Abstract
The fusion of vesicles in the secretory pathway involves the interaction of t-soluble N-ethylmaleimide-sensitive factor attachment protein receptors (t-SNAREs) on the target membrane and v-SNAREs on the vesicle membrane. AtSNAP33 is an Arabidopsis homolog of the neuronal t-SNARE SNAP-25 involved in exocytosis and is localized at the cell plate and at the plasma membrane. In this paper, the expression of AtSNAP33 was analyzed after different biotic and abiotic stresses. The expression of AtSNAP33 increased after inoculation with the pathogens Plectosporium tabacinum and virulent and avirulent forms of Peronospora parasitica and Pseudomonas syringae pv tomato. The expression of PR1 transcripts encoding the secreted pathogenesis-related protein 1 also increased after inoculation with these pathogens and the expression of AtSNAP33 preceded or occurred at the same time as the expression of PR1. AtSNAP33 was also expressed in npr1 plants that do not express PR1 after pathogen inoculation as well as in cpr1 plants that overexpress PR1 in the absence of a pathogen. The level of AtSNAP33 decreased slightly in leaves inoculated with P. parasitica in the NahG plants, and eds5 and sid2 mutants that are unable to accumulate salicylic acid (SA) after pathogen inoculation, indicating a partial dependence on SA. AtSNAP33 was also expressed in systemic noninoculated leaves of plants inoculated with P. syringae. In contrast to the situation in infected leaves, the expression of AtSNAP33 in systemic leaves was fully SA dependent. Thus, the expression of AtSNAP33 after pathogen attack is regulated by SA-dependent and SA-independent pathways. Mechanical stimulation also led to an increase of AtSNAP33 transcripts.
The plant secretory pathway comprises different organelles including the endoplasmic reticulum, the Golgi apparatus, the plasma membrane, and the vacuole. Proteins destined to the extracellular space or the vacuole enter the secretory pathway at the endoplasmic reticulum and are then transported through the Golgi apparatus. At the trans-Golgi network, secreted proteins are sorted from vacuolar proteins and packaged into secretory vesicles. Transport between the organelles of the secretory pathway occurs by budding of vesicles from a donor membrane and fusion with an acceptor membrane. Exocytosis is the fusion of secretory vesicles with the plasma membrane, permitting the release of their content outside the cell. The fusion of vesicles involves the interaction of vesicle (v)-soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein receptors (SNAREs) localized on the vesicle membrane and t-SNAREs localized on the target membrane (Rothman, 1996; Hay and Scheller, 1997). A four-helical bundle of SNAREs is formed. One helix of this SNARE complex is provided by a v-SNARE and three helices are provided by t-SNAREs, which always include a member of the syntaxin family contributing one helix. The remaining two helices are contributed from a single SNAP-25-like protein or from two separate t-SNARE light chains. Trimeric SNARE complexes have been described for the fusion of vesicles at the plasma membrane, whereas endomembrane fusion processes involve tetrameric SNARE complexes (Fukuda et al., 2000). Two cytosolic proteins, general components, α-soluble NSF-attachment protein (SNAP) and NSF disassemble this complex after fusion. The machinery of membrane fusion is highly conserved among eukaryotes (for review, see Blatt et al., 1999; Jahn and Südhof, 1999). Several plant SNAREs have been isolated, and the sequence of the Arabidopsis genome contains many SNAREs (for review, see Sanderfoot and Raikhel, 1999; Sanderfoot et al., 2000). The function of most of these proteins remains to be demonstrated. However, they often show a high degree of conservation with their yeast or mammalian counterparts. Thus, they may have similar functions in the plant cell.
Plants respond to bacterial, fungal, and viral pathogen attack by the synthesis of several pathogenesis-related (PR) proteins, including PR1 (van Loon and van Strien, 1999). Many of them are synthesized at the endoplasmic reticulum and are transported to the extracellular space or the vacuole by the secretory pathway. For example, infection of Arabidopsis by Pseudomonas syringae pv tomato DC 3000 leads to the induction of the expression of PR1, PR2 (a β-1, 3-glucanase), and PR5 (a thaumatin-like protein), which are secreted to the extracellular space (Uknes et al., 1992). The accumulation of the signaling molecule salicylic acid (SA) correlates with the expression of PR proteins and resistance (Malamy et al., 1990; Métraux et al., 1990; Yalpani et al., 1991). Transgenic tobacco (Nicotiana tabacum) and Arabidopsis plants that express a bacterial NahG gene encoding a SA hydroxylase degrade SA to catechol (Gaffney et al., 1993; Delaney et al., 1994) and express very low amounts of PR proteins after pathogen attack (Delaney et al., 1994). In the Arabidopsis eds5 and sid2 mutants, there is no accumulation of SA after pathogen attack, and a strong reduction of PR1 expression, but PR2 and PR5 are still expressed (Nawrath and Métraux, 1999).
Many plants respond to mechanical stimulation such as touch and wind by reduced growth and stocky morphology. These developmental changes called thigmomorphogenesis make the plant more resistant to environmental challenges such as blasts of wind. The effect of mechanical strain on plant growth has been well documented (Jaffe and Forbes, 1993; Mitchell and Myers, 1995). However, the molecular mechanisms underlying these effects are not well understood. The expression of several genes from Arabidopsis and other plant species has been shown to be regulated by mechanical stimulation. They include touch (TCH) and protein kinase genes (Braam and Davis, 1990; Mizoguchi et al., 1996; Braam et al., 1997). TCH genes include various enzymes such as xyloglucan endotransglycosylase, Ca2+-binding proteins, 1-aminocyclopropane-1-carbo-xylic acid synthases, calmodulins, or lipoxygenase (Botella et al., 1995; Takezawa et al., 1995; Xu et al., 1995; Mauch et al., 1997; Depège et al., 1999; van der Luit et al., 1999).
