Abstract
Here, we report the systematic exploration and modeling of interactions between light and sugar signaling. The data set analyzed explores the interactions of sugar (sucrose) with distinct light qualities (white, blue, red, and far-red) used at different fluence rates (low or high) in etiolated seedlings and mature green plants. Boolean logic was used to model the effect of these carbon/light interactions on three target genes involved in nitrogen assimilation: asparagine synthetase (ASN1 and ASN2) and glutamine synthetase (GLN2). This analysis enabled us to assess the effects of carbon on light-induced genes (GLN2/ASN2) versus light-repressed genes (ASN1) in this pathway. New interactions between carbon and blue-light signaling were discovered, and further connections between red/far-red light and carbon were modeled. Overall, light was able to override carbon as a major regulator of ASN1 and GLN2 in etiolated seedlings. By contrast, carbon overrides light as the major regulator of GLN2 and ASN2 in light-grown plants. Specific examples include the following: Carbon attenuated the blue-light induction of GLN2 in etiolated seedlings and also attenuated the white-, blue-, and red-light induction of GLN2 and ASN2 in light-grown plants. By contrast, carbon potentiated far-red-light induction of GLN2 and ASN2 in light-grown plants. Depending on the fluence rate of far-red light, carbon either attenuated or potentiated light repression of ASN1 in light-grown plants. These studies indicate the interaction of carbon with blue, red, and far-red-light signaling and set the stage for further investigation into modeling this complex web of interacting pathways using systems biology approaches.
Light is an important environmental signal that is directly perceived by the plant through photoreceptors and is essential for driving photosynthesis. As such, light provides the reducing power for carbon fixation, nitrogen assimilation, amino acid biosynthesis, and other necessary metabolic pathways. Information about light quality, intensity, and duration is measured through numerous photoreceptors (Mancinelli, 1994; Smith, 1994). Phytochromes are the primary red-light photoreceptors. The blue-light, UV-A/B photoreceptors include the cryptochromes, phototropin, and other yet unidentified photoreceptors (for review, see Briggs and Huala, 1999). The various qualities of light perceived through these photoreceptors control diverse developmental programs in plants such as seed germination, hypocotyl elongation, shade avoidance, circadian rhythms, flowering, chloroplast differentiation, and cotyledon expansion (for review, see Fankhauser and Chory, 1997; Briggs and Huala, 1999; Neff et al., 2000). The most well-characterized photoreceptors are the phytochromes. In Arabidopsis, five different phytochromes exist (phyA–E), each containing both overlapping and unique biological functions. PhyA is predominately involved in physiological responses to continuous far-red light, whereas phyB is involved in responses to red light. Additionally, phyA mediates responses to very low fluences of red, blue, and far-red light. At different stages of plant development the influence of each photoreceptor may change (for review, see Moller et al., 2002).
Light perception and signaling through various photoreceptors has been intensely investigated. The identification of downstream components of photoreceptor-signaling pathways has revealed cross-talk between pathways of different light qualities as well as with other seemingly unrelated pathways (for review, see Moller et al., 2002; Nagy and Schafer, 2002). For example, SUB1 is both a component of a cryptochrome-signaling pathway and a modulator of a phytochrome-signaling pathway (Guo et al., 2001). Auxin, brassinosteroid, gibberellic acid, cytokinin, and ethylene signal transduction pathways are all influenced by light-signaling pathways either directly or indirectly (for review, see Moller et al., 2002). Additionally, sugars that serve as growth and signaling molecules have been shown to modulate phytochrome sensing and signaling pathways (Barnes et al., 1996; Dijkwel et al., 1997; Short, 1999).
Sugars initiate changes in the expression of genes involved in diverse functions such as embryogenesis, flowering, seedling development, and senescence. Some genes encoding proteins involved in or relating to photosynthesis are strongly induced by light yet repressed by carbon (e.g. chlorophyll a/b binding protein, plastocyanin, and small subunit of Rubisco; Koch, 1996) or induced by both light and sugar (e.g. Gln synthetase, nitrate reductase, and Asn synthetase 2; Koch, 1996; Lam et al., 1998; Oliveira and Coruzzi, 1999). More specific interactions between carbon and light have been observed by the ability of Suc to suppress a phyA-specific, far-red-light-induced block of greening. Hence, Suc may antagonize or suppress the phyA-signaling pathway(s) in this case (Barnes et al., 1996). A class of Suc-uncoupled (sun) mutants have been identified that exhibit reduced Suc repression of light-induced genes and are defective in the ability of Suc to inhibit the far-red block of greening (Dijkwel et al., 1997). One of the SUN genes is identical to ABI4, a gene involved in abscisic acid signal transduction, suggesting the involvement of abscisic acid in sugar responses (Huijser et al., 2000). Additionally, use of a phyA or phyB pathway may be influenced by Suc where it may modify or change the preference of common downstream components of a phyA or phyB pathway (Short, 1999).
In contrast to the perception and transduction of light, our understanding of sugar perception and signaling is less well studied (for recent review, see Rolland et al., 2002). Although there is evidence that hexokinases HXK1 and -2 may act as sensors involved in sugar sensing and transduction in plants (Jang et al., 1997; Sheen et al., 1999), other mechanisms have also been proposed (Halford et al., 1999; for review, see Rolland et al., 2002). Many sugar-regulated genes have been categorized as being regulated through an HXK-dependent or -independent pathway, where other sugar-sensing pathways most likely also exist (Sheen et al., 1999). Although not much is known about downstream components of carbon-signaling pathways, a number of components are proposed to play a role including protein phosphatases, transcription factors, and numerous kinases (calcium-dependent, mitogen-activated, and SNF1-related; Sheen et al., 1999; Smeekens, 2000; Rolland et al., 2002). Mutants defective in sugar sensing and signaling have been isolated and demonstrate interactions with hormone signal transduction pathways (see Rolland et al., 2002). For example, Glc-insensitive mutants gin5 and gin6 show the involvement of abscisic acid in sugar responses (Arenas-Huertero et al., 2000), where they are allelic to genes (aba3 and abi4) involved in abscisic acid biosynthesis or signal transduction, respectively (for review, see Rolland et al., 2002). As more information is gleaned about any signal transduction pathway, signaling pathways can no longer be studied in a “linear” manner, but the influence and integration of numerous pathways must be considered.
