Abstract
In this paper, we report the cloning and characterization of the plastid-located glutamine synthetase (GS) of Medicago truncatula Gaertn (MtGS2). A cDNA was isolated encoding a GS2 precursor polypeptide of 428 amino acids composing an N-terminal transit peptide of 49 amino acids. Expression analysis, by Westerns and by northern hybridization, revealed that MtGS2 is expressed in both photosynthetic and non-photosynthetic organs. Both transcripts and proteins of MtGS2 were detected in substantial amounts in root nodules, suggesting that the enzyme might be performing some important role in this organ. Surprisingly, about 40% of the plastid GS in nodules occurred in the non-processed precursor form (preGS2). This precursor was not detected in any other organ studied and moreover was not observed in non-fixing nodules. Cellular fractionation of nodule extracts revealed that preGS2 is associated with the plastids and that it is catalytically inactive. Immunogold electron microscopy revealed a frequent coincidence of GS with the plastid envelope. Taken together, these results suggest a nodule-specific accumulation of the GS2 precursor at the surface of the plastids in nitrogen-fixing nodules. These results may reflect a regulation of GS2 activity in relation to nitrogen fixation at the level of protein import into nodule plastids.
Glutamine synthetase (GS, EC 6.3.1.2) is an essential enzyme in nitrogen metabolism of higher plants (Miflin and Lea, 1980). In conjunction with Glu synthase (EC 1.4.1.14 and EC 1.4.7.1), it catalyzes the assimilation of ammonium into Gln and Glu, which then serve as the nitrogen donors for the biosynthesis of all nitrogenous organic compounds in the plant. GS is an octameric enzyme represented by a number of isoenzymes located both in the cytosol (GS1) and in the plastids (GS2). These isoenzymes are derived from the differential expression of a small family of nuclear genes (Forde and Cullimore, 1989; McGrath and Coruzzi, 1991). In legumes, GS plays a key role in root nodules being responsible for the assimilation of ammonia that is released at high rates by nitrogen-fixing rhizobia (Atkins 1987).
The legume Medicago truncatula is being extensively used for studies on symbioses due to its small genome and ease of manipulation, and a variety of genetic and genomic tools have been developed for this model plant (Barker et al., 1990; Cook, 1999; Bell et al., 2001; Journet et al., 2002; Thoquet et al., 2002). Studies on GS in M. truncatula have revealed only two expressed genes, MtGSa and MtGSb, encoding cytosolic polypeptides; a third cytosolic GS gene, MtGSc, appears not to be expressed. Both MtGSa and MtGSb are induced during symbiotic root nodule development, although to different extents (Stanford et al., 1993). Cellular expression studies have shown that they have different but partially overlapping patterns of expression in nodules (Carvalho et al., 1997, 2000a, 2000b). MtGSa is highly expressed in infected cells and is presumed to play the major role in the assimilation of ammonium derived from dinitrogen fixation (Carvalho et al., 2000a).
Studies on GS isoezymes in M. truncatula have revealed that a considerable proportion (about 20%) of the plant GS activity in nodules is attributed to the plastid form (Carvalho et al., 1997). Work on other higher plants has shown that this form, which is expressed predominantly in leaves, is responsible for the reassimilation of photorespiratory ammonia (Wallsgrove et al., 1987; Migge and Becker, 2000; Orea et al., 2002), and it has also been implicated in the assimilation of ammonia reduced from nitrate and nitrite (Vézina et al., 1987). In root nodules, its role is unknown.
Like most plastid proteins, GS2 is a nuclear-encoded protein initially synthesized in the cytosol as a higher molecular mass precursor polypeptide containing a cleavable N-terminal extension, the transit peptide (Lightfoot et al., 1988; Tingey et al., 1988). The transit peptide mediates routing to the inside of the organelle where it is cleaved off by stromal processing peptidases (Keegstra and Cline, 1999; May and Soll, 1999). Inside the organelles, the GS2 polypeptides presumably assemble to form the catalytically active octameric enzyme.
In this work, we have extended our knowledge on the GS gene family of M. truncatula by the cloning and characterization of the plastid-located GS. Special attention was devoted to its regulation and potential role in root nodules. Surprisingly, this work revealed an accumulation of the GS2 precursor specifically in root nodules. We have evaluated the accumulation of this precursor protein as it relates to nitrogen fixation and nodule development.
RESULTS
Isolation and Characterization of a cDNA Encoding M. truncatula Plastid GS
To complete the characterization of the GS multigene family of M. truncatula, we set out to isolate clones coding for the plastid-located isoenzyme. A root nodule cDNA library of M. truncatula (Gamas et al., 1996) was screened for GS2 clones by hybridization with a heterologous probe prepared from the plastid GS cDNA clone pcGS-δ1 from bean (Phaseolus vulgaris; Lightfoot et al., 1988). A clone containing a cDNA of about 1.6 kb was isolated (GenBank accession no. AY225150), and sequencing revealed a complete open reading frame encoding a polypeptide of 428 amino acids (47.1 kD). This sequence shows less than 70% similarity to the two cytosolic GS polypeptides of M. truncatula and matches more closely the plastid-located GS of alfalfa (Medicago sativa; Zozaya-Garza and Sengupta-Gopalan, 1999), pea (Pisum sativum; Tingey et al., 1988), and bean (Lightfoot et al., 1988) with 98%, 96%, and 91% similarity, respectively, at the amino acid level. The encoded polypeptide contains a 56-amino acid N-terminal extension relative to the start of the cytosolic GS polypeptides (Carvalho et al., 1997), which by comparison with other plastid-located GS sequences corresponds to the plastid targeting presequence. It is notable that this presequence is rich in Ser and Thr residues and contains no acidic amino acids, typical of the transit peptide of nuclear-encoded plastid precursor proteins (Karlin-Neumann and Tobin, 1986). An alignment of the M. truncatula GS2 protein with the plastid-located GS precursor of pea (Fig. 1) suggests a point of cleavage at amino acid 49 of the N-terminal extension (Tingey et al., 1988) leading to a molecular mass of the mature polypeptide (without the transit peptide) of 41.7 kD.
Figure 1.
Comparison of the N-terminal transit peptide sequences of GS2 from M. truncatula (MtGS2) and pea (PsGS2). The deduced transit peptides are shown in bold.