We have previously characterized AtSNAP33, a homolog of the t-SNARE SNAP-25, in Arabidopsis (Heese et al., 2001). AtSNAP33 was localized at the plasma membrane and at the forming cell plate. It interacts with the syntaxin KNOLLE localized at the cell plate (Heese et al., 2001), which functions in cell plate formation and Nt-Syr1 (NtSyp121), a tobacco syntaxin localized at the plasma membrane (Leyman et al., 2000; Kargul et al., 2001), which functions in secretion (Geelen et al., 2002). The null mutant atsnap33 develops large necrotic lesions on cotyledons and rosette leaves as well as cytokinetic defects, and eventually dies before flowering (Heese et al., 2001). We postulate that AtSNAP33 could be involved in diverse membrane fusion processes, including exocytosis and the formation of the cell plate.
It has been shown that the expression of several endoplasmic-reticulum-resident proteins, including the luminal binding-protein (BiP), calreticulin, protein disulfide isomerase, and the barley (Hordeum vulgare) Glc-regulated protein GRP94 is induced upon infection, treatment with plant cell wall degrading enzymes, or SA (Walther-Larsen et al., 1993; Denecke et al., 1995; Jelitto-van Dooren et al., 1999).
Here, we show that the expression of AtSNAP33, a component of a later step of the secretory pathway, i.e. transport to the plasma membrane, was induced after inoculation with pathogens in inoculated leaves and in systemic uninoculated leaves. The expression was compared with that of PR1, a defense-related protein secreted into the extracellular space and a marker for systemic acquired resistance (Uknes et al., 1992). The expression of AtSNAP33 was determined in NahG transgenic plants as well as in eds5, sid2, npr1, and cpr1 mutants. Finally, AtSNAP33 induction was also studied after mechanical stimulation, wind, and wounding.
RESULTS
The Expression of AtSNAP33 Is Increased by Inoculation with Plectosporium tabacinum, Peronospora parasitica Isolates NOCO and EMWA, and P. syringae pv tomato Carrying or Not avrRpt2
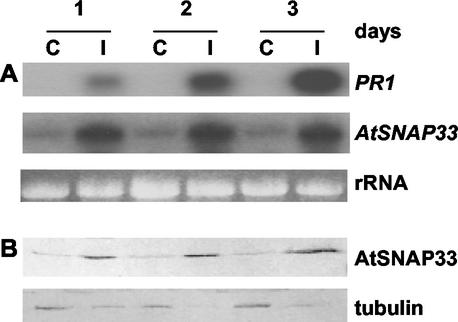
Figure 1 shows a time course of expression of AtSNAP33 and PR1 after inoculation of Arabidopsis accession Col-0 with the ascomycete P. tabacinum. The hyphae grew into the plant tissue and 3 d after inoculation, the leaves were yellow and the fungus had sporulated. Four days after the inoculation, the leaves were completely macerated. The level of AtSNAP33 transcripts in infected leaves increased up to a steady level 1 d post inoculation (dpi) compared with the level in control plants treated with water, which showed very low expression of AtSNAP33. As a comparison, PR1 underwent a steady increase up to 3 dpi.
Figure 1.
Time course of expression of the AtSNAP33 and PR1 transcripts (A) and AtSNAP33 protein (B) after inoculation with P. tabacinum. Leaves were inoculated with P. tabacinum (I) or water as a control (C) and were collected after different times in days indicated above A. A, The RNA was extracted and AtSNAP33 and PR1 mRNA levels were detected by RNA-blot hybridization. The RNA gel stained with ethidium bromide is shown as a control for loading. B, The proteins were extracted, separated on SDS-PAGE, and transferred to a nitrocellulose membrane. The membrane was then probed with anti-AtSNAP33 antibodies or anti-tubulin antibodies as a control for loading.
The expression of AtSNAP33 protein in infected leaves as visualized on immunoblots was already induced 1 dpi, and remained high until 3 dpi, compared with the low levels observed in uninfected controls (Fig. 1B). The level of AtSNAP33 protein reflected the level of transcripts in Figure 1A and many other independent experiments (data not shown). Therefore, only the level of AtSNAP33 transcripts is shown in further experiments.
Figure 2 shows the kinetics of expression of AtSNAP33 and PR1 after inoculation of Arabidopsis Col-0 with the downy mildew pathogen P. parasitica isolate NOCO or EMWA. The isolate NOCO forms a compatible interaction and the isolate EMWA forms an incompatible interaction with Arabidopsis accession Col-0 (Parker et al., 1993; Holub et al., 1994). In the compatible interaction with the isolate NOCO, the hyphae grew into the plant tissues, forming numerous haustoria, and no symptoms were visible before 7 dpi when the conidiophores bearing the asexual conidia grew out of the stomata. During colonization, sexual spores formed as well. In the incompatible interaction with the isolate EMWA, the hyphae grew for 3 d, forming haustoria. The growth of the fungus then stopped and trailing necroses of host cells along the length of the hyphae were observed. There was no asexual sporulation.
Figure 2.
Time course of expression of AtSNAP33 and PR1 transcripts (A) and accumulation of PR1 protein in the intercellular space (B) after inoculation with P. parasitica isolates NOCO and EMWA. Leaves were inoculated with P. parasitica isolates NOCO (N) or EMWA (E) or water as control (C) and were collected after different times in days as indicated. A, The RNA was extracted and AtSNAP33 and PR1 mRNA levels were detected by RNA-blot hybridization. The RNA gel stained with ethidium bromide is shown as a control for loading. B, Proteins in the intercellular fluids were separated on SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was probed with anti-PR1 antibodies. The experiments in A and B were performed independently.
In the compatible interaction with the isolate NOCO, induction of the expression of AtSNAP33 and PR1 was slower than in the incompatible interaction with the isolate EMWA where the increase in expression was faster and transient. The increase in PR1 transcripts occurred 1 d after the increased levels of AtSNAP33.
To verify that the PR1 protein was secreted to the extracellular space, intercellular fluids of leaves inoculated with P. parasitica were obtained. Figure 2B shows that PR1 was found in the intercellular space 4 to 7 dpi with isolate NOCO and 7 dpi with isolate EMWA. No PR1 was detectable in the intercellular fluids at earlier time points (data not shown).