Light- and sugar-signaling pathways have been shown to regulate the transcription of genes involved in metabolism. For example, the assimilation of nitrogen into amino acids is a process partially controlled at the transcriptional level by light and sugar signaling. Gln synthetase (GLN2) and ferredoxin-Glu synthase, Fd-GOGAT (GLU1) are two enzymes involved in the assimilation of ammonia into Gln and Glu, whose genes are induced by light (Coschigano et al., 1998; Oliveira et al., 1999). In contrast, light represses transcription of the genes ASN1 and GDH1, which encode the enzymes Asn synthetase and Glu dehydrogenase (Melo-Oliveira et al., 1996; Lam et al., 1998). Interestingly, exogenously supplied Suc has been shown to mimic the effect of light. Suc supplied to dark-adapted plants increases expression of GLN2 and represses ASN1 expression (Lam et al., 1998; Oliveira et al., 1999). Reciprocal regulation of the GLN2/ASN1 genes by light reflects changes in the levels of their cognate amino acids, Gln and Asn. Gln levels are high in light-grown plants, and Asn levels are high in dark-grown/adapted plants (Lam et al., 1995; Ngai and Coruzzi, 1998). ASN1 is the major gene controlling Asn synthesis in Arabidopsis, where expression of ASN1 in the dark serves to enable the conversion of Gln to Asn, which is used for N-transport when C-skeletons are limiting (Lam et al., 1995). However, light activates the expression of ASN2, another member of the ASN gene family. ASN2 is induced by both light and Suc (Lam et al., 1998). It is postulated that ASN2 serves to synthesize low levels of Asn in the light, used for protein synthesis, whereas ASN1 is responsible for making high levels of Asn for N-transport/storage in the dark (Lam et al., 1998). It has been shown that light regulation of the genes GLN2, ASN1, and ASN2 are mediated in part via phytochrome in etiolated seedlings (Lam et al., 1994; for review, see Lam et al., 1996; Hsieh, 1999) and also through the indirect effects of light (e.g. light-induced increases in carbon).
This study represents the first systematic approach to investigate the interactions between light- and carbon-signaling pathways. ASN1, ASN2, and GLN2 serve as sentinel genes for the examination of light and carbon interactions. Expression profiles of these genes were analyzed in plants treated with different wavelengths of light at low- or high-fluence rates in the presence or absence of a carbon source. Etiolated and light-grown plants were analyzed to investigate possible differences in light- and carbon-signaling cross-talk in these very different stages of development. Because these pathways are expected to be complex, their interactions were analyzed and modeled using Boolean circuits. Depending on the developmental stage of the plant and the gene analyzed, it is shown carbon can attenuate, potentiate or enhance light responses at specific wavelengths and fluence rates.
RESULTS
“Experimental Space” for Investigating Light and/or Carbon Signaling
The experiments represented in Table I were designed in a systematic manner (a) to further investigate the individual light qualities and quantities regulating genes involved in nitrogen assimilation and (b) to investigate the influence of carbon on these specific light-signaling pathways in an attempt to further our understanding of light- and carbon-signaling interactions. The experimental setup consisted of using carbon (supplied exogenously as Suc) as a binary input (±) combined with various light qualities. Experiments 1 through 8 were designed to investigate the influence of individual light qualities in the absence of an exogenously supplied carbon source. Experiments 9 through 16 were designed to investigate the influence of carbon on the individual light qualities.
Table I.
Experimental space for investigating light and/or carbon signaling
Variables or inputs for each experiment are indicated as present (Y) or absent (N). The carbon source was 1% (w/v) Suc, and fluence rates for white light were 70 μE m-2 s-1 and 2 or 100 μE m-2 s-1 for blue, red, and far-red light.
| Experiment No.
|
Carbon
|
White Light
|
Blue Light
|
Red Light
|
Far-Red Light
|
|||
|---|---|---|---|---|---|---|---|---|
| Low-Fluence | High-Fluence | Low-Fluence | High-Fluence | Low-Fluence | High-Fluence | |||
| 1 | N | Y | N | N | N | N | N | N |
| 2 | N | N | Y | N | N | N | N | N |
| 3 | N | N | N | Y | N | N | N | N |
| 4 | N | N | N | N | Y | N | N | N |
| 5 | N | N | N | N | N | Y | N | N |
| 6 | N | N | N | N | N | N | Y | N |
| 7 | N | N | N | N | N | N | N | Y |
| 8 | N | N | N | N | N | N | N | N |
| 9 | Y | N | N | N | N | N | N | N |
| 10 | Y | Y | N | N | N | N | N | N |
| 11 | Y | N | Y | N | N | N | N | N |
| 12 | Y | N | N | Y | N | N | N | N |
| 13 | Y | N | N | N | Y | N | N | N |
| 14 | Y | N | N | N | N | Y | N | N |
| 15 | Y | N | N | N | N | N | Y | N |
| 16 | Y | N | N | N | N | N | N | Y |
Light Overrides Carbon Regulation of ASN1 and GLN2 But Not ASN2 in Etiolated Seedlings
Quantitative real-time PCR was used to monitor the transcript abundance of ASN1, ASN2, and GLN2 in etiolated plants treated with light and/or Suc (Fig. 1). Control transcripts from a putative clathrin coat assembly protein (At4g24550; CLH) were also detected and used as a normalization control. CLH was chosen as a control gene because these transcripts remained unchanged in plants at different developmental stages (etiolated versus light grown) and in response to light, carbon, or nitrogen (G.M. Coruzzi laboratory, unpublished data). Plants were grown in the absence or presence of 1% (w/v) Suc for 7 d in continuous darkness. After this growth period, plants were maintained in continuous darkness or illuminated with white light (WL) at 70 μE m–2 s–1 or with blue, red, or far-red light separately at 2 or 100 μE m–2 s–1 for an additional 3 h (Fig. 1; see Table I for experimental design). The results shown in Figures 1 and 2 are from five replicates.