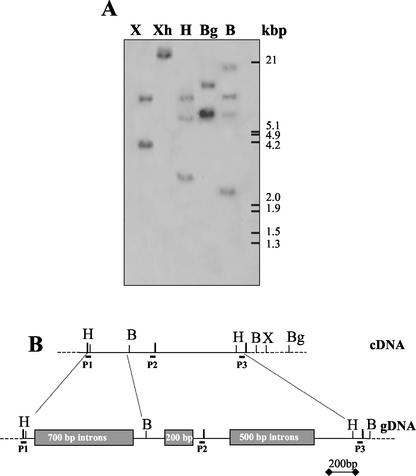
To determine the gene copy number of the plastid-located GS in M. truncatula, a Southern-blot analysis of genomic DNA was performed under stringent hybridization conditions using the M. truncatula GS2 cDNA as a probe (Fig. 2A). M. truncatula genomic DNA was digested with different restriction enzymes selected according to the cDNA restriction map. Considering the number of times that each restriction enzyme cuts the cDNA (Fig. 2B) and the number and size of the hybridizing genomic fragments, the results suggest either a single (or very few) intron-containing genes coding for M. truncatula GS2. The existence of introns in the M. truncatula GS2 gene was confirmed by PCR amplification of the genomic DNA. Two sets of primers were used to amplify this DNA, and after the digestion of the amplification products with HincII and BamHI, the existence of at least three introns in the gene was confirmed located between positions 200 and 1,080 of the 1,600-bp cDNA (Fig. 2B).
Figure 2.
Structure of the GS2 gene(s) of M. truncatula. A, Southern hybridization of M. truncatula DNA digested with different enzymes using a complete MtGS2 cDNA as probe. The positions of size markers are shown on the right. B, Restriction map of the MtGS2 cDNA and the corresponding genomic region (gDNA) deduced by PCR using primers P1, P2, and P3. The approximate sizes but not the number of introns are shown boxed. The enzymes are X, XbaI; Xh, XhoI; H, HincII; Bg, BglII; and B, BamHI.
Analysis of the M. truncatula expressed sequence tag (EST) databases (Bell et al., 2001; Journet et al., 2002) revealed that all of the current ESTs closely related to the M. truncatula GS2 cDNA cluster as a single gene entity (data not shown), further supporting the idea that M. truncatula contains a single expressed GS2 gene, which we have called MtGS2.
Expression of MtGS2 in Different Organs of M. truncatula
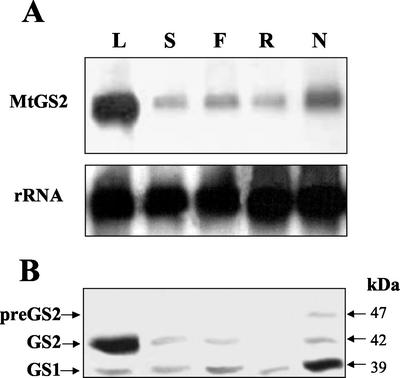
The expression of MtGS2 was examined in various organs of M. truncatula by northern analysis of total RNA isolated from leaves, stems, flowers, roots, and mature nodules (13 d postinoculation). The GS2 probe was shown to be highly specific for GS2 because it showed very poor cross-hybridization to the cytosolic GS cDNAs in Southern blots (data not shown) and because the pattern of expression in different organs using the GS2 probe was found to be different from those using the two cytosolic GS gene probes (see Stanford et al., 1993). The blots were also hybridized with a rRNA probe to evaluate the RNA loading. The GS2 hybridization signals were quantified, and the values were standardized against the rRNA hybridization data. The results (Fig. 3A) show that MtGS2 mRNA accumulated to more than 3-fold higher levels in leaves than in the other organs. Surprisingly, this mRNA was found to be more than 2.5-fold more abundant in nodules than in roots and was also higher than in stems and flowers. The high expression in leaves and expression in nodules, roots, and stems is moreover supported by a statistical analysis (Journet et al., 2002) of the distribution of GS2 ESTs in different cDNA libraries (data not shown).
Figure 3.
Expression analysis of MtGS2 in different organs of M. truncatula. A, Total RNA (15 μg lane−1) isolated from different organs was fractionated on formaldehyde-agarose gels. The gels were blotted onto nitrocellulose and hybridized to a 32P-labeled GS2 fragment (MtGS2) or a rRNA gene fragment (rRNA). B, Total soluble protein extracted from the same organs (30 μg lane−1) was fractionated by SDS-PAGE and transferred to nitrocellulose, and the membranes were probed with a GS antibody. GS1, GS2, and preGS2 designate cytosolic, plastid, and plastid precursor GS, respectively. L, Leaves; S, stems; F, flowers; R, roots; and N, nodules. Molecular masses are indicated.
Western analysis of protein extracts from the same organs was then used to evaluate the expression of the GS proteins (a representative blot is shown in Fig. 3B). The GS immunoreactive bands were quantified in three different membranes. The results revealed that all of the organs contained two types of GS polypeptide, one of around 39 kD corresponding to the cytosolic GS (GS1) and another of about 42 kD corresponding to the mature plastidic GS2. The amount of GS2 protein in each of these organs could be generally correlated with the amount of the corresponding transcript. However, in roots, GS2 polypeptides were hardly detected even though the amount of transcripts was considerable, and in leaves, the amount of the GS2 polypeptide accumulated to almost 5-fold higher levels compared with the other organs.
Interestingly, in root nodule extracts, the anti-GS antibody was able to detect a third polypeptide with a higher molecular mass than the cytosolic and mature plastidic GS polypeptides. This polypeptide was not detected in other organs of the plant (Fig. 3B; H.G. Carvalho, L.M. Lima, and J.V. Cullimore, unpublished data). Western experiments on roots and leaves of nodulated and non-nodulated plants suggest that it cannot be induced in these other organs by growth in symbiotic conditions (data not shown). Because the antiserum does not cross-react with rhizobial glutamine synthetase I encoded by a glnA gene or glutamine synthetase II encoded by a glnII gene (Cullimore and Miflin, 1984), this polypeptide must be related to M. truncatula GS. It has a molecular mass of around 47 kD, similar to the predicted size of the GS2 precursor, and it represents about 40% of the total GS2 polypeptides in the nodules (Fig. 3B).
Expression of GS2 Mature and Precursor Polypeptides in Escherichia coli
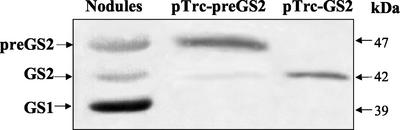
To examine whether this nodule-specific polypeptide could indeed be the GS2 precursor (preGS2), we expressed both the complete GS2 open reading frame encoding the precursor (pTrc-preGS2) and a cDNA encoding a mature GS polypeptide lacking the transit peptide (pTrc-GS2) in an E. coli mutant strain deficient for GS (glnA deletion).