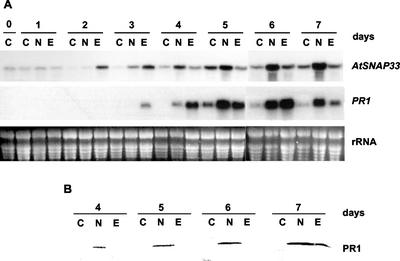
Figure 3A shows a time course of the expression of AtSNAP33 and PR1 after inoculation with the bacteria P. syringae pv tomato DC 3000 carrying or not the avirulence gene Rpt2. The expression of AtSNAP33 was strongly induced 1 dpi with both races of bacteria. In the compatible interaction, the level of expression remained high until 3 dpi, whereas in the incompatible interaction the level decreased somewhat after 2 dpi compared with mock-inoculated controls that showed little expression of AtSNAP33. In comparison, the expression of PR1 similarly increased at 1 dpi and remained strongly induced up to 3 dpi in all cases compared with uninfected controls.
Figure 3.
Time course of expression of AtSNAP33 and PR1 transcripts in inoculated (A) and uninoculated systemic (B) leaves after inoculation with P. syringae pv tomato DC3000 (D) or DC3000 avrRpt2 (R) or water as a control (C). Leaves were inoculated with the bacteria or water, and inoculated as well as uninoculated systemic leaves were collected after different times in days as indicated. The RNA was extracted and AtSNAP33 and PR1 mRNA levels were detected by RNA-blot hybridization. The RNA gel stained with ethidium bromide is shown as a control for loading.
Interestingly, the expression of AtSNAP33 was also induced in systemic noninoculated leaves of plants inoculated with P. syringae pv tomato DC3000 carrying or not avrRpt2 (Fig. 3B). As expected (Uknes et al., 1992), PR1 was also expressed systemically in inoculated plants.
The Local Expression of AtSNAP33 in Infected Leaves Is Partially Dependent on SA, and the Systemic Expression Is Fully SA Dependent
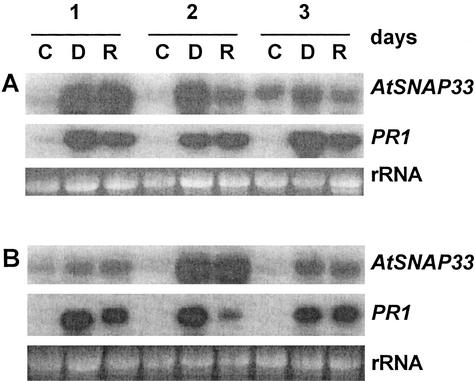
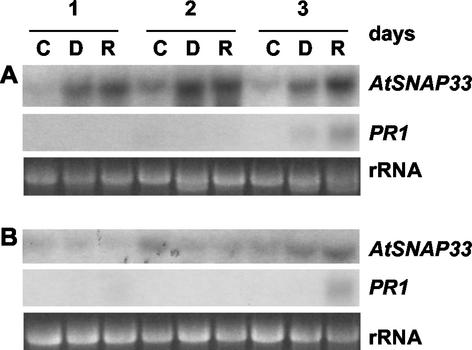
The dependence of the systemic expression of AtSNAP33 on the signaling molecule SA was tested in NahG-transgenic plants that degrade SA to catechol (Gaffney et al., 1993). The plants were inoculated with P. syringae pv tomato DC 3000 carrying or not avrRpt2. The expression of AtSNAP33 was slightly decreased in inoculated leaves of NahG compared with wild-type plants, but was absent in systemic uninoculated leaves (Figs. 3,A and B and 4, A and B, which represent experiments performed in parallel). Thus, the local induction of AtSNAP33 is partially SA independent in contrast to the systemic induction, which requires SA. In comparison, PR1 was not expressed locally or systemically in NahG plants, in agreement with previously published results (Gaffney et al., 1993).
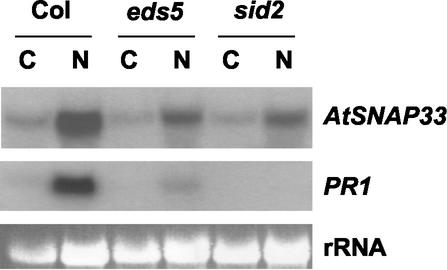
The expression of AtSNAP33 and PR1 was analyzed in eds5 and sid2, two mutants unable to accumulate SA (Nawrath and Métraux, 1999) and in wild-type Arabidopsis Col-0 inoculated with P. parasitica isolate NOCO (Fig. 5). Six days after inoculation with P. parasitica isolate NOCO, AtSNAP33 was still expressed but to a lower level than in wild-type Col-0 plants (Fig. 5). Thus, the partial SA independence of AtSNAP33 expression could be confirmed. PR1 was not expressed in eds5 or sid2 in agreement with previous reports (Nawrath and Métraux, 1999).
Figure 5.
Expression of AtSNAP33 and PR1 transcripts after inoculation of Arabidopsis Col-0 plants (Col), eds5, and sid2 mutants with P. parasitica isolate NOCO. Leaves were inoculated with P. parasitica isolate NOCO (N) or water as a control (C) and were collected after 6 d. The RNA was extracted and AtSNAP33 and PR1 mRNA levels were detected by RNA-blot hybridization. The RNA gel stained with ethidium bromide is shown as a control for loading.
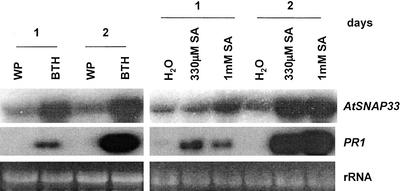
In agreement with the partial sensitivity of AtSNAP33 expression to endogenous SA after pathogen inoculation, soil-drench application of 330 μm or 1 mm SA increased the level of expression of AtSNAP33 in the absence of a pathogen (Fig. 6). This increase occurred already after 1 d but was higher after 2 d. Similarly, the expression of AtSNAP33 and PR1 was also induced 1 and 2 d after soil-drench application of 330 μm benzo(1, 2, 3) thiadiazole-7 carbonic acid S-methyl ester (BTH), an inducer of defense reactions (Fig. 6).
Figure 6.
Expression of AtSNAP33 and PR1 transcripts 1 and 2 d after treatment with 330 μm BTH or 330 μm or 1 mm SA. Wetting powder (WP) or water were used as a control for BTH or SA, respectively. Leaves were collected, the RNA was extracted, and AtSNAP33 and PR1 mRNA levels were detected by RNA-blot hybridization. The RNA gel stained with ethidium bromide is shown as a control for loading.