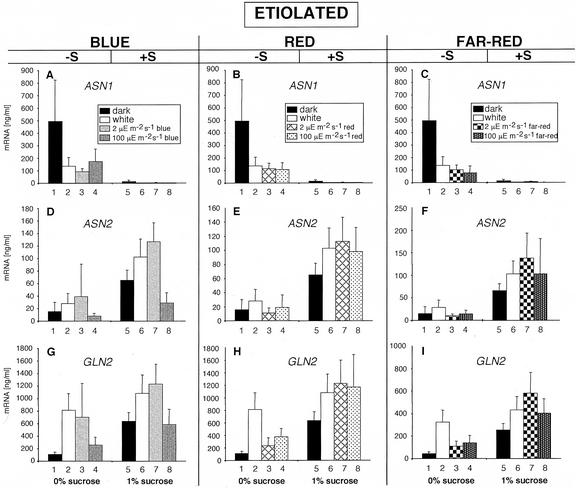
Figure 1.
Analysis of ASN1, ASN2, and GLN2 transcript accumulation in etiolated seedlings. A through C, ASN1 transcript levels in 7-d-dark-grown plants in the presence or absence of carbon and illuminated with WL, blue (A), red (B), or far-red (C) light. D through F, ASN2 transcript levels in 7-d-dark-grown plants in the presence or absence of carbon and illuminated with WL, blue (D), red (E), or far-red (F) light. G through I, GLN2 transcript levels in 7-d-dark-grown plants in the presence or absence of carbon and illuminated with WL, blue (G), red (H), or far-red (I) light. The carbon source used was 1% (w/v) Suc. All transcripts were measured using real-time quantitative PCR and normalized to a putative clathrin coat-assembly protein (At4g24550). The data represent the mean and sd of at least five separate experiments.
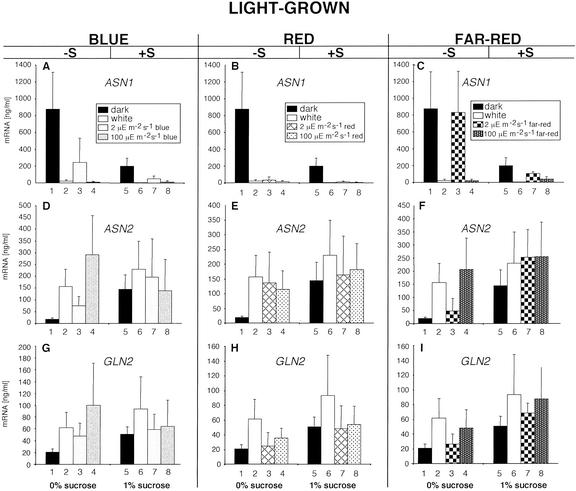
Figure 2.
Analysis of ASN1, ASN2, and GLN2 transcript accumulation in light-grown plants. A through C, ASN1 transcript levels in 14-d-light-/dark-grown plants in the presence or absence of carbon and illuminated with WL, blue (A), red (B), or far-red (C) light. D through F, ASN2 transcript levels in 14-d-light-/dark-grown plants in the presence or absence of carbon and illuminated with WL, blue (D), red (E), or far-red (F) light. G through I, GLN2 transcript levels in 14-d-light-/dark-grown plants in the presence or absence of carbon and illuminated with WL, blue (G), red (H), or far-red (I) light. The carbon source used was 1% (w/v) Suc. All transcripts were measured using real-time quantitative PCR and normalized to a putative clathrin coat-assembly protein (At4g24550). The data represent the mean and sd of at least five separate experiments.
Figure 1A, 1 shows the high level accumulation of ASN1 transcripts in dark-grown plants in the absence of Suc. Illumination of these plants with WL, blue, red, or far-red light at either a low- or high-fluence rate decreased ASN1 transcript levels (Fig. 1, A–C, 1 versus 2–4). The presence of Suc in the media caused a dramatic decrease in the amount of ASN1 transcripts in the absence of light (Fig. 1A, 5). Illumination of these plants in all light conditions in the presence of Suc further reduced ASN1 transcripts to almost undetectable levels (Fig. 1, A–C, 5 versus 6–8).
ASN2 transcript levels in dark Suc-free-grown plants were low, and illumination with any of the light qualities or fluency rates used in this study had no significant effect on ASN2 transcript abundance (Fig. 1, D–F, 1 versus 2–4). The presence of Suc on etiolated seedlings increased ASN2 transcripts in the absence of light (Fig. 1D, 5). Illumination of these plants with most light qualities and quantities increased the level of ASN2 transcripts (Fig. 1, D–F, 5 versus 6–8). Interestingly, illumination with blue light at 100 μE m–2 s–1 resulted in a decrease of ASN2 transcripts below that observed for dark-grown plants (Fig. 1D).
Consistent with the reported reciprocal regulation of ASN1 and GLN2 (Lam et al., 1994), GLN2 mRNA levels were low in etiolated seedlings grown in the absence of Suc (Fig. 1G, 1). Illumination of these plants with any of the light qualities and quantities in this study increased GLN2 transcripts, albeit to varying degrees (Fig. 1, G–I, 1 versus 2–4). GLN2 mRNA levels increased in etiolated plants grown in the presence of Suc (Fig. 1G, 1 versus 5). Illumination of these plants with all light qualities and quantities except for blue light at 100 μE m–2 s–1, increased GLN2 transcripts (Fig. 1, G–I, 5 versus 6–8). An effect of high-fluence blue light in the presence of Suc but not in the absence can also be observed for ASN2.
Carbon Overrides Light Regulation of GLN2 and ASN2 But Not ASN1 in Light-Grown Plants
As with the analysis for etiolated seedlings, quantitative real-time PCR was used to characterize Suc and/or light-modulated changes in ASN1, ASN2, and GLN2 transcript abundance in 14-d-light-/dark-grown plants (Fig. 2). After dark adaptation, plants were maintained in continuous darkness or illuminated with WL at 70 μE m–2 s–1, or with blue, red, or far-red light separately at 2 or 100 μE m–2 s–1 for an additional 3 h.
ASN1 mRNA levels were high in dark-adapted plants in the absence of Suc (Fig. 2A, 1), and illumination of these plants with most light qualities and fluence rates used in this study decreased ASN1 transcripts to varying degrees (Fig. 2, A–C, 1 versus 2–4). One exception is the illumination with 2 μE m–2 s–1 of far-red light, which appears to be unable to repress ASN1. The presence of Suc on plants in the dark, resulted in a decrease in ASN1 transcript levels compared with those observed for ASN1 in plants in the absence of Suc (Fig. 2A, 5). Illumination of dark-adapted plants in the presence of Suc with all of the light qualities and quantities further reduced ASN1 mRNA levels (Fig. 2, A–C, 5 versus 6–8).