The bacteria carrying the construct expressing the mature form of GS2 (pTrc-GS2) was able to complement the E. coli glnA mutation for growth on minimal medium containing ammonium as nitrogen source, whereas the construct expressing the precursor form (pTrc-preGS2) could only grow in the presence of a Gln supplement. These results indicate that the precursor polypeptides were not functional in E. coli, and GS activity determinations of the extracts revealed that the mature form was catalytically active, whereas no GS activity was found associated with the precursor. It was notable also that the precursor expressed poorly in E. coli, attaining only about 10% of the level of GS protein as the mature GS2 (see legend to Fig. 4).
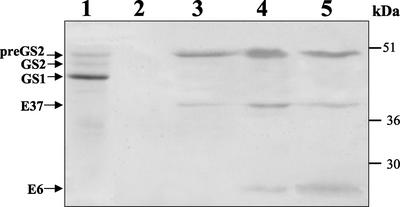
Figure 4.
Expression of GS2 and preGS2 in E. coli and comparison with nodule GS. Western immunodetection of GS polypeptides from a root nodule extract and from E. coli expressing pTrc-preGS2 and pTrc-GS2. GS1, GS2, and preGS2 designate cytosolic, plastidic, and plastid precursor GS, respectively. Due to the different levels of expression of GS in E. coli, different amounts of protein were loaded on the gel (nodule, 40 μg; pTrc-preGS2, 200 μg; and pTrc-GS2, 20 μg). Molecular masses are indicated.
The E. coli-produced polypeptides were analyzed by western blot. The bacteria harboring pTrc-preGS2 produced a 47-kD GS product that migrated in an identical position to the third GS polypeptide detected in the nodule extract, thus confirming that this nodule-specific polypeptide is the GS2 precursor (Fig. 4). A minor band with molecular mass of around 42 kD was also seen in this extract similar to native plastid mature GS2 from nodules and to the GS polypeptide expressed in E. coli harboring pTrc-GS2. This minor polypeptide probably results from a partial processing of the precursor polypeptide by the bacterial proteases in a way similar to the cleavage performed by the plant stromal processing peptidase.
Expression of MtGS2 during Nodule Development
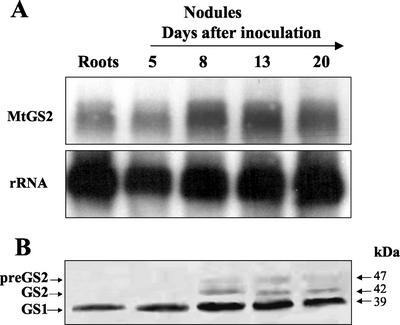
To examine more fully the high levels of MtGS2-specific mRNA and proteins detected in mature nodules and the nodule-specific accumulation of the plastid GS precursor, the expression of the GS2 gene was examined over a time course of nodule development. RNA and proteins were isolated from roots and from nodules harvested at different times after inoculation (5–20 d) and were analyzed and quantified by northern and western analysis as described above.
The results show that the abundance of MtGS2 mRNA increased about 2.5-fold during nodulation from a basal level detected in roots and at d 5 to a maximal level at d 8 after inoculation (Fig. 5A). At d 13 and 20 postinoculation, a slight and progressive decrease of the level of the GS2 mRNA was observed, such that at d 20, the level was about 85% of the maximum level.
Figure 5.
Expression of MtGS2 during nodule development. A, Total RNA (15 μg lane−1) isolated from roots and from nodules harvested from 5 to 20 d after inoculation with S. meliloti was fractionated on formaldehyde-agarose gels followed by RNA-blot hybridization using a 32P-labeled GS2 fragment (MtGS2) and a rRNA gene fragment (rRNA). B, Total soluble protein extracted from similar roots and nodules (30 μg lane−1) was fractionated by SDS-PAGE and transferred to nitrocellulose, and the membranes were probed with a GS antibody. GS1, GS2, and preGS2 designate cytosolic, plastid, and plastid precursor GS, respectively. Molecular masses are indicated.
Western-blot analysis of the same nodule samples revealed a general correlation between the levels of the GS2 polypeptide with the levels of its corresponding mRNA (Fig. 5B); the abundance of the GS2 polypeptides also increased markedly in abundance by d 8 followed by a slight reduction at d 13 and 20. It is noteworthy that qualitatively the temporal changes in abundance of the GS2 polypeptide were very similar to the GS1 polypeptides, however the quantitative analysis suggest that the latter are about 5-fold more abundant. The precursor polypeptide showed a slightly different pattern of expression; it was first observed at d 8 after inoculation and increased in abundance by d 13, followed by a reduction in level by d 20.
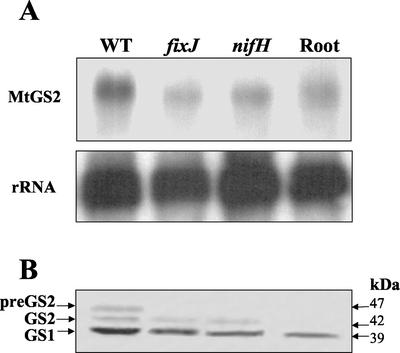
To evaluate whether the expression of MtGS2 and the accumulation of the GS2 precursor could be related to active nitrogen fixation, the expression of this gene was compared in both nitrogen-fixing (effective) and non-nitrogen-fixing (ineffective) nodules. RNA and proteins were extracted from 15-d-old nodules of plants inoculated with three different Sinorhizobium meliloti strains, an essentially wild-type strain and two Fix− strains (nifH and fixJ). The nifH gene encodes a subunit of nitrogenase, whereas fixJ encodes a regulator of nif and fix gene expression. The MtGS2 mRNA and polypeptides were quantified as before, and the results are shown in Figure 6.
Figure 6.
Expression of MtGS2 in effective and ineffective root nodules. A, Total RNA (15 μg lane−1) isolated from nodules inoculated with S. meliloti, wild-type (WT), fixJ, and nifH mutants, and from noninoculated roots was fractionated on formaldehyde-agarose gels followed by RNA-blot hybridization using a 32P-labeled GS2 fragment (MtGS2) and an rRNA gene fragment (rRNA). B, Total soluble protein extracted from roots and nodules produced by S. meliloti, wild-type (WT), fixJ, and nifH mutants (30 μg lane−1) was fractionated by SDS-PAGE and transferred to nitrocellulose, and the membrane was probed with a GS antibody. GS1, GS2, and preGS2 refer to the cytosolic, plastid, and plastid precursor GS, respectively. Molecular masses are indicated.