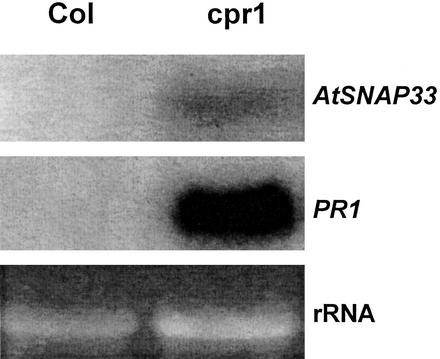
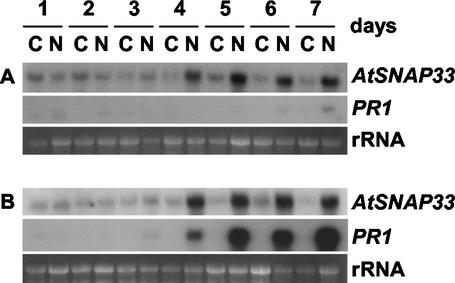
In uninfected cpr1 mutants that overexpress PR1, PR2, and PR5 constitutively (Bowling et al., 1994), AtSNAP33 was expressed constitutively compared with wild-type Col-0 plants (Fig. 7). However, the expression of AtSNAP33 could also be dissociated from that of PRs. AtSNAP33 was induced in the npr1 mutants infected with P. parasitica isolate NOCO (Fig. 8). NPR1 is a transcriptional regulator of SA-induced PR gene expression, and npr1 mutants show no expression of PR1 and a reduced level of PR2 and PR5 after pathogen infection (Cao et al., 1994; Delaney et al., 1995; Fig. 8).
Figure 7.
Expression of AtSNAP33 and PR1 transcripts in leaves of Arabidopsis Col-0 plants (Col), and cpr1 mutants. Leaves were collected, the RNA was extracted, and AtSNAP33 and PR1 mRNA levels were detected by RNA-blot hybridization. The RNA gel stained with ethidium bromide is shown as a control for loading.
Figure 8.
Time course of expression of AtSNAP33 and PR1 transcripts after inoculation of Arabidopsis npr1 mutants (A) and Col-0 plants (B) with P. parasitica isolate NOCO. Leaves were inoculated with P. parasitica isolate NOCO (N) or water as a control (C) and were collected after different times in days as indicated. The RNA was extracted and AtSNAP33 and PR1 mRNA levels were detected by RNA-blot hybridization. The RNA gel stained with ethidium bromide is shown as a control for loading.
The Expression of AtSNAP33 Is Induced after Mechanical Stimulation and Wounding
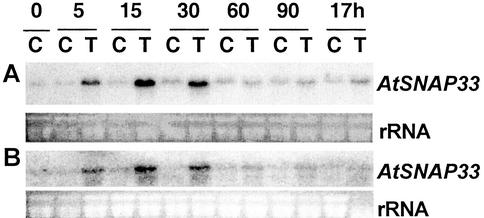
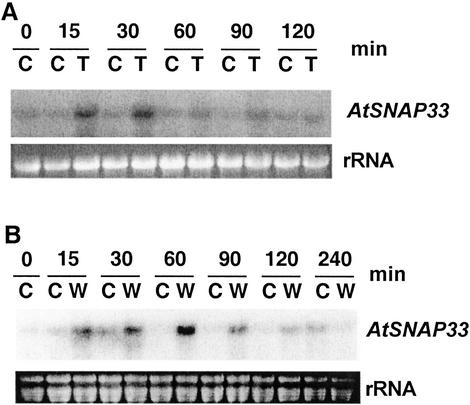
The level of AtSNAP33 transcripts increased transiently 5 min after a mechanical stimulation that consisted in gently rubbing the leaves 10 times between the thumb and forefinger, reached a maximum between 15 to 30 min and decreased to control levels after 60 min (Fig. 9A). To test if the enhanced expression was due to breakage of trichomes, which occurs by touching the leaves, the expression of AtSNAP33 was analyzed after mechanical stimulation in Arabidopsis glabrous1 mutant plants that bear almost no trichomes (Koornneef et al., 1982). The expression of AtSNAP33 in glabrous1 followed a similar transient increase as in wild-type Col-0 plants (Fig. 9B). The expression of AtSNAP33 was not increased in upper systemic leaves after mechanical stimulation of the lower leaves (data not shown).
Figure 9.
Time course of expression of AtSNAP33 transcripts after mechanical stimulation of Arabidopsis Col-0 (A) and glabrous1 mutant (B) plants. Leaves were touched (T) or not touched as a control (C) and were collected at time 0 and after 5, 15, 30, 60, and 90 min, and 17 h. The RNA was extracted and the AtSNAP33 mRNA level was detected by RNA-blot hybridization. The RNA gel stained with ethidium bromide is shown as a control for loading.
The effect of two other mechanical stimulations on the expression of AtSNAP33 was also tested. Wind, blown by a hair drier, caused a transient increase of AtSNAP33 expression between 15 min and 30 min after beginning the treatment compared with control plants; the expression returned to control levels after 60 min (Fig. 10A). AtSNAP33 expression was stimulated transiently after wounding with pliers. It increased after 15 min, reached a maximum after 60 min, and returned to levels of unwounded leaves 240 min after the beginning of the experiment (Fig. 10B).
Figure 10.
Time course of expression of AtSNAP33 transcripts after exposure to wind (A) and wounding (B). A, Leaves were exposed to a cool stream of air from a hair drier (T) or not exposed as a control (C) and were collected after different times in minutes as indicated. B, Leaves were wounded (W) or not wounded as a control (C) and were collected after different times in minutes as indicated. A and B, The RNA was extracted and the AtSNAP33 mRNA level was detected by RNA-blot hybridization. The RNA gel stained with ethidium bromide is shown as a control for loading.
DISCUSSION
In this paper, we show that the expression of the t-SNARE AtSNAP33 increased after inoculation with virulent pathogens such as P. tabacinum or virulent and avirulent forms of P. parasitica or P. syringae. In addition, we describe induction of AtSNAP33 after mechanical stimulation, wind, and wounding.