Transcript levels of ASN2 were low in dark-adapted plants in the absence of Suc and could be increased, albeit at varying levels, by illumination with most light qualities except far-red at 2 μE m–2 s–1 (Fig. 2, D–F, 1 versus 2–4). Dark-adapted plants in the presence of Suc had higher levels of ASN2 mRNA compared with those in the absence of Suc (Fig. 2D, 5). Illumination of dark-adapted plants with WL or far-red light (2 or 100 μE m–2 s–1) was able to significantly increase ASN2 transcript levels, where blue and red light had minimal effects (Fig. 2, D–F, 5 versus 6–8).
GLN2 mRNA levels were low in dark-adapted plants in the absence of Suc (Fig. 2G, 1), and illumination with most light qualities and quantities except red and far-red light at 2 μE m–2 s–1 increased GLN2 transcript abundance (Fig. 2, G–I, 1 versus 2–4). As observed with ASN2, albeit more modest, the presence of Suc in dark-adapted plants resulted in an increase of GLN2 transcripts (Fig. 2G, 5). Only illumination of these plants with WL or far-red light at 2 or 100 μE m–2 s–1 was able to increase GLN2 transcript levels above those observed for dark-adapted plants in the presence of Suc alone (Fig. 2I, 5 versus 7 and 8). Red- or blue-light illumination of plants in the presence of Suc was unable to alter GLN2 transcript levels beyond those observed for plants in the presence of Suc (Fig. 2, G and H, 5 versus 6–8).
Boolean Circuits Determine Significant Regulators of ASN1, ASN2, and GLN2
To model the interactions of light and carbon signaling, we use Boolean logic to analyze the data generated from the experiments shown in Table I. In brief, two base conditions, no light/no carbon (Table I, experiment 8) and no light/carbon (Table I, experiment 9) were used for comparison against all non-base conditions (all other experiments). Specific thresholds were assigned where expression levels relative to the base condition were categorized as inductive, super-inductive, repressive, or super-repressive. On the basis of an unpaired t test (P = 0.05), a particular Boolean input is deemed statistically significant and affects the output, whereas the absence of a Boolean input is due to either statistical insignificance or due to no effect of the input.
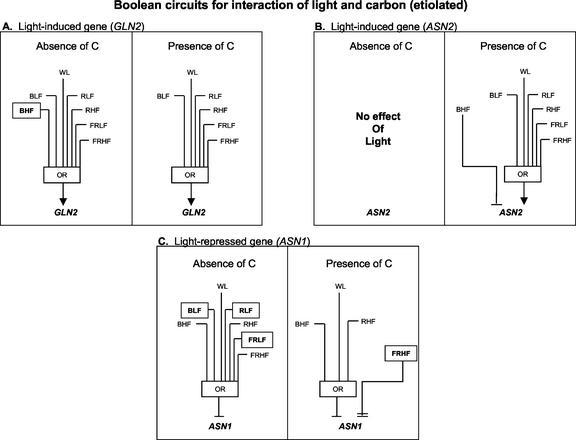
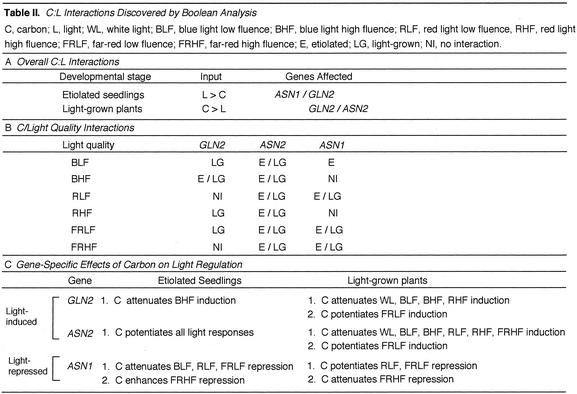
Figure 3 shows Boolean circuits for GLN2, ASN2, and ASN1 in etiolated plants in the absence or presence of carbon. Figure 3A shows that WL OR red light low fluence (RLF) OR red light high fluence (RHF) OR far-red light low fluence (FRLF) OR far-red light high fluence (FRHF) OR blue light low fluence (BLF) OR blue light high fluence (BHF), singly, in the absence of carbon each induce expression of GLN2. In the presence of carbon, all light qualities at different fluence rates induce GLN2, except for BHF. Interestingly, light has no significant effect on ASN2 expression levels in the absence of carbon, whereas in the presence of carbon, light becomes inductive for all light qualities with the exception of BHF, where it is repressive (Fig. 3B). ASN1 is repressed by all light qualities at any quantity in the absence of carbon, whereas only WL, RHF, and BHF are repressive in the presence of carbon, and FRHF becomes super-repressive (Fig. 3C).
Figure 3.
Boolean circuits model ASN1, ASN2, and GLN2 regulation by light and carbon in etiolated seedlings. A through C, Boolean circuits based on 16 experiments represented in Table I. A, GLN2 regulation by WL, blue, red, or far-red light when compared against a base case of no light, no carbon, or no light, carbon (see “Materials and Methods”). B, ASN2 regulation by WL, blue, red, or far-red light when compared against a base-case of no light, no carbon, or no light, carbon. C, ASN1 regulation by WL, blue, red, or far-red light when compared against a base case of no light, no carbon, or no light, carbon. The inputs are WL, BLF, BHF, RLF, RHF, FRLF, or FRHF. Low fluence is 2 μE m–2 s–1; high fluence is 100 μE m–1 s–1. The arrow or barred lines indicate the function of the inputs as either inductive or repressive. Double arrows or double bars denote super-induction or super-repression, respectively. For a Boolean OR, if any one of the inputs is active, the output will also be active. Differences in the input for Boolean circuits when comparing “absence of carbon” to “presence of carbon” are shown by boxed inputs except for ASN2 where everything is different in the presence versus absence of carbon.
