table 2). In Msh2+/− cells, the number of Hprt mutants was not increased upon transient down regulation of MSH2. To exclude the possibility that 6-TG-resistant colonies are lost due to metabolic co-operation by wild-type cells, we also plated cells at a density of 106 per 300 cm2. Except for Msh2−/− cells, this did not lead to increased numbers of colonies (Table 2, exp. 4). The large fluctuation in the number of 6-TG-resistant Msh2−/− colonies is most likely due to variations in the appearance of the first mutant cell in each culture.
Table 2.
Spontaneous mutation frequency at the Hprt locus
| Genotype | Number of Hprt mutants/106 cells | |||
|---|---|---|---|---|
| Exp. 1a | Exp. 2a | Exp. 3a | Exp. 4b | |
| WT pS | 0 | 0 | 1 | 0 |
| WT pS-MSH2 | 0 | 1 | 2 | 2 |
| Msh2+/− pS | 0 | 2 | 0 | 0 |
| Msh2+/− pS-MSH2 | 0 | 1 | 0 | 0 |
| Msh2−/− pS | 15 | 7 | 5 | 36 |
aCells were plated onto 85 cm2.
bCells were plated onto 300 cm2.
These results indicate that the accumulation of spontaneous mutations is increased upon transient disabling of the MMR machinery but remains well below that in constitutive MMR-deficient cells.
Oligonucleotide-mediated codon substitution in Rb
Our results indicate that transient suppression of MSH2 activity allows effective substitution of 4 nt and may be used to introduce codon substitutions in non-selectable genes. As an example, we present an oligonucleotide-mediated codon substitution (N750F) in the retinoblastoma gene Rb. We designed a 38-mer oligonucleotide with four centrally located substitutions in order to replace the asparagine at position 750 by a phenylalanine (Figure 4a). In this experiment, we used Msh2+/− cells, although we later realized that wild-type cells perform equally well (Figure 1c). pS-MSH2-treated Msh2+/− ES cells were exposed to oligonucleotide Rb-CTTT and seeded into four 96-well plates at a density of 5000 cells per well. Genomic DNA was isolated and a 738 bp product was amplified by PCR using primers 1 and 2 (Figure 4a). Pools containing cells with the oligonucleotide-mediated modification were identified by a nested mutation-specific PCR (Figure 4b). In this second PCR, primers 3/4 or primers 5/6 could only amplify a product when the CTTT mutation was present. One well was identified giving a PCR product with primers 3/4 and primers 5/6. From this positive pool, a single cell clone was isolated in three subsequent screening rounds, in which cells were seeded in pools of 100, 10 and 1 cells per well, respectively. The Rb+/N750F ES cell clone was verified by sequencing of mRNA (Figure 4c) and used for the generation of mouse chimeras. In contrast to Msh2−/− ES cells, pS-MSH2 transfected Msh2+/− cells were able to contribute to the germ line of chimeric mice. The RbN750F allele was transmitted to their offspring as indicated by the presence of the 213-bp mutation-specific PCR band amplified from genomic mouse DNA (Figure 4d).
DISCUSSION
The results presented here establish a generally applicable procedure for the generation of subtle gene modifications in mouse ES cells. Oligonucleotide-mediated gene targeting in combination with a temporary knockdown of the MMR system allowed effective substitution of 4 nt, which makes it possible to substitute virtually any codon in any gene. We have successfully created a codon substitution (N750F) in the Rb gene of MMR-proficient ES cells and used these cells to generate mutant mice. The targeting procedure was cost-effective and rapid: already 2 weeks after oligonucleotide transfection, a single modified cell could be identified in a pool of 5000 unmodified cells by a mutation-specific PCR. This example illustrates the potential power of oligonucleotide-mediated gene targeting as an effective novel tool for mouse genetics.