The expression of AtSNAP33 increased after inoculation with the pathogen P. parasitica isolates NOCO and EMWA, as well as P. tabacinum. There was a faster increase in the expression of AtSNAP33 in the incompatible interaction with P. parasitica isolate EMWA than in the compatible interaction with isolate NOCO.
We propose that this reflects the overall secretory activity during the infection process. In the incompatible interaction with isolate EMWA, the hyphae grew into the tissue and formed haustoria until 3 dpi and then growth stopped. The expression of AtSNAP33 increased transiently within the same time frame. In the compatible interaction with isolate NOCO, the tissue was increasingly colonized from 2 to 7 dpi. This was accompanied by a steady increase in the expression of AtSNAP33. Haustoria were continuously formed by the growing hyphae. These specialized structures grow into plant cells without rupturing the plasma membrane and are surrounded by an invagination of a modified form of the host plasma membrane, the extrahaustorial matrix. The growth of the plasma membrane that surrounds the developing haustoria most likely occurs by fusion of vesicles, the membrane of the vesicle becoming part of the extrahaustorial membrane (Callow and Green, 1996). This may require an increased level of components of the vesicle fusion machinery such as AtSNAP33.
Similarly, an enhanced level of the t-SNARE AtSNAP33 can be expected during pathogen infection when secretion of specific proteins is increased, probably leading to more vesicle fusion events with the plasma membrane. Analysis by western blots of intercellular fluids showed that there was an increased secretion of PR1 after inoculation.
In the compatible interaction with P. parasitica isolate NOCO, the expression of PR1 increased steadily starting 4 dpi until 6 dpi. It is interesting to note that the increase in the expression of AtSNAP33 in the same plants preceded the increase in the expression of PR1, indicating that it does not result from a feedback mechanism whereby an increased secretion would lead to an increased expression of AtSNAP33. Moreover, in the incompatible interaction with P. parasitica isolate EMWA, the level of AtSNAP33 transcripts returned to control levels when the level of PR1 transcripts was still high. These differences in the expression of AtSNAP33 and PR1 suggest that the expression of the two genes is regulated by different signaling pathways.
The local expression of AtSNAP33 in infected leaves was slightly decreased in NahG plants, as well as in eds5 and sid2 mutants, whereas systemic induction was abolished. This demonstrates a partial independence of AtSNAP33 induction on SA and stands in contrast with the expression of PR1 that is fully SA dependent (Delaney et al., 1994; Nawrath and Métraux, 1999). This most likely reflects different signaling processes that take place in infected leaves versus uninfected systemic leaves.
It was found serendipitously that the expression of AtSNAP33 increased after mechanical stimulation. The expression of AtSNAP33 shows the same characteristics as the expression of other genes induced by mechanical stimulation, including the TCH genes (Braam et al., 1997), i.e. the increase in the level of transcript was high, rapid, and transient. When the leaves were mechanically stimulated once every day for 35 d, there was a decrease in the height of the inflorescence stem (reaching 26% of control, untouched plants; data not shown). Thus, mechanical stimulation leads to changes in development that may be related to cell wall modifications. The increased expression of AtSNAP33 after mechanical stimulation may reflect an increase in secretion of cell wall components. TCH4, which is induced after mechanical stimulation, encodes a xyloglucan endotransglycosylase and possibly functions in cell wall modifications after mechanical stimulation, leading to changes in development (Xu et al., 1995; Braam et al., 1997).
It is unlikely that breakage of trichomes is the perceived stimulus leading to wound-induced expression of AtSNAP33 because mechanical stimulation of glabrous1 mutants that bear almost no trichomes (Koorneef et al., 1982) gave a similar result to wild-type plants.
The expression of AtSNAP33 increased transiently after wounding, similar to the increase after mechanical stimulation. Because the expression of AtSNAP33 was back to control levels 240 min after wounding, it may be a consequence of the mechanical stimulation occurring during wounding rather than a response to the damage provoked by the wound.
It was shown previously that the expression of the endoplasmic reticulum resident calreticulin, BiP, and protein disulfide isomerase was induced after different treatments, including SA (Denecke et al., 1995). Moreover, HvGRP94, a barley endoplasmic reticulum resident protein is induced by inoculation with Erysiphe graminis (Walther-Larsen et al., 1993). In a detailed study of the effect of cell wall-degrading enzymes mimicking bacterial inoculation on BiP expression, Jelitto-van Dooren et al. (1999) showed that the expression of BiP increases locally and systemically and is not dependent on SA. The expression of BiP was also induced by wounding (Kalinski et al., 1995; Leyman et al., 2000). Thus, components of the entry point of the secretory pathway are up-regulated by different stresses. Our study shows that AtSNAP33, a SNARE component of the exit point of the secretory pathway, transport to the plasma membrane and to the phragmoplast, is also up-regulated by biotic stresses, mechanical stimulation, and wounding. Whether this is a general response of components of the secretory pathway remains to be determined. In this regard, it would be interesting to analyze the expression of other SNAREs after biotic and abiotic stresses. The syntaxin Nt-Syr1 was shown to be regulated by abscisic acid, jasmonic acid, SA, salt stress, and wounding (Leyman et al., 2000). The increased expression of BiP (Kalinski et al., 1995), Nt-Syr1 (Leyman et al., 2000), and AtSNAP33 after wounding may reflect an increased secretion of protective compounds at the site of repair (Blatt et al., 1999).
As AtSNAP33 is a t-SNARE localized at the cell plate and at the plasma membrane (Heese et al., 2001), we hypothesize that it is involved in vesicle fusion for the formation of the cell plate after cell division and for secretion of extracellular components. After pathogen attack, increased vesicle fusion may be required to permit increased secretion of PR proteins. In addition, increased vesicle fusion may also be required for repair of damage to the plasma membrane provoked by reactive oxygen species. Reactive oxygen species are produced after pathogen attack and mechanical stimulation (Yahraus et al., 1995). It has been shown in sea urchin eggs that SNAP-25 is required for membrane resealing after injury (Steinhardt et al., 1994). Moreover, overexpression of AtVAMP7, an Arabidopsis v-SNARE, prevented H2O2-induced apoptosis in yeast and Arabidopsis cells (Levine et al., 2001). It is not known if the induction of expression of AtSNAP33 after pathogen inoculation and mechanical stimulation is linked to its function in cell division.