The quantitative analysis shows that there was no increase in the level of MtGS2 mRNA in nodules produced by the two S. meliloti Fix− strains compared with roots; the level of the GS2 mRNA in the nifH and fixJ nodules was 16% and 37% lower, respectively, than in roots. However, in contrast to roots, the mature GS2 polypeptide was easily detected in the ineffective nodules, although its abundance was almost 2-fold lower compared with wild-type nodules (Fig. 6B). The GS2 precursor polypeptide was not detected in the Fix− nodules.
Subcellular Location of the GS2 Precursor by Cell Fractionation
To determine the subcellular location of the GS2 precursor in root nodules, a M. truncatula nodule extract was fractionated by Percoll density gradient centrifugation. The fractions corresponding to the plastids and symbiosomes were identified using antibodies to two membrane markers: E37, a plastid outer envelope polypeptide (Joyard et al., 1982); and E6, a nodulin-26 polypeptide from the symbiosome membrane (Weaver et al., 1991).
After centrifugation on a self-generated Percoll density gradient, the fractions were collected and analyzed for GS, E6, and E37 immunoreactivity by western blots (Fig. 7). Although it is possible to detect the precursor polypeptide in the soluble fraction (fraction 1), most of this form of GS2 was located in the denser fractions of the nodules, suggesting an association with membranes. The plastid marker E37 was detected in fractions 3, 4, and 5 with relative amounts in these three fractions of 1:2.6:1.4. Previous studies have shown that the majority of nodule plastids are amyloplasts and that their starch content can be highly variable between cell types (Carvalho et al., 2000a). Thus the presence of the plastid membrane marker in the three fractions probably corresponds to amyloplasts with variable starch content and therefore with different densities. The symbiosome membrane marker (E6) was detected in fractions 4 and 5 but not in fraction 3. The GS precursor was detected in fractions 3, 4, and 5 with relative amounts of 1:2.3:1.3, respectively, thus showing less than 13% variation in comparison with the distribution of the plastid membrane marker. This close correlation and the lack of correlation with the symbiosome marker suggests an association of the GS2 precursor with the plastids. GS activity was determined in each of the fractions using the highly sensitive and highly specific GS transferase assay. Only fraction 1 contained detectable GS activity (data not shown), suggesting that the precursor is catalytically inactive.
Figure 7.
Localization of the GS2 precursor in nodule extracts fractionated by Percoll gradient centrifugation. Western immunodetection of GS, symbiosome membrane nodulin-26 (E6), and plastid membrane polypeptide (E37) in fractions of a nodule extract separated on a self-generated Percoll gradient. The fractions are numbered from the top to the bottom of the gradient. Equal amounts of total protein (50 μg) were loaded on a SDS-PAGE gel and transferred to nitrocellulose, and the membrane was probed with the different antibodies. Molecular mass markers are indicated.
Immunolocalization of GS Polypeptides
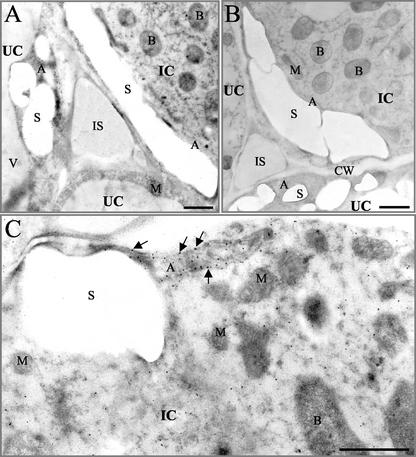
To determine whether the GS precursor could be visualized in association with the plastid membranes, GS was localized in root nodules by immunogold electron microscopy. As ascertained by western blot, the GS antibody recognizes both the cytosolic and plastidic GS polypeptides present in M. truncatula nodules. GS was found to be highly abundant in the amyloplasts of both infected and uninfected cells (Fig. 8A) and was also evident in the ground cytosol of both cell types as previously described by Carvalho et al. (2000a). In control sections, where nonimmune rabbit serum was used instead of the anti-GS primary antiserum, the general background of nonspecific labeling was very low (Fig. 8B). Gold particles were frequently observed at the periphery of the plastids on top of the membranes, indicating an association of GS with the plastid envelope (Fig. 8C).
Figure 8.
Immunogold localization of GS in root nodules of M. truncatula. A, Section through the central tissues of the nitrogen-fixing zone showing infected (IC) and uninfected (UC) cells and GS localization using immunogold (black dots). B, Similar nonimmune control section. C, High magnification using the GS antiserum. The amyloplast in an infected cell (IC) shows an association of the gold particles with the plastid membrane (arrows). IC, infected cell; UC, uninfected cell; B, rhizobial bacteroids; A, amyloplast; S, starch grain; M, mitochondria; IS, intercellular space; CW, cell wall; V, vacuole. Bar = 1 μm.
DISCUSSION
The importance of GS in plant nitrogen metabolism justifies the numerous studies performed on this enzyme. Particular interest has been devoted to the study of GS in leguminous plants where the enzyme plays a major role in the assimilation of symbiotically fixed nitrogen. In this paper, we report on the cloning and characterization of the plastid-located GS of the model legume M. truncatula and studies on the regulation of expression of this enzyme particularly in root nodules.
A combination of Southern and PCR analyses (Fig. 2) and examination of the EST databases is consistent with GS2 being encoded by a single intron-containing expressed gene in M. truncatula. Similar conclusions were obtained by molecular analysis of GS2 in pea (Tingey et al., 1988), bean (Cock et al., 1991), and Arabidopsis (Peterman and Goodman, 1991) and by genetic analysis of barley (Hordeum vulgare) and Lotus japonicus (Wallsgrove et al., 1987; Orea et al., 2002). However, in soybean (Glycine max) and alfalfa, several GS2 genes have been identified (Zozaya-Garza and Sengupta-Gopalan, 1999).
The identification of the plastid-located GS gene augments the characterization of the GS gene family of M. truncatula. Three cytosolic GS genes and one plastid GS gene have now been identified. To distinguish between plastid GS- and cytosolic GS-encoding genes we propose to rename the MtGSa, MtGSb, and MtGSc genes MtGS1a, MtGS1b, and MtGS1c, respectively, and the plastid GS encoding gene becomes MtGS2 (referred to as MtGSd in a previous publication; Carvalho et al., 2000b). MtGS1a, MtGS1b, and MtGS2 are clearly the three most highly expressed GS genes in M. truncatula, but whether there are additional, functional GS genes in this model legume must await further genomic analysis.