We demonstrated that transient knockdown of MSH2 rendered cells permissive for oligonucleotide-mediated nucleotide substitutions. Yet, not all substitutions were equally efficient: oligonucleotides 3N, 4n, 4M and 4N performed best with targeting frequencies comparable to those in Msh2−/− ES cells (Figure 1c). Interestingly, three of these oligonucleotides (3N, 4n and 4N) already showed a slightly higher targeting frequency in untreated wild-type cells than the other oligonucleotides (26), indicating that the mismatches that are created by these oligonucleotides are not very well recognized by the MMR machinery. On the other hand, oligonucleotides substituting 1 (1m), 2 (mxm and 2M) or 3 nt (3n and 3M) performed moderately in MSH2 knockdown cells and very poorly in untreated wild-type cells. This indicates that these mismatches are strongly recognized by the MMR machinery.
We showed that nucleotide substitutions were mainly suppressed by MSH2/MSH6 complexes, since they were only efficient in Msh2−/− and Msh6−/− ES cells, but not in Msh3−/− ES cells (Figure 1c). Remarkably, targeting efficiencies in Msh2−/− cells were ∼5-fold higher than in Msh6−/− cells, suggesting that nucleotide substitutions were also suppressed by MSH2/MSH3 complexes.
Single or 4 nt insertions were only marginally improved in MSH2 knockdown cells compared to untreated wild-type cells. High efficiencies in Msh2−/− and Msh3−/− ES cells indicate that nucleotide insertions are primarily suppressed by MSH2/MSH3 complexes (26). Again, Msh2−/− cells showed ∼2-fold higher efficiencies than Msh3−/− cells, suggesting a slight suppression of nucleotide insertions by MSH2/MSH6 complexes.
Transient down regulation of MSH2 resulted in an increased mutation frequency (Table 1). Yet, we have good reasons to believe that this should not hamper general application of our targeting approach. First, simple oligonucleotide-mediated gene modifications were still suppressed in pS-MSH2 transfected wild-type cells, indicating that many spontaneous mutations may be suppressed by residual MMR activity as well. Second, temporary knockdown of MSH2 only modestly affected the slippage frequency at the (CA)15C reporter (Table 1) and the mutation frequency at the endogenous Hprt gene in both wild-type and Msh2+/− cells (Table 2). Only the slippage frequency at the (G)10 reporter was strongly increased (although still 7-fold lower than in Msh2−/− cells). This seems in contrast to the strong suppression of a single nucleotide insertion by oligonucleotide +1 (Figure 2) and may reflect a particularly high sensitivity of the (G)10 reporter to MMR defects as has been observed in yeast (27,28). Moreover, poly(G/C) repeats are less abundant than poly(A/T) tracts and seem to be excluded from the coding regions of various eukaryotic genomes (29,30). Third, pS-MSH2-treated Rb+/N750F ES cells were used to generate chimeric mice that were able to transmit the modified allele through the mouse germ line, while our constitutively MSH2-deficient ES cell line did not contribute to the mouse germ line. Finally, inadvertent mutations that may have arisen during the brief period of MSH2 down regulation will be crossed out in subsequent generations of mice. This backcrossing will avoid confounding effects of unlinked mutations on the phenotype of the oligonucleotide-mediated modification.
We have demonstrated that MMR raises a strong barrier to successful oligonucleotide-mediated gene targeting in ES cells. Similar observations have been made in E. coli, where loss of MMR improved oligonucleotide-mediated gene targeting by Red-mediated recombination by ∼100-fold (12). However, constitutive MMR deficiency may lead to an unacceptably high level of inadvertent spontaneous mutations. The experiments presented here disclose a dual approach to circumvent this problem and yet achieve reasonable targeting frequencies: a temporary down regulation of MMR activity and the use of oligonucleotides that escape detection by residual MMR. During the brief period of MMR disabling, residual mismatch recognition capacity still largely suppresses spontaneous mutagenesis caused by simple mismatches. However, complex mismatches are not recognized allowing effective 4 nt substitutions. We are now routinely using this procedure to introduce codon substitutions into the germ line of mice. We have targeted three different loci and found targeting efficiencies between 0.25 and 1.5 oligonucleotide-modified cell per 106 cells, which is comparable to the Rb+/N750F mutant presented here. Targeting efficiencies appear to be locus dependent, although more experiments are needed to confirm this observation.