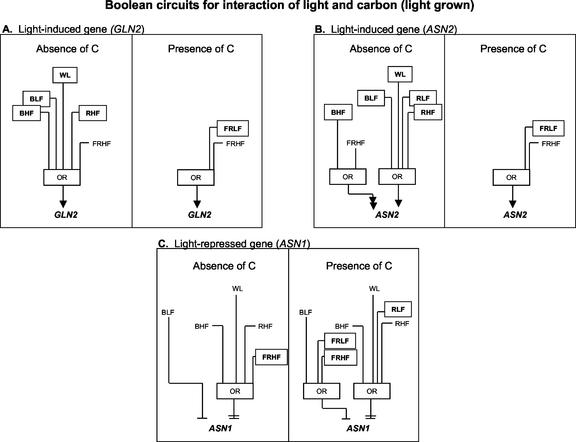
Figure 4 shows Boolean circuits for GLN2, ASN2, and ASN1 in 14-d-light-grown plants in the absence or presence of carbon. In the absence of carbon, WL, RHF, FRHF, BLF, and BHF each induce GLN2 expression (Fig. 4A). In the presence of carbon, FRHF remains and FRLF becomes inductive. ASN2 is super-induced by BHF and FRHF and induced by WL, RLF, RHF, or BLF in the absence of carbon (Fig. 4B). In the presence of carbon, ASN2 is induced only by FRLF or FRHF, as shown also for GLN2. ASN1 is repressed by BLF and super-repressed by WL, BHF, RHF, or FRHF in the absence of carbon (Fig. 4C). In the presence of carbon, the super-repression of ASN1 by WL, BHF, and RHF remains and RLF becomes super-repressive. BLF remains repressive and FRLF and FRHF become repressive for ASN1 in the presence of carbon.
Figure 4.
Boolean circuits model ASN1, ASN2, and GLN2 regulation by light and carbon in light-grown plants. A through C, Boolean circuits based on 16 experiments represented in Table I. A, GLN2 regulation by WL, blue, red, or far-red light when compared against a base case of no light, no carbon, or no light, carbon (see “Materials and Methods”). B, ASN2 regulation by WL, blue, red, or far-red light when compared against a base case of no light, no carbon, or no light, carbon. C, ASN1 regulation by WL, blue, red, or far-red light when compared against a base case of no light, no carbon, or no light, carbon. The inputs are WL, BLF, BHF, RLF, RHF, FRLF, and FRHF. Low fluence is 2 μE m–2 s–1; high fluence is 100 μE m–1 s–1. The arrow or barred lines indicate the function of the inputs as either inductive or repressive. Double arrows or double bars denote super-induction or super-repression, respectively. For a Boolean OR, if any one of the inputs is active, the output will also be active. Differences in the input for Boolean circuits when comparing “absence of carbon” to “presence of carbon” are shown as boxed inputs.
DISCUSSION
In this study, we employed a systematic approach to investigate and model the interactions between light- and carbon-signaling pathways. Because very little is known about the interactions between these two pathways, all possible combinations of light (WL, BLF, BHF, RLF, RHF, FRLF, and FRHF) and carbon were examined in both etiolated and light-grown seedlings in an attempt to cover a systematic experimental space. The analysis and modeling of these results as Boolean circuits represents a novel method to investigate complex interactions of carbon and light signaling and to identify the major regulatory signals. This analysis revealed interactions between carbon and light that are distinct in etiolated versus green plants, and ones that are specific to a gene or condition. A summary of all results can be found in Table II. In etiolated seedlings, light was generally able to override carbon as a major regulator of ASN1 and GLN2 expression. By contrast, in light-grown plants, carbon was shown to override light as the major regulator of GLN2 and ASN2 expression. Additionally, carbon was shown to interact with blue, red, or far-red light-signaling pathways in both etiolated and light-grown plants, where carbon was shown to either potentiate or attenuate specific light responses. The significance of these major findings in this study are addressed below. This initial analysis of light and carbon interactions provides the framework for further experiments that we have designed using “combinatorial design” to understand how interactions of distinct light qualities may also be affected by interactions with carbon.
Table II.
C:L Interactions Discovered by Boolean Analysis
Boolean Circuits for Analysis of Complex Interactions
Modeling the cellular activity of a set of genes/proteins as a functional network permits researchers to devise predictive models that may eventually permit intervention in pathways for diagnostic and therapeutic purposes. Two major kinds of network circuits are possible: discrete and continuous. The simplest discrete model is a Boolean network model in which input variables such as light (used here) can be set to one of several values 0/LF/HF, and gene regulation results from a Boolean function, possibly augmented by continuous elements such as amplifiers (Davidson et al., 2002). By contrast, continuous models may be based on stochastic kinetics (Arkin et al., 1998; Goss and Peccoud, 1998) and may include hidden environmental variables (Weaver et al., 1999). Many researchers believe that continuous models are more faithful to nature than discrete ones. For example D'haeseleer et al. (2000) note that Boolean networks do not provide a suitable framework to model feedback and other elements of biological control. Whereas Boolean models are incomplete compared with continuous models, they are more robust to biological noise and are easier to understand. They must be augmented with amplifiers as Davidson et al. (2002) have done (Yuh et al., 1998) enabling one to model, for example, a situation in which input A alone produces no change, input B gives induction, but A and B together give super-induction.
Boolean analysis requires inputs to be described in absolute terms of either having an effect or not having an effect (based on statistical analysis), which may not accurately represent biological systems, as discussed above. In our study, gene responses were deemed significant or not significant based on an unpaired t test at a P value of 0.05, where the presence of a particular Boolean input in the model represents cases where the input(s) had a statistically significant effect on the output (gene expression; Figs. 3 and 4). The absence of a Boolean input is either due to statistical insignificance of the effect or due to no effect of the input on gene expression. To address the biological relevance of this approach, we also carried out Boolean analyses at a lower P value of 0.1 (data not shown). A comparative analysis showed that few minor differences in the Boolean circuits were observed between data analyzed at P = 0.05 versus P = 0.1.
Boolean analyses for the modeling of plant-signaling networks have previously been described (Genoud and Metraux, 1999; Genoud et al., 2001). Here, we used this type of Boolean analysis to investigate how light and/or carbon interactions affected the regulation of three genes involved in nitrogen assimilation. Figures 3 and 4 give the simplest and most likely Boolean circuits to explain our results thus far. This analysis indicates that any one of a set of light conditions has the effect shown (either inductive or repressive; Figs. 3 and 4). The “OR” indicates that any of the light qualities tested give similar results. The simplicity of these circuits reflects the fact that, so far, our data do not explore combinations of light quality. We propose methods to test such combinations at the end of this discussion. Further experiments involving multiple inputs can be integrated into these already existing circuits to observe their effect on the output, or on regulation of GLN2, ASN2, and ASN1. Finally, our description of these light and carbon interactions in a binary manner serves as a precursor for the eventual computer modeling of more complex signaling pathways, with dose and kinetic parameters included.