Expression analysis of MtGS2 in several organs of M. truncatula by western and northern hybridization (Fig. 3) revealed that the plastid GS is expressed in both photosynthetic and non-photosynthetic organs. Although highly abundant in leaves, the plastid GS was also found to be present in root nodules and to be more highly expressed in this organ than in roots. A more detailed study of nodule development (Fig. 5) revealed that the MtGS2-specific mRNA and polypeptide accumulated rapidly from basal levels in the root to more than 2-fold higher levels by d 8 after inoculation with S. meliloti. This pattern of expression parallels the kinetics of expression of the cytosolic GS gene MtGS1a (Stanford et al., 1993) and the late nodulin leghemoglobin Mtlb1 (Gallusci et al., 1991) from M. truncatula. These results suggest that the three genes may respond to the same physiological signals. Moreover, the level of MtGS2 mRNA was found to be enhanced only during effective rather than ineffective nodule development (Fig. 6), implying that the level of nitrogen fixation at least partially regulates the expression of this gene in root nodules of M. truncatula.
In general, in all of the experiments, the analysis of plastid GS polypeptides showed a similar pattern of changes in abundance to that observed for the mRNA, suggesting that the regulation of MtGS2 expression occurs primarily at the mRNA level. However, it is noteworthy that in roots, MtGS2 expression seems to be regulated posttranscriptionally because the GS2 polypeptide was hardly detected in this organ despite the accumulation of its mRNA (Fig. 3).
Although studies on other higher plants have established clearly a role of the plastid GS in green tissues in assimilating ammonium from photorespiration and also from nitrate/nitrite reduction, it is noteworthy that the plastid isoenzyme represents a considerable proportion of total GS activity in root nodules of M. truncatula (Carvalho et al., 1997). Careful examination of the literature reveals that plastid GS is also expressed well in nodules of other legume species such as pea (Tingey et al., 1987), bean (Bennett and Cullimore, 1989), and soybean (Brangeon et al., 1989), suggesting that this isoform is performing an important role within the nodule. Because cytosolic GS generally constitutes more than 80% of total nodule GS activity, it has generally been assumed that this isoform is responsible for the assimilation of the ammonia released by nitrogen fixation in legume root nodules (Forde et al., 1989; Datta et al., 1991; Carvalho et al., 2000a). However, we cannot exclude that this function is partly fulfilled by GS2 and that these two forms have partially redundant functions. Another possibility is that GS2 is performing some additional function in the plastids, presumably related to nitrogen fixation because of its expression pattern. Because GS2 is in the same subcellular compartment as porphobilinogen (PBG) deaminase, one likely function could be the assimilation of ammonia released by this enzyme during heme biosynthesis. In addition to acting as cofactors for cytochromes and other proteins, heme is required in nodules to produce leghemoglobin, the most abundant protein in infected cells. Although it was initially suggested that the heme moiety is synthesized by the bacteroids, there is now convincing evidence that this function is performed by the plant host (Santana et al., 1998). PBG deaminase has been shown to be up-regulated in nodules of both soybean and pea, and immunohistochemical studies have demonstrated that this enzyme is preferentially located in the central infected cells (Santana et al., 1998). GS2 shows a similar pattern of distribution in root nodules of M. truncatula (Carvalho et al., 2000a) and thus could conceivably play a role in the assimilation of ammonia produced by PBG deaminase during activation of the heme biosynthetic pathway during effective nodulation.
In addition to the cytosolic and mature plastidic GS, a higher Mr GS polypeptide was detected in nodules that by comparison with the size of the protein expressed in E. coli (Fig. 4) corresponds to the precursor to the plastid GS (preGS2). The accumulation of this precursor was specific to root nodules and moreover was not observed in non-fixing nodules formed with rhizobial Fix− mutants. The GS2 precursor appears to be bound to the plastid envelope, which was demonstrated both by cellular fractionation studies (Fig. 7) and by electron microscopy (Fig. 8).
The physiological relevance of the accumulation of the GS2 precursor in relation to nodule nitrogen metabolism is not clear. The precursor seems to be catalytically inactive both in the plant and when expressed in E. coli, thus its accumulation on the surface of the plastids is unlikely to provide a mechanism for channeling Gln into the plastids for Glu synthase activity. It may, however, represent a regulatory mechanism for controlling GS activity inside the plastids, and the fact that we only observed the precursor in nitrogen-fixing nodules suggests that this form of regulation is related to the nitrogen fixation process. We cannot, however, eliminate the possibility that the accumulation of the precursor is due to a “translocation failure,” perhaps due to the levels of ATP being too low to complete this process. Moreover, the absence of this protein in non-fixing nodules could be due to a higher protease activity in these early-senescing nodules rather than a more efficient translocation.
To our knowledge, this is the first report of an accumulation of a plastid-targeted precursor protein in planta. The precursors of plastid-located proteins are normally transient forms that are rapidly transported and processed to the mature forms within the organelle (Keegstra and Cline, 1999; May and Soll, 1999). However, it is well documented that some yeast and mammalian nuclear proteins are retained in the cytosol before import to the nuclei, and it has recently been shown that their import is regulated by protein phosphorylation (Hood and Silver, 1999). In vitro studies have shown that the transit peptides of chloroplast precursors can be phosphorylated and that this modification affects the efficiency of precursor import into plastids (Waegemann and Soll, 1996; Su et al., 2001). Waegemann and Soll (1996) have postulated that a cycle of phosphorylation/dephosphorylation might be a regulatory switch in the translocation pathway of plastid precursor proteins in vivo. By consequence, phosphorylated precursors might accumulate outside the plastids if the cycle is blocked. Our observation that the plastid GS precursor is accumulating in root nodules may represent the in vivo proof of the hypothesis raised by Waegemann and Soll (1996), and studies are currently being undertaken to evaluate the involvement of phosphorylation in the regulation of GS2 import into the plastids.
In conclusion, the work presented here has provided new insights into the functional significance and regulation of plastid-located GS in root nodules. An important role of GS2 for nodule functioning is postulated by this work, unequivocal proof requiring either silencing GS2 gene expression or isolating mutants lacking GS2 activity. The possibility of a regulation of GS2 activity at the level of protein import into the plastids opens the way to new experiments aimed at understanding the importance of this process for regulating the nitrogen assimilatory pathways and defining whether the mechanism of GS2 precursor accumulation involves protein phospho-rylation.