As we have shown previously, generation of knockout mice can readily be achieved by 4 nt insertion oligonucleotides in Msh3−/− ES cells, which are germ line competent (26).
More insight into the mechanisms of oligonucleotide-mediated gene targeting may extend its use to in vivo applications. Stable correction of gene mutations by chemically modified oligonucleotides in muscle fibers (31) and retinas (14) of mice holds great promise for therapeutic applications. A major advantage of oligonucleotide-mediated gene targeting is the opportunity of correcting gene mutations without integration of exogenous DNA, potentiating this technique for gene therapy strategies and treatment of human disease.
Acknowledgments
We thank E. Delzenne for maintaining mouse strains and F. Foijer, T. Vormer, C. Wielders, and E. Wielders for valuable comments on the manuscript. This work was supported by grants from the Dutch Cancer Society (NKI 2000–2233) and the Netherlands Genomics Initiative (Horizon Breakthrough project 050-71-007). Funding to pay the Open Access publication charges for this article was provided by the Netherlands Cancer Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Capecchi M.R. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 2.Campbell C.R., Keown W., Lowe L., Kirschling D., Kucherlapati R. Homologous recombination involving small single-stranded oligonucleotides in human cells. New Biol. 1989;1:223–227. [PubMed] [Google Scholar]
- 3.Igoucheva O., Alexeev V., Yoon K. Targeted gene correction by small single-stranded oligonucleotides in mammalian cells. Gene Ther. 2001;8:391–399. doi: 10.1038/sj.gt.3301414. [DOI] [PubMed] [Google Scholar]
- 4.Kmiec E.B., Ye S., Peng L. Targeted gene repair in mammalian cells using chimeric oligonucleotides. Genet. Eng. (N.Y.) 2000;22:23–31. doi: 10.1007/978-1-4615-4199-8_3. [DOI] [PubMed] [Google Scholar]
- 5.Liu L., Rice M.C., Kmiec E.B. In vivo gene repair of point and frameshift mutations directed by chimeric RNA/DNA oligonucleotides and modified single-stranded oligonucleotides. Nucleic Acids Res. 2001;29:4238–4250. doi: 10.1093/nar/29.20.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang G., Seidman M.M., Glazer P.M. Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science. 1996;271:802–805. doi: 10.1126/science.271.5250.802. [DOI] [PubMed] [Google Scholar]
- 7.Liu L., Rice M.C., Drury M., Cheng S., Gamper H., Kmiec E.B. Strand bias in targeted gene repair is influenced by transcriptional activity. Mol. Cell. Biol. 2002;22:3852–3863. doi: 10.1128/MCB.22.11.3852-3863.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Igoucheva O., Alexeev V., Pryce M., Yoon K. Transcription affects formation and processing of intermediates in oligonucleotide-mediated gene alteration. Nucleic Acids Res. 2003;31:2659–2670. doi: 10.1093/nar/gkg360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X.T., Costantino N., Lu L.Y., Liu D.P., Watt R.M., Cheah K.S., Court D.L., Huang J.D. Identification of factors influencing strand bias in oligonucleotide-mediated recombination in Escherichia coli. Nucleic Acids Res. 2003;31:6674–6687. doi: 10.1093/nar/gkg844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrara L., Kmiec E.B. Camptothecin enhances the frequency of oligonucleotide-directed gene repair in mammalian cells by inducing DNA damage and activating homologous recombination. Nucleic Acids Res. 2004;32:5239–5248. doi: 10.1093/nar/gkh822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dekker M., Brouwers C., te Riele H. Targeted gene modification in mismatch-repair-deficient embryonic stem cells by single-stranded DNA oligonucleotides. Nucleic Acids Res. 2003;31:e27. doi: 10.1093/nar/gng027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costantino N., Court D.L. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc. Natl Acad. Sci. USA. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olsen P.A., Randol M., Krauss S. Implications of cell cycle progression on functional sequence correction by short single-stranded DNA oligonucleotides. Gene Ther. 2005;12:546–551. doi: 10.1038/sj.gt.3302454. [DOI] [PubMed] [Google Scholar]