Biological Significance of the Major Conclusions of Light/Carbon Interactions
Light Overrides Carbon as the Major Regulator of ASN1 and GLN2 in Etiolated Plants
In general, light overrides carbon regulation of ASN1 and GLN2 expression in etiolated seedlings (Fig. 3, A and C). This regulation may occur because these plants are not yet photosynthetically active, and because light of all wavelengths is required for the induction of genes encoding proteins involved in chloroplast development, metabolism, and the further development of etiolated plants. The primary regulation of GLN2 expression by light in etiolated seedlings makes sense physiologically, because such pre-induction by light (before photosynthate) will make Gln synthetase available for primary nitrogen assimilation using the energy generated through photosynthesis. Because Asn synthetase (ASN1) catalyzes the synthesis of Asn from Gln in the dark when C-skeletons are limiting, the presence of this enzyme in illuminated seedlings is unnecessary, hence the repressive effect of light dominates. However, the repressive effects of some qualities of light on ASN1 expression are enhanced (FRHF)/or attenuated (BLF, RLF, and FRLF) by Suc, suggesting that some light and carbon interactions exist, requiring further investigation. Interestingly, regulation of ASN2 expression is an exception to the observed general effect of light overriding C-regulation of ASN1/GLN2, where in fact, the induction of ASN2 by light requires a carbon interaction in etiolated seedlings (Fig. 3B).
Carbon Overrides Light as the Major Regulator of GLN2 and ASN2 in Light-Grown Plants
Carbon appears to attenuate the light regulation of GLN2 and ASN2 expression in light-grown plants. In the absence of carbon, light has a major influence on regulation of ASN2 and GLN2 expression. By contrast, in the presence of carbon, the effect of light becomes negligible (Fig. 4, A and B). The ability of a plant to sense and respond to light in the absence of carbon assures that at the onset of photosynthesis, the assimilation of nitrogen onto C-skeletons will be immediate via GLN2/ASN2 activation. At the onset of photosynthesis, however, the predominant regulation of these genes by carbon enables the plants to regulate nitrogen assimilation in response to levels of photosynthate.
By contrast to GLN2/ASN2, the expression of ASN1, appears to be equally regulated by both light and carbon at this stage of development. Because ASN1 is most likely involved in the dark synthesis of Asn, the preferred amino acid for the transport of nitrogen in dark-adapted plants, repression by both light and carbon guarantees the absence of this enzyme in plants in the light whether or not they are photosynthesizing. The different regulation of these genes in etiolated versus light-grown plants may be due to different regulatory pathways or that some of the signaling components regulating these genes in light-grown plants are not yet present in etiolated seedlings.
Gene-Specific Interactions of Light and Carbon
Carbon Attenuates Blue-Light Induction of GLN2 in Etiolated Seedlings
Boolean analysis of the data from Table I showed that BHF was able to induce GLN2 expression in the absence of Suc, but not in the presence of Suc. This indicates that BHF perception or signaling may be antagonized by carbon in etiolated seedlings (Fig. 3A). These results suggest a fluence-rate dependence of the carbon/blue-light interaction, because carbon affected the BHF response and not the BLF response. A detailed fluence rate study needs to be carried out for blue light in the presence or absence of carbon to confirm these preliminary findings.
ASN2 Is Repressed by BHF in the Presence of Carbon in Etiolated Seedlings
Interactions between blue-light and carbon signaling were further observed to regulate expression of the ASN2 gene. We found that repression of ASN2 by blue light occurs only in the presence of Suc (Fig. 3B). This suggests a Suc dependence of blue-light signaling. Because the presence of Suc potentiates all light responses for ASN2 (Fig. 3B), the interaction between BHF and Suc may be a general effect of carbon. However, the finding that BHF represses ASN2 expression is surprising because, as shown in this study, all other light qualities are inductive for ASN2. Thus, this appears to be a specific interaction of BHF light and Suc.
Carbon Affects Far-Red Light Repression of ASN1 in Etiolated Seedlings
ASN1 expression is repressed by FRHF and FRLF in the absence of carbon, whereas FRHF becomes super-repressive in the presence of carbon (Fig. 3C). These results suggest that the fluence rate of far-red light influences its interaction with carbon. Carbon attenuates the repression of ASN1 by FRLF and yet it enhances repression of ASN1 by FRHF. The additive effect of FRHF and carbon results in super-repression of ASN1, suggesting that they are two separate pathways converging on this gene. Alternatively, carbon and far-red-light may affect the same pathway, where the level of either signal by itself was not high enough to maximize the effect. Carbon and phyA interactions have been documented, where carbon antagonizes a phyA pathway in etiolated seedlings (Barnes et al., 1996). PhyA is the predominant red/far-red light-absorbing phytochrome present in etiolated seedlings where it most likely plays a role in the repression of ASN1.
For ASN1 repression, carbon seems to antagonize only FRLF but not FRHF (Fig. 3C). Carbon may interfere with FRLF repression, or because this is observed for light at all low fluences in this study, it is more likely a general effect and not specific to any wavelength of light. This suggests that it may be the number but not wavelength of photons that is important in carbon/light interactions. Carbon attenuation of ASN1 repression by FRLF may be due to carbon overriding or masking the repression of low fluences of light, or it could be that the differences between low- and high-fluence light at any wavelength are not large enough to distinguish between the two effects on ASN1 repression.