MATERIALS AND METHODS
Plant Material
Plants of Medicago truncatula Gaertn. cv Jemalong J5 were grown in aeroponic conditions at 20°C with a relative humidity of 75% and a 14-h light period. For nodule induction, the growth medium described by Lullien et al. (1987) was replaced with fresh medium lacking ammonium nitrate 2 d before inoculation with Sinorhizobium meliloti 2011 effective wild-type (GMI 708) and ineffective fixJ (GMI 347) and nifH (GMI 296) strains. Nodules were harvested at 0, 5, 8, 13, and 20 d after inoculation. Flowers, leaves, stems, and roots were collected from noninoculated plants. All plant material was immediately frozen in liquid nitrogen and stored at −70°C.
Isolation and Sequencing of MtGS2 cDNAs
A M. truncatula nodule cDNA library (Gamas et al., 1996) was screened by plaque hybridization using a 32P-labeled probe prepared from the EcoRV/XhoI fragment from the plastid-located GS cDNA clone pcGS-δ1 of bean (Phaseolus vulgaris; Lightfoot et al., 1988), essentially as described by Carvalho et al. (1997). The cDNA in pBluescript SK− were excised in vivo from the λ-ZAPII vector (Stratagene, La Jolla, CA) and sequenced on both strands using an automatic sequencer (Applied Biosystems, Foster City, CA).
Sequence and Database Analyses
Sequence analyses used the GCG programs (Genetic Computer Group, Madison, WI) and the PC gene computer program (IntelliGenetics, Mountain View, CA). Homology searches used the National Center for Biotechnology Information BLAST server (Altschul et al., 1997). Searches for GS2 EST sequences used both the National Center for Biotechnology Information EST database and the two M. truncatula EST databases: The Institute for Genomic Research Medicago truncatula gene index, v5.0 (http://www.tigr.org/tdb/tgi/mtgi/) and the Functional Genomics in Medicago truncatula (September 2001) database (Toulouse, France; http://medicago.toulouse.inra.fr/Mt/EST/DOC/MtB.html; Journet et al., 2002). The electronic northern analysis used the GS2 MtC50790_GC cluster at the latter EST database.
Southern and PCR Analysis of Genomic DNA
All molecular biology techniques were carried out essentially as described (Sambrook et al., 1989). Southern hybridization used a probe corresponding to the GS2 cDNA. PCR analysis of GS2 genes used primers P1 (5′-CAGAGCCATGGCAGTCAAC-3′), P2 (5′-GCCATTTCACATCGTTTG-3′) and P3 (5′-TGTCTCTTCCCACACGG-3′).
Expression of MtGS2 cDNA in Escherichia coli
Two constructs were prepared to express the M. truncatula GS2 precursor and mature polypeptide in E. coli using the expression vector pTrc99A (Amersham Biosciences, Uppsala). The cloning procedure involved the introduction by PCR of NcoI restriction sites both at the start codon of the complete cDNA and at the predicted start of the mature polypeptide (after the transit peptide region) to allow in frame ligation into the NcoI site of the vector. The PCR was performed over the first 250 bp of the cDNA or 100 bp of the non-transit peptide coding region using VentR DNA polymerase (New England Biolabs, Beverly, MA), and the amplified fragments were fully sequenced to assure that no mistakes had been introduced. The rest of the cDNA was then introduced from a ClaI restriction site. In this way, the N termini of the precursor and mature proteins commenced with MAQI and MAVN, respectively.
For the complementation analysis and protein induction the E. coli glnA mutant, strain ET8894 (rbs lacZ::IS1 gyrA hutCx [glnA- ntrC]) was used as host (McNeil, 1981). Recombinant colonies were grown on M9 minimal medium plates, containing ammonium as nitrogen source and 2 mm isopropyl-thio-β-d-galactopyranoside, with or without a Gln supplement (250 μg mL−1). For protein induction, the cells were grown in liquid M9 medium in the presence of 2 mm isopropyl-thio-β-d-galactopyranoside.
Protein Extraction, GS Activity Determinations, and Western Immunoblotting
Plant material was homogenized by grinding the tissues with insoluble polyvinylpyrrolidone in a mortar and pestle at 4°C, with 2 volumes of extraction buffer (10 mm Trizma [pH 7.5], 5 mm sodium Glu, 10 mm MgSO4, 1 mm dithiothreitol, 10% [v/v] glycerol, and 0.05% [v/v] Triton X-100). E. coli cells were collected by centrifugation, and the pellets were frozen in liquid nitrogen and ground with alumina type V (Sigma-Aldrich, St. Louis) in a mortar and pestle with 2 volumes of the same buffer. The homogenates were centrifuged at 12,000g for 15 min at 4°C.
GS transferase activity was determined as described by Cullimore and Sims (1980). Soluble protein concentration was measured by the method of Bradford (1976) using the Bio-Rad dye reagent (Bio-Rad, Hercules, CA) and bovine serum albumin as a standard. For western analysis, protein samples (30 μg lane−1) were separated by SDS-PAGE and electroblotted onto nitrocellulose membranes. The blots were incubated with primary antibodies: anti-GS antibody (Cullimore and Miflin, 1984), antibody E6 against nodulin 26, (Weaver et al., 1991), and antibody E37 against a plastid envelope polypeptide (Joyard et al., 1982) and detected with secondary peroxidase-conjugated goat IgG (Vector Laboratories, Peterborough, UK). The intensity of the immunoreactive bands from three replicate experiments were quantified using the ImageQuant software on a Typhoon Molecular Imager (Molecular Dynamics, Sunnyvale, CA), and the average values were plotted.
RNA Isolation and Northern Analysis
Total RNA was isolated from plant material using TRIzol (Invitrogen, Carlsbad, CA) reagent as described by the manufacturer. RNA samples (15 μg lane−1) were separated by electrophoresis on formaldehyde gels and were transferred to positively charged nylon membranes by capillary action. Hybridization was performed as described before, using a specific probe constituted by a BamHI internal fragment of MtGS2 cDNA, corresponding to nucleotide 445 to 1,222 of the coding sequence. The blots were washed with 2× SSC and 0.1% (w/v) SDS at 57°C.
The blots were also probed with an rRNA gene probe to evaluate equal loading of total RNA. The hybridization signals were quantified on a Typhoon PhosphorImager using ImageQuant software (Molecular Dynamics), and the values for the GS2 gene probe were standardized against the hybridization signals with the rRNA gene probe.