- 14.Andrieu-Soler C., Casas M., Faussat A.M., Gandolphe C., Doat M., Tempe D., Giovannangeli C., Behar-Cohen F., Concordet J.P. Stable transmission of targeted gene modification using single-stranded oligonucleotides with flanking LNAs. Nucleic Acids Res. 2005;33:3733–3742. doi: 10.1093/nar/gki686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pierce E.A., Liu Q., Igoucheva O., Omarrudin R., Ma H., Diamond S.L., Yoon K. Oligonucleotide-directed single-base DNA alterations in mouse embryonic stem cells. Gene Ther. 2003;10:24–33. doi: 10.1038/sj.gt.3301857. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Muyrers J.P., Rientjes J., Stewart A.F. Phage annealing proteins promote oligonucleotide-directed mutagenesis in Escherichia coli and mouse ES cells. BMC Mol. Biol. 2003;4:1. doi: 10.1186/1471-2199-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 18.Genschel J., Littman S.J., Drummond J.T., Modrich P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J. Biol. Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 19.Umar A., Risinger J.I., Glaab W.E., Tindall K.R., Barrett J.C., Kunkel T.A. Functional overlap in mismatch repair by human MSH3 and MSH6. Genetics. 1998;148:1637–1646. doi: 10.1093/genetics/148.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Wind N., Dekker M., Berns A., Radman M., te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 21.de Wind N., Dekker M., Claij N., Jansen L., van Klink Y., Radman M., Riggins G., van der Valk M., van't Wout K., te Riele H. HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nature Genet. 1999;23:359–362. doi: 10.1038/15544. [DOI] [PubMed] [Google Scholar]
- 22.Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 23.de Wind N., Dekker M., van Rossum A., van der Valk M., te Riele H. Mouse models for hereditary nonpolyposis colorectal cancer. Cancer Res. 1998;58:248–255. [PubMed] [Google Scholar]
- 24.Thomas K.R., Capecchi M.R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 25.te Riele H., Maandag E.R., Berns A. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc. Natl Acad. Sci. USA. 1992;89:5128–5132. doi: 10.1073/pnas.89.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dekker M., Brouwers C., Aarts M., van der Torre J., de Vries S., van de Vrugt H., te Riele H. Effective oligonucleotide-mediated gene disruption in ES cells lacking the mismatch repair protein MSH3. Gene Ther. 2006;13:686–694. doi: 10.1038/sj.gt.3302689. [DOI] [PubMed] [Google Scholar]
- 27.Sia E.A., Kokoska R.J., Dominska M., Greenwell P., Petes T.D. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gragg H., Harfe B.D., Jinks-Robertson S. Base composition of mononucleotide runs affects DNA polymerase slippage and removal of frameshift intermediates by mismatch repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 2002;22:8756–8762. doi: 10.1128/MCB.22.24.8756-8762.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katti M.V., Ranjekar P.K., Gupta V.S. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol. Biol. Evol. 2001;18:1161–1167. doi: 10.1093/oxfordjournals.molbev.a003903. [DOI] [PubMed] [Google Scholar]
- 30.Toth G., Gaspari Z., Jurka J. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 2000;10:967–981. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertoni C., Morris G.E., Rando T.A. Strand bias in oligonucleotide-mediated dystrophin gene editing. Hum. Mol. Genet. 2005;14:221–233. doi: 10.1093/hmg/ddi020. [DOI] [PubMed] [Google Scholar]