Carbon Attenuates Light Induction of GLN2 and ASN2 in Light-Grown Plants
The induction of GLN2 in the absence of carbon requires WL, BLF, BHF, or RHF (Fig. 4A). By contrast, in the presence of Suc, these wavelengths of light no longer induce GLN2 expression in light-grown plants. This regulation of GLN2 is similar to that observed for ASN2 (Fig. 4B). Carbon may attenuate the WL, blue-, or red-light induction of ASN2 and GLN2, or it is possible that carbon overrides the induction of these wavelengths of light. Some physiological responses require co-action between light qualities, specifically between red and blue light (Chon and Briggs, 1966; Mohr, 1994). Because GLN2 and ASN2 are not modulated by blue or red monochromatic illumination in the presence of Suc, it would be interesting to investigate dichromatic illumination in the presence of Suc to see whether these wavelengths together are able to significantly induce the expression of ASN2 and GLN2. Because the coaction between wavelengths of light should be observed by illumination with WL, it is possible that the WL used in this study did not encompass the appropriate fluence rates of blue and red light to observe this interaction.
Carbon Potentiates Far-Red-Light Induction of GLN2 and ASN2 in Light-Grown Plants
FRLF induces GLN2 and ASN2 in the presence of carbon and has no effect on the expression of these genes in the absence of carbon (Fig. 4, A and B). This suggests that the presence of carbon permits regulation by far-red light of these genes. The fact that in the presence of Suc, far-red is the only wavelength of light able to induce the expression of ASN2 and GLN2 suggests that phytochrome is involved in the regulation of these genes in photosynthetically active plants. Exploitation of different phytochrome mutants in this response will be useful to identify the photoreceptor (s) involved in this response. Additionally, it is interesting that GLN2 and ASN2 both retain the same regulation by far-red light in the presence of Suc. It is possible that these genes share a similar pathway for far-red-light regulation in the presence of Suc in light-grown plants.
Carbon Affects Far-Red-Light Repression of ASN1 in Light-Grown Plants
ASN1 is super-repressed by FRHF in the absence of carbon, whereas in the presence of carbon, FRHF is only repressive (Fig. 4C). This suggests that carbon may antagonize the far-red super-repression of ASN1. FRLF is only repressive in the presence of carbon, indicating that carbon may potentiate the far-red repression of ASN1. The interaction between carbon and far-red light suggests carbon interacts with a phytochrome-signaling pathway. By contrast, blue-light repression of ASN1 occurs in the presence or absence of carbon (Fig. 4C), indicating the involvement of a blue-light photoreceptor or signaling pathway that is carbon independent.
Interactions between Distinct Light Qualities and Carbon
Previous studies have shown interactions between carbon and phytochrome signaling (Barnes et al., 1996; Dijkwel et al., 1997; Short, 1999). Our studies extend this analysis to describe models for specific interactions of carbon and red- versus far-red-light signaling. In addition, we discovered that blue-light perception or signaling is influenced by the carbon status of the plants. This is of new and particular interest, suggesting an interaction between blue-light and sugar signaling. The photoreceptor, cryptochrome 1 is primarily involved in BHF-light responses (Cashmore, 1997; Cashmore et al., 1999), phototropin is involved in BLF-light responses (Liscum and Briggs, 1995), and cryptochrome 2 plays a role in both BHF- and BLF-light responses (Lin, 2000). Experiments are under way in our lab to identify the blue-light-signaling pathway affected by Suc through the analysis of these blue-light photoreceptor mutants.
Combinatorial Design for Further Analysis of Complex Interactions
This initial, systematic investigation into the interactions of light and/or carbon signaling was investigated by looking at monochromatic light of different wavelengths and fluences independent of each other, in the absence or presence of carbon. This work sets the stage for further investigation into light and carbon signaling using photoreceptor mutants and downstream light-signaling mutants. It is also known that complex interactions exist between different qualities of light, where some physiological responses require dichromatic wavelengths of lights to achieve their maximum effects (Chon and Briggs, 1966; Mohr, 1994). These interactions of light qualities and the possible influence of Suc on these interactions are of interest to us. Table III shows additional experiments designed using combinatorial design to investigate how combinations of multiple light quality inputs, and their interactions with each another and with carbon can ultimately affect gene expression (Shasha et al., 2001; L.V. Lejay, D.E. Shasha, A.Y. Kouranov, P.M. Palenchar, A.A. Cruikshank, M. Chou, and G.M. Coruzzi, unpublished data). Combinatorial design allows a minimal number of experiments to be designed that cover a large experimental space of treatment conditions (Shasha et al., 2001; L.V. Lejay, D.E. Shasha, A.Y. Kouranov, P.M. Palenchar, A.A. Cruikshank, M. Chou, and G.M. Coruzzi, unpublished data). The design of experiments in this manner should allow us to study and model a large web of light and carbon interactions, using a minimal number of samples, amenable to genome scale analyses. Furthermore, such studies that include the analysis of genome-scale data should allow us to model networks of genes that are the downstream targets of converging light- and carbon-signaling pathways in plants. Such information should enable us to predict how changes in light quality, and photosynthesis will affect many processes involved in metabolism and plant development.
Table III.
Prediction of additional experiments using combinatorial design (Shasha et al., 2001) to further investigate interactions between light qualities alone and their interaction with sugar
Input variables are indicated as present (Y) or absent (N) or present at a low fluence (LF) or high fluence (HF).
| Experiment No. | Carbon | Blue Light | Red Light | Far-red Light |
|---|---|---|---|---|
| 1 | N | LF | HF | HF |
| 2 | N | HF | N | HF |
| 3 | N | N | LF | HF |
| 4 | N | LF | LF | N |
| 5 | N | HF | LF | LF |
| 6 | N | N | HF | LF |
| 7 | N | N | N | N |
| 8 | N | LF | N | LF |
| 9 | N | HF | HF | N |
| 10 | Y | LF | HF | HF |
| 11 | Y | HF | N | HF |
| 12 | Y | N | LF | HF |
| 13 | Y | LF | LF | N |
| 14 | Y | HF | LF | LF |
| 15 | Y | N | HF | LF |
| 16 | Y | N | N | N |
| 17 | Y | LF | N | LF |
| 18 | Y | HF | HF | N |
MATERIALS AND METHODS
Plant Growth and Treatment for Analysis
All experiments were carried out at least five times using the ecotype Columbia of Arabidopsis. Seeds were surface-sterilized, plated on designated media, and vernalized for 48 h at 8°C. For studies on etiolated seedlings, approximately 150 seeds plate–1 were grown on media containing 1× basal Murashige and Skoog (Invitrogen, Carlsbad, CA) and 0.9% (w/v) bactoagar, pH adjusted to 5.7 with KOH, supplemented with 2 mm KNO3, and either 0% or 1% (w/v) Suc. Plants were grown vertically in the dark at 23°C for 7 d, after which seedlings grown on 0% or 1% (w/v) Suc-containing media were maintained in the darkness or illuminated with either red (2 or 100 μE m–2 s–1), blue (2 or 100 μE m–2 s–1), far-red (2 or 100 μE m–2 s–1), or WL (70 μE m–2 s–1) for an additional 3 h. For experiments carried out on light-grown plants, approximately 30 seeds plate–1 were grown on the same media used for etiolated seedlings, except the media contained 0.5% (w/v) Suc. Plants were grown vertically under 16-h-light (70 μE m–2 s–1)/8-h-dark cycles at a constant temperature of 23°C. After growth for 14 d, all plants were transferred to fresh media containing either 0% or 1% (w/v) Suc and dark-adapted for 48 h, after which the plants were treated with different light treatments as described for etiolated seedlings. After light treatments, whole plants were harvested, immediately frozen in liquid nitrogen, and stored at –80°C until further use.