Cell Fractionation
A sample of 0.5 g fresh weight of nodules was gently crushed in a mortar and pestle with 1 mL of breaking medium (Atkins et al., 1997). The homogenate was filtered through Miracloth (Calbiochem, San Diego), and the filtrate was centrifuged at 200g for 2 min. The supernatant was loaded onto 2 mL of 30% to 60% (v/v) Percoll density gradient and centrifuged for 30 min at 4°C and 25,000g in an Optima centrifuge with a TLA-100.4 fixed-angle rotor (Beckman Coulter, Fullerton, CA). The gradient was fractionated in 500-μL aliquots.
Immunogold Electron Microscopy
Eight-day-old root nodules were fixed and embedded in LR-white resin (London Resin Company, Berkshire, UK), sectioned, and processed as described by Carvalho et al. (1992).
Materials
The MtGS2 cDNA clone sequence is GenBank accession number AY225150. Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
ACKNOWLEDGMENTS
We thank Dr. P. Gamas (Institut des Interactions Plantes Microorganismes, Toulouse, France) for making available the cDNA library and Dr. D.M. Roberts (University of Tennessee, Knoxville), Dr. M. Block, and Dr. J. Joyard (Centre National de la Recherche Scientifique/Université de Grenoble, France) for providing the E6 and E37 antibodies, respectively.
Footnotes
This work was supported by the Fundação para a Ciëncia e Tecnologia (project no. POC/PI/41433/BC1/2001), and the cooperation between the French and Portuguese laboratories was supported by the Luso-Français Program for Scientific and Technical Cooperation.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.016675.
LITERATURE CITED
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins CA. Metabolism and translocation of fixed nitrogen in the nodulated legumes. Plant Soil. 1987;100:157–169. [Google Scholar]
- Atkins CA, Smith PMC, Store PJ. Reexamination of the intracellular localization of de novo purine synthesis in cowpea nodules. Plant Physiol. 1997;113:127–135. doi: 10.1104/pp.113.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D, Bianchi S, Blondon F, Dattée Y, Duc G, Essad S, Flament P, Gallusci P, Génier G, Guy P et al. Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol Biol Rep. 1990;8:40–49. [Google Scholar]
- Bell CJ, Dixon RA, Farmer AD, Flores R, Inman J, Gonzales RA, Harrison MJ, Paiva NL, Scott AD, Weller JW et al. The Medicago Genome Initiative: a model legume database. Nucleic Acids Res. 2001;29:114–117. doi: 10.1093/nar/29.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MJ, Cullimore JV. Glutamine synthetase isoenzymes of Phaseolus vulgaris L.: subunit composition in developing root nodules and plumules. Planta. 1989;179:433–440. doi: 10.1007/BF00397582. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein dye-binding. Anal Biochem. 1976;72:248–252. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brangeon J, Hirel B, Forchioni A. Immunolocalization of glutamine synthetase in soybean leaves, roots and nodules. Protoplasma. 1989;151:88–97. [Google Scholar]
- Carvalho H, Lescure N, de Billy F, Chabaud M, Lima L, Salema R, Cullimore J. Cellular expression and regulation of the Medicago truncatula cytosolic glutamine synthetase genes in root nodules. Plant Mol Biol. 2000a;42:741–756. doi: 10.1023/a:1006304003770. [DOI] [PubMed] [Google Scholar]
- Carvalho M, Lima L, Lescure N, Camut S, Salema R, Cullimore J. Differential expression of the two cytosolic glutamine synthetase genes in various organs of Medicago truncatula. Plant Sci. 2000b;159:301–312. doi: 10.1016/s0168-9452(00)00360-5. [DOI] [PubMed] [Google Scholar]
- Carvalho H, Pereira S, Sunkel C, Salema R. Detection of a cytosolic glutamine synthetase in leaves of Nicotiana tabacum L. by immunocytochemical methods. Plant Physiol. 1992;100:1591–1594. doi: 10.1104/pp.100.3.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho H, Sunkel C, Salema R, Cullimore JV. Heteromeric assembly of the cytosolic glutamine synthetase polypeptides of Medicago truncatula: complementation of a glnA Escherichia coli mutant with a plant domain-swapped enzyme. Plant Mol Biol. 1997;35:623–632. doi: 10.1023/a:1005884304303. [DOI] [PubMed] [Google Scholar]
- Cock JM, Brock JW, Watson AT, Swarup R, Morby AP, Cullimore JV. Regulation of glutamine synthetase genes in leaves of Phaseolus vulgaris. Plant Mol Biol. 1991;17:671–771. doi: 10.1007/BF00037059. [DOI] [PubMed] [Google Scholar]
- Cook D. Medicago truncatula: a model in the making! Curr Opin Plant Biol. 1999;2:301–304. doi: 10.1016/s1369-5266(99)80053-3. [DOI] [PubMed] [Google Scholar]
- Cullimore JV, Miflin BJ. Immunological studies on glutamine synthetase using antisera raised to the two plant forms of the enzyme from Phaseolus vulgaris nodules. J Exp Bot. 1984;35:581–587. [Google Scholar]
- Cullimore JV, Sims AP. An association between photorespiration and protein catabolism: studies with Chlamydomonas. Planta. 1980;150:392–396. doi: 10.1007/BF00390175. [DOI] [PubMed] [Google Scholar]
- Datta DB, Cai X, Wong P, Triplett EW. Immunocytochemical localisation of glutamine synthetase in organs of Phaseolus vulgaris L. Plant Physiol. 1991;96:507–512. doi: 10.1104/pp.96.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG, Cullimore JV. Glutamine synthetase in higher plants. Oxf Surv Plant Mol Cell Biol. 1989;6:247–298. [Google Scholar]
- Forde BG, Day HM, Turton JF, Shen WJ, Cullimore JV, Oliver JE. Two glutamine synthetase genes from Phaseolus vulgaris L. display contrasting developmental and spatial patterns of expression in transgenic Lotus corniculatus plants. Plant Cell. 1989;1:391–401. doi: 10.1105/tpc.1.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallusci P, Dedieu A, Journet EP, Huguet T, Barker DG. Synchronous expression of leghaemoglobin genes in Medicago truncatula during nitrogen fixing root nodule development and response to exogenously supplied nitrate. Plant Mol Biol. 1991;17:335–349. doi: 10.1007/BF00040629. [DOI] [PubMed] [Google Scholar]
- Gamas P, de Carvalho-Niebel F, Lecsure N, Cullimore JV. Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol Plant-Microbe Interact. 1996;9:233–242. doi: 10.1094/mpmi-9-0233. [DOI] [PubMed] [Google Scholar]