In the future, it will be interesting to analyze the signal transduction pathway leading to the expression of AtSNAP33 after pathogen inoculation and mechanical stimulation, as well as to determine the implication of this SNARE in defense mechanisms.
MATERIALS AND METHODS
Plant Material, Mechanical Stimulation, Wind Treatment, and Wounding
Arabidopsis accession Col-0 plants were grown in growth chambers under short-day conditions (12 h of light/12 h of dark) at 20°C. For the mechanical stimulation, six leaves of 4-week-old plants were gently rubbed with ungloved fingers 10 times over the entire leaf surface, unless otherwise stated. For wind treatment, 4-week-old plants were exposed to a cool stream of air from a hair drier at a distance of 20 cm for 1 min. Plants were wounded by squeezing the leaves between flat-bladed pliers at two different sites. The npr1 and cpr1 mutants were kindly provided by Dr. X. Dong (Duke University, Durham, NC), and the eds5 and sid2 mutants were kindly provided by Dr. C. Nawrath (University of Fribourg, Fribourg, Switzerland).
Inoculation with Peronospora parasitica Isolates NOCO and EMWA, Pseudomonas syringae pv tomato DC 3000, and Pseudomonas syringae pv tomato DC 3000 avrRtp2, and Plectosporium tabacinum
Three-week-old Arabidopsis plants, grown in pots (20–30 plants per pot) were inoculated by spraying with a conidial suspension (105–106 conidia mL−1 tap water) of P. parasitica isolate NOCO or EMWA prepared as described (Mauch-Mani and Slusarenko, 1994). After the treatment, the plants were kept in a growth chamber under short-day conditions, high air humidity, and at 18°C. The control plants were treated with water.
P. syringae pv tomato DC 3000 and P. syringae pv tomato DC 3000 avrRpt2 were grown overnight in Miller's Luria broth base liquid medium (Sigma, Buchs, Switzerland) at 28°C. A dilution of 2 × 108 bacteria mL−1 water was introduced by infiltration with a syringe through the lower side of leaves of 4- to 5-week-old Arabidopsis plants grown as one plant per pot. Five leaves per plant were inoculated, subsequently harvested, and analyzed. The control plants were infiltrated with water. After the inoculation, the plants were kept in the growth chamber until collection of the inoculated leaves, and respectively, the uninoculated systemic leaves.
P. tabacinum, kindly provided by Dr. B. Mauch-Mani (University of Neuchâtel, Neuchâtel, Switzerland), was kept in petri plates on potato-dextrose-agar medium (Difco' Chemie Brunschwig, Basel). To obtain conidia, the surface was scraped with an inoculation loop and the conidia were suspended in water. Conidia were counted under a microscope, and a suspension of 2 × 106 conidia mL−1 was prepared in water and sprayed onto 3-week-old plantlets grown in pots containing 20 to 30 plantlets per pot. After the treatment, the plants were kept in a growth chamber under short-day conditions, high air humidity, and at 18°C. The control plants were treated with water.
Chemical Treatments
Three-week-old Arabidopsis Col-0 plants grown in pots with 20 to 30 plants per pot were soil-drenched with a final concentration of 330 μm BTH (Bion 50 WG; Syngenta, Research Triangle Park, NC) and 330 μm or 1 mm SA, and for controls, plants were treated with wetting powder or H2O, respectively.
RNA Preparation, RNA-Blot Analysis, and Probes
For RNA extraction, plant tissues were harvested, frozen, and pulverized in liquid nitrogen. One volume of 2 m Tris, pH 8, containing 0.5 m EDTA, pH 8, and 20% (v/v) SDS (1:2:1) at 95°C was added, followed by one volume of saturated phenol:chloroform:isoamyl alcohol (v/v; 25:24:1) at 40°C. The two phases were separated by centrifugation, and the aqueous phase containing the RNA was mixed with 1 volume of chloroform and separated by centrifugation. The RNA was precipitated overnight with 1 volume of 6 m LiCl at 4°C, washed with 70% (v/v) ethanol, and resuspended in water. For RNA-blot analysis, 6 μg of total RNA was applied on a 1% (w/v) formaldehyde-agarose gel, separated, and transferred to a nylon membrane (Hybond-N; Amersham, Piscataway, NJ) as described by Sambrook et al. (1989). The membranes were hybridized overnight at 65°C with a DNA probe made by random primed labeling in the presence of α-32P dCTP with the RadPrimed DNA Labeling System according to manufacturer's instructions (Invitrogen, Basel). The membranes were washed subsequently for 5 min with 2× SSC containing 0.1% (w/v) SDS and then three times for 20 min with 0.2× SSC containing 0.1% (w/v) SDS at 65°C before being exposed to film (X-Omat; Kodak, Denges, Switzerland). For the PR1 probe, the cDNA was used for the labeling reaction. For the AtSNAP33 probe, the set of primers 5′-GAGTCGTCTCCGGGTTA-3′ and 5′-ACTACGGAAATTGTT-CTTG-3′ were used to amplify 500 bp in the 5′ region of the AtSNAP33 cDNA by PCR and cloned in pGEM-T (Promega, Wallisellen, Switzerland) according to manufacturer's instructions.
Protein Extraction, Intercellular Fluid, and Protein-Blot Analysis
Arabidopsis tissues were harvested, frozen, and ground at 4°C with a mortar and a pestle in 2 mL of SDS-PAGE sample buffer g−1 fresh weight. The homogenate was centrifuged for 10 min at 14,000 rpm in an Eppendorf 5415C centrifuge at 4°C and the supernatant was collected.
To obtain the intercellular fluid, leaves were infiltrated under vacuum for 2 min in 50 mm Tris, pH 7.4, containing 150 mm NaCl. The surface of the leaves was dried and the rolled leaves were centrifuged in a syringe placed in a centrifuge tube for 20 min at 150g. The fluid was collected and volumes corresponding to equal amounts of leaf fresh weight were analyzed by SDS-PAGE.