- Hood JK, Silver PA. In or out? Regulating nuclear transport. Curr Opin Cell Biol. 1999;11:241–247. doi: 10.1016/s0955-0674(99)80032-5. [DOI] [PubMed] [Google Scholar]
- Journet EP, van Tuinen D, Gouzy J, Carreau V, Farmer MJ, Niebel A, Schiex T, Crespeau H, Jaillon O, Chatagnier O et al. Exploring root symbiotic programs of the model legume Medicago truncatula using EST analysis. Nucleic Acids Res. 2002;30:5579–5592. doi: 10.1093/nar/gkf685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyard J, Grossman A, Bartlett SG, Douce R, Chua NH. Characterization of envelope membrane polypeptides from spinach chloroplasts. J Biol Chem. 1982;257:1095–1101. [PubMed] [Google Scholar]
- Karlin-Neumann GA, Tobin EM. Transit peptides of nuclear-encoded chloroplast proteins share a common amino acid framework. EMBO J. 1986;5:9–13. doi: 10.1002/j.1460-2075.1986.tb04170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K, Cline K. Protein import and routing system of chloroplasts. Plant Cell. 1999;11:557–570. doi: 10.1105/tpc.11.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot DA, Green NK, Cullimore JV. The chloroplast-located glutamine synthetase of Phaseolus vulgaris L.: nucleotide sequence, expression in different organs and uptake into isolated chloroplast. Plant Mol Biol. 1988;11:191–202. doi: 10.1007/BF00015671. [DOI] [PubMed] [Google Scholar]
- Lullien V, Barker DG, da Lajudie P, Huguet T. Plant gene expression in effective and ineffective root nodules of alfalfa (Medicago sativa) Plant Mol Biol. 1987;9:469–478. doi: 10.1007/BF00015878. [DOI] [PubMed] [Google Scholar]
- May T, Soll J. Chloroplast precursor protein translocon. FEBS Lett. 1999;452:52–56. doi: 10.1016/s0014-5793(99)00527-x. [DOI] [PubMed] [Google Scholar]
- McGrath RB, Coruzzi GM. A gene network controlling glutamine and asparagine biosynthesis in plants. Plant J. 1991;1:275–280. doi: 10.1046/j.1365-313x.1991.00999.x. [DOI] [PubMed] [Google Scholar]
- McNeil D. General method, using Mu-Mud1 dilysogens, to determine the direction of transcription of the generate deletions in the glnA region of Escherichia coli. J Bacteriol. 1981;146:260–268. doi: 10.1128/jb.146.1.260-268.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miflin BJ, Lea PJ. Ammonia assimilation. In: Miflin BJ, editor. The Biochemistry of Plants. Vol. 5. New York: Academic Press; 1980. pp. 169–202. [Google Scholar]
- Migge A, Becker T. Greenhouse-grown conditionally lethal tobacco plants obtained by expression of plastidic glutamine synthetase antisense RNA may contribute to biological safety. Plant Sci. 2000;153:107–112. doi: 10.1016/s0168-9452(99)00232-0. [DOI] [PubMed] [Google Scholar]
- Orea A, Pajuelo P, Pajuelo E, Quidiello C, Romero JM, Marquez AJ. Isolation of photorespiratory mutants from Lotus japonicus deficient in glutamine synthetase. Physiol Plant. 2002;115:352–361. doi: 10.1034/j.1399-3054.2002.1150304.x. [DOI] [PubMed] [Google Scholar]
- Peterman TK, Goodman HM. The glutamine synthetase gene family of Arabidopsis thaliana: light-regulation and differential expression in leaves, roots and seeds. Mol Gen Genet. 1991;230:145–154. doi: 10.1007/BF00290662. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Santana MA, Pihakaski-Maunsbach K, Sandal N, Marcker KA, Smith AG. Evidence that the plant host synthesizes the heme moiety of the leghemoglobin in root nodules. Plant Physiol. 1998;116:1259–1269. doi: 10.1104/pp.116.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford AC, Larsen K, Barker DG, Cullimore JV. Differential expression within the glutamine synthetase gene family of the model legume Medicago truncatula. Plant Physiol. 1993;103:73–81. doi: 10.1104/pp.103.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q, Schmid K, Schild C, Boschetti A. Effect of protein phosphorylation on import into isolated chloroplasts from Chlamydomonas. FEBS Lett. 2001;508:165–169. doi: 10.1016/s0014-5793(01)03012-5. [DOI] [PubMed] [Google Scholar]
- Thoquet P, Ghéradi M, Journet EP, Kereszt A, Ané JM, Prosperi JM, Huguet T. The molecular genetic linkage map of the model legume Medicago truncatula: an essential tool for comparative legume genomics and the isolation of agronomically important genes. BioMed Central Plant Biol. 2002;2:1. doi: 10.1186/1471-2229-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingey SV, Tsai FY, Edwards JW, Walker EL, Coruzzi GM. Chloroplast and cytosolic glutamine synthetase are encoded by homologous nuclear genes which are differentially expressed in vivo. J Biol Chem. 1988;263:9651–9657. [PubMed] [Google Scholar]
- Tingey SV, Walker EL, Coruzzi GM. Glutamine synthetase genes of pea encode distinct polypeptides which are differentially expressed in leaves, roots and nodules. EMBO J. 1987;6:1–9. doi: 10.1002/j.1460-2075.1987.tb04710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vézina LP, Hope HJ, Joy KW. Isoenzymes of glutamine synthetase in roots of pea (Pisum sativum L. cv Little Marvel) and alfalfa (Medicago media Pers. cv Saranac) Plant Physiol. 1987;83:58–63. doi: 10.1104/pp.83.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waegemann K, Soll J. Phosphorylation of the transit sequence of chloroplast precursor proteins. J Biol Chem. 1996;271:6545–6554. doi: 10.1074/jbc.271.11.6545. [DOI] [PubMed] [Google Scholar]
- Wallsgrove RM, Turner JC, Hall NP, Kendall AC, Bright SWJ. Barley mutants lacking chloroplast glutamine synthetase: biochemical and genetic analysis. Plant Physiol. 1987;83:155–158. doi: 10.1104/pp.83.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CD, Crombie B, Stacey G, Roberts DM. Calcium-dependent phosphorylation of symbiosome membrane proteins from nitrogen-fixing soybean nodules. Plant Physiol. 1991;95:222–227. doi: 10.1104/pp.95.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zozaya-Garza M, Sengupta-Gopalan C. Glutamine synthetase gene isolation from an alfalfa leaf cDNA library. Plant Physiol. 1999;119:1568. [Google Scholar]