Proteins were fractionated on a 13% (w/v) SDS-PAGE gel according to Sambrook et al. (1989) using a Mini-Protean II apparatus (Bio-Rad, Reinach, Switzerland). Gels were blotted onto a BA85-S nitrocellulose membrane (Schleicher & Schuell, Bottmingen, Switzerland) and membranes were incubated with antibodies as described (Sambrook et al., 1989). Rabbit anti-AtSNAP33 serum was diluted 1:2,000. Goat anti-PR1 immunopurified serum kindly provided by L. Friedrich (Syngenta) was diluted 1:1,000. Alkaline phosphatase- or peroxidase-conjugated secondary antibodies were diluted according to the manufacturer's instructions (Dako, Zug, Switzerland). For alkaline phosphatase, the reaction was visualized in 10 mm Tris buffer, pH 9.6, containing 100 mm NaCl, 5 mm MgCl2, 0.33 mg mL−1 sodium 5-bromo-4-chloro-3-indolyl-phosphate (Eurobio, Chemie Brunschwig, Basel) dissolved in dimethylformamide, and 0.66 mg mL−1 4-nitroblue tetrazolium chloride (Fluka, Buchs, Switzerland) dissolved in 50% (w/v) dimethylformamide. For peroxidase, the reaction was visualized in 50 mm Tris, pH 7.6, containing 0.6 mg mL−1 3,3′-diaminobenzidine tetrahydrochloride (Sigma, Buchs, Switzerland) and 0.03% (w/v) H2O2.
Figure 4.
Time course of expression of AtSNAP33 and PR1 transcripts in inoculated (A) and uninoculated systemic (B) leaves of nahG plants after inoculation with P. syringae pv tomato DC3000 (D) or DC3000 avrRpt2 (R) or water as a control (C). Leaves were inoculated with the bacteria or water, and inoculated as well as uninoculated systemic leaves were collected after different times in days. The RNA was extracted and AtSNAP33 and PR1 mRNA levels were detected by RNA-blot hybridization. The RNA gel stained with ethidium bromide is shown as a control for loading.
ACKNOWLEDGMENTS
We thank Laurence Charrier and Christophe Folly for excellent technical assistance. We also thank Xinnian Dong (Duke University) for kindly providing npr1 and cpr1, Christiane Nawrath (University of Fribourg) for kindly providing eds5 and sid2, Leslie Friedrich (Syngenta) for kindly providing the PR1 cDNA and anti-PR1 antibodies, and Brigitte Mauch-Mani (University of Neuchâtel) for kindly providing Plectosporium tabacinum. We thank Jean-Pierre Métraux (University of Fribourg) for critical reading of the manuscript and stimulating discussions.
Footnotes
This work was supported by the Swiss National Foundation for Scientific Research (grant no. 31–39595.93 to L.S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.012633.
LITERATURE CITED
- Blatt MR, Leyman B, Geelen D. Tansley review no. 108: molecular events of vesicle trafficking and control by SNARE proteins in plants. New Phytol. 1999;144:389–418. doi: 10.1046/j.1469-8137.1999.00546.x. [DOI] [PubMed] [Google Scholar]
- Botella JR, Arteca RN, Frangos JA. A mechanical strain-induced 1-aminocyclopropane-1-carboxylic acid synthase gene. Proc Natl Acad Sci USA. 1995;92:1595–1598. doi: 10.1073/pnas.92.5.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell. 1994;6:1845–1857. doi: 10.1105/tpc.6.12.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam J, Davis RW. Rain-, wind-, and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell. 1990;60:357–364. doi: 10.1016/0092-8674(90)90587-5. [DOI] [PubMed] [Google Scholar]
- Braam J, Sistrunk ML, Polisensky DH, Xu W, Purugganan MM, Antosiewicz DM, Campbell P, Johnson KA. Plant responses to environmental stress: regulation and functions of the Arabidopsis TCH genes. Planta. 1997;203:S35–S41. doi: 10.1007/pl00008113. [DOI] [PubMed] [Google Scholar]
- Callow JA, Green JR. The plant plasma membrane in fungal disease. In: Smallwood M, Knox J-P, Bowles DJ, editors. Membranes: Specialized Functions in Plants. Oxford: Bios Scientific Publishers; 1996. pp. 543–562. [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney TP, Friedrich L, Ryals JA. Arabidopsis signal transduction mutant defective in chemically and biologically induced resistance. Proc Natl Acad Sci USA. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E et al. A central role of salicylic acid in plant disease resistance. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- Denecke J, Carlsson LE, Vidal S, Höglund A-S, Ek B, van Zeijl MJ, Sinjorgo KMC, Palva ET. The tobacco homolog of mammalian calreticulin is present in protein complexes in vivo. Plant Cell. 1995;7:391–406. doi: 10.1105/tpc.7.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depège N, Thonat C, Julien J-L, Boyer N. Thigmomorphogenesis: modification of calmodulin mRNA and protein levels in tomato plants. J Plant Physiol. 1999;155:561–567. [Google Scholar]
- Fukuda R, McNew JA, Weber T, Parlati F, Engel T, Nickel W, Rothman JE, Söllner TH. Functional architecture of an intracellular membrane t-SNARE. Nature. 2000;407:198–202. doi: 10.1038/35025084. [DOI] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. Requirement of salicylic acid for the induction of systemic acquired resistance. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- Geelen D, Leyman B, Batoko H, Di Sansabastiano G-P, Moore I, Blatt I. The abscisic acid-related SNARE homolog NtSyr1 contributes to secretion and growth: evidence from competition with its cytosolic domain. Plant Cell. 2002;14:387–406. doi: 10.1105/tpc.010328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay JC, Scheller RH. SNAREs and NSF in targeted membrane fusion. Curr Opin Cell Biol. 1997;9:505–512. doi: 10.1016/s0955-0674(97)80026-9. [DOI] [PubMed] [Google Scholar]
- Heese M, Gansel X, Sticher L, Wick P, Grebe M, Granier F, Jürgens G. Functional characterization of the KNOLLE-interacting t-SNARE AtSNAP33 and its role in plant cytokinesis. J Cell Biol. 2001;155:239–249. doi: 10.1083/jcb.200107126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holub EB, Beynon JL, Crute IR. Phenotypic and genotypic characterization of interactions between isolates of Peronospora parasitica and accessions of Arabidopsis thaliana. Mol Plant-Microbe Interact. 1994;7:223–229. [Google Scholar]
- Jaffe MJ, Forbes S. Thigmomorphogenesis: the effect of mechanical perturbation on plants. Plant Growth Regul. 1993;12:313–324. doi: 10.1007/BF00027213. [DOI] [PubMed] [Google Scholar]
- Jahn R, Südhof TC. Membrane fusion and exocytosis. Annu Rev Biochem. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863. [DOI] [PubMed] [Google Scholar]
- Jelitto-van Dooren EPWM, Vidal S, Denecke J. Anticipating endoplasmic reticulum stress: a novel early response before pathogenesis-related gene induction. Plant Cell. 1999;11:1935–1943. doi: 10.1105/tpc.11.10.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinski A, Rowley DL, Loer DS, Foley C, Buta G, Herman E. Binding-protein expression is subject to temporal, developmental and stress-induced regulation in terminally differentiated soybean organs. Planta. 1995;195:611–621. doi: 10.1007/BF00195722. [DOI] [PubMed] [Google Scholar]
- Kargul J, Gansel X, Tyrrell, Sticher L, Blatt MR. Protein-binding partners of the tobacco syntaxin NtSyr1. FEBS Lett. 2001;16:253–258. doi: 10.1016/s0014-5793(01)03089-7. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Dellaert LWM, van der Keen JH. EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. Mutat Res. 1982;93:109–123. doi: 10.1016/0027-5107(82)90129-4. [DOI] [PubMed] [Google Scholar]
- Levine A, Belenghi B, Damari-Weisler, Granot D. Vesicle-associated membrane protein of Arabidopsis suppresses Bax-induced apoptosis in yeast downstream of oxidative burst. J Biol Chem. 2001;276:46284–46289. doi: 10.1074/jbc.M107375200. [DOI] [PubMed] [Google Scholar]
- Leyman B, Geelen D, Blatt MR. Localization and control of expression of Nt-Syr1, a tobacco snare protein. Plant J. 2000;24:369–381. doi: 10.1046/j.1365-313x.2000.00886.x. [DOI] [PubMed] [Google Scholar]
- Malamy J, Carr JP, Klessig DF, Raskin I. Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science. 1990;250:1002–1004. doi: 10.1126/science.250.4983.1002. [DOI] [PubMed] [Google Scholar]
- Mauch F, Kmecl A, Schaffrath U, Volrath S, Görlach J, Ward E, Ryals J, Dudler R. Mechanosensitive expression of a lipoxygenase gene in wheat. Plant Physiol. 1997;114:1561–1566. doi: 10.1104/pp.114.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch-Mani B, Slusarenko AJ. Systemic acquired resistance in Arabidopsis thaliana induced by a predisposing infection with a pathogenic isolate of Fusarium oxysporum. Mol Plant-Microbe Interact. 1994;7:378–383. [Google Scholar]
- Métraux J-P, Signer H, Ryals J, Wyss-Beuz M, Gaudin J, Raschdorf K, Schmid E, Blum W, Inverardi B. Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science. 1990;250:1004–1006. doi: 10.1126/science.250.4983.1004. [DOI] [PubMed] [Google Scholar]
- Mitchell CA, Myers PN. Mechanical stress regulation of plant growth and development. Hort Rev. 1995;17:1–42. doi: 10.1002/9780470650585.ch1. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi-Shinozaki K, Matsumoto K, Shinozaki K. A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:765–769. doi: 10.1073/pnas.93.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Métraux J-P. Salicylic acid induction-deficient mutants of Arabidopsis express PR2 and PR5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JE, Szabo V, Staskawicz BJ, Lister C, Dean C, Daniels MJ, Jones JDG. Phenotypic characterization and molecular mapping of the Arabidopsis thaliana locus RPP5, determining disease resistance to Peronospora parasitica. Plant J. 1993;4:821–831. [Google Scholar]
- Rothman JE. The protein machinery of vesicle budding and fusion. Protein Sci. 1996;5:185–194. doi: 10.1002/pro.5560050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanderfoot AA, Assaad FF, Raikhel NV. The Arabidopsis genome: an abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 2000;124:1558–1569. doi: 10.1104/pp.124.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Raikhel NV. The specificity of vesicle trafficking: coat proteins and SNAREs. Plant Cell. 1999;11:629–641. doi: 10.1105/tpc.11.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhardt RA, Bi G, Alderton JM. Cell membrane resealing by a vesicular mechanism similar to neurotransmitter release. Science. 1994;263:390–393. doi: 10.1126/science.7904084. [DOI] [PubMed] [Google Scholar]
- Takezawa D, Liu ZH, An G, Poovaiah BW. Calmodulin gene family in potato: developmental and touch-induced expression of the mRNA encoding a novel isoform. Plant Mol Biol. 1995;27:693–703. doi: 10.1007/BF00020223. [DOI] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. Acquired resistance in Arabidopsis. Plant Cell. 1992;4:645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Luit AH, Olivari C, Haley A, Knight MR, Trewavas AJ. Distinct calcium signaling pathways regulate calmodulin gene expression in tobacco. Plant Physiol. 1999;121:705–714. doi: 10.1104/pp.121.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, van Strien EA. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR1 type proteins. Physiol Mol Plant Pathol. 1999;55:85–97. [Google Scholar]
- Walther-Larsen H, Brandt J, Collinge DB, Thordal-Christensen H. A pathogen-induced gene of barley encodes a HSP90 homologue showing striking similarity to vertebrate forms resident in the endoplasmic reticulum. Plant Mol Biol. 1993;21:1097–1108. doi: 10.1007/BF00023606. [DOI] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J. Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell. 1995;7:1555–1567. doi: 10.1105/tpc.7.10.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahraus T, Chandra S, Gendre L, Low P. Evidence for a mechanically induced oxidative burst. Plant Physiol. 1995;109:1259–1266. doi: 10.1104/pp.109.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalpani N, Silverman P, Wilson MA, Kleier DA, Raskin Y. Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco. Plant Cell. 1991;3:809–818. doi: 10.1105/tpc.3.8.809. [DOI] [PMC free article] [PubMed] [Google Scholar]