Light Sources
Photon fluence rates of WL, red, and blue light were measured with a quantum photometer (LI-1800, LI-COR, Lincoln, NE). WL was obtained from fluorescent light tubes (F72T12/CW; Philips, Eindhoven, The Netherlands). Blue light was obtained using actinic blue-light tubes (peak at 420 nm, Coralife, Pembroke Pines, FL). Red and far-red light was obtained using an SNAP-LITE light-emitting diode array from Quantum Devices (Barneveld, WI). All light experiments were carried out in light-tight boxes maintained in a dark, temperature-controlled environmental growth chamber.
RNA Isolation and Quantitative PCR
RNA was isolated from whole plants according to Kim et al. (1993). cDNA synthesis from total RNA was carried out according to Invitrogen (catalog no. 11146-024). Subsequent real-time quantitative PCR was carried out with a LightCycler (Roche Diagnostics, Mannheim, Germany). PCR amplification in a 20-μL reaction volume consisted of a master mixture containing Taq DNA polymerase, dNTP mixture and buffer (LightCycler DNA Master Hybridization probes, Roche Diagnostics), 4 mm MgCl2, 0.9 μm of each primer, 0.2 μm of each hybridization probe, and cDNA in a glass capillary tube. Primers and hybridization probes spanned at least one intron for each gene analyzed and were designed using the LightCycler probe design software (Roche Diagnostics). The primers were synthesized at Invitrogen and the fluorescent-labeled hybridization probes were synthesized and HPLC purified by TIB Molbiol LLC (Adelphia, NJ). Anchor probes were labeled at the 3′ end with fluorescein, and sensor probes were labeled at the 5′ end with LC-Red 640 and phosphorylated at the 3′ end. The following primers and probes were used for amplification and detection: ASN1, 5′-TCACGCTGCTCAAAATGT-3′ (forward primer), 5′-AGCTTGCATCCCACTC-3′ (reverse primer), 5′-AGAACTCTGCGAGACTAACGG-3′ (anchor probe), and 5′-CCTGGAGGTGCCACCG-3′ (sensor probe); ASN2, 5′-GAGCGACTGTACCAGG-3′ (forward primer), 5′-ACAACGTGTATCACTTGC-3′ (reverse primer), 5′-ATGGGATGCAACTTGGTCAAAG-3′ (anchor probe), and 5′-TCTTGATCCGTCAGGCCGT-3′ (sensor probe); GLN2, 5′-AGCTAGTATTGACCAGTTCT-3′ (forward primer), 5′-GCTGCAAGGGCTTCAG-3′ (reverse primer), 5′-AACCGTGGATGCTCTATTCGT-3′ (anchor probe), and 5′-GGGACGTGACACCGAGG-3′ (sensor probe); and At4g24550 (putative clathrin coat assembly protein), 5′-ATACACTGCGTGCAAAG-3′ (forward primer), 5′-TTCGCCTGTGTCACAT-3′ (reverse primer), 5′-AAGGAAGCAGGGCCAGT-3′ (anchor probe), and 5′-AAGGAAGCAGGGCCAGT-3′ (sensor probe). Thermal cycling was performed as follows: initial denaturation at 95°C for 2 min, followed by 35 cycles of denaturation at 95°C for 0 s, annealing at 55°C for 5 s (GLN2) or 10 s (ASN1, ASN2, and At4g24550), and extension at 72°C for 10 s (GLN2) or 15 s (ASN1, ASN2, and At4g24550). Standards were prepared with a 10-fold serial dilution (10–4 to 10 pg) of the PCR products and were run under the same PCR conditions used for the samples. The amount of ASN1, ASN2, and GLN2 was corrected/normalized according to the amount of At4g24550.
Boolean Analysis
For Boolean analysis (Nelson and Nagle, 1995), the two base conditions (a) no carbon, no light and (b) carbon, no light were used as a comparison against non-base conditions. For every non-base condition, the expression of the target gene was compared with the expression of that target gene in the base condition. If the expression was significantly different based on an unpaired t test at the 5% level, the values of all input variables for that non-base condition and the expression level relative to the base condition were recorded. For the experiments in this study, the input variables are carbon, WL, BLF, BHF, RLF, RHF, FRLF, and FRHF. The expression annotation relative to the base condition was (a) super-inductive if the average expression value was more than 10 times greater than the level for the base condition; (b) inductive if the average expression value was less than or equal to 10 times greater than the level for the base condition but remains significantly inductive; (c) super-repressive if the average expression value was more than 10 times less than the level for the base condition; or (d) repressive if the average expression value was less than or equal to 10 times less than the level for the base condition but still significantly repressive. The set of all recorded input values at a certain annotation level constitutes a Boolean conjunction, where Boolean circuit reduction techniques reduced this set to fewer conjunctions having “don't care” elements.
Acknowledgments
We thank Michael Shin for his comments and critical reading of the manuscript and Shelly Davidor for her technical assistance.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.022780.
This work was supported by the Department of Energy (grant no. DEFG02–92–20071 to G.M.C.), the National Science Foundation (grant nos. 11S-9988636 and N2010-0115586 to D.E.S.), and by the National Institutes of Health, National Research Service Award (no. GM63350 to K.E.T.).
References
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, Leon P (2000) Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev 14: 2085–2096 [PMC free article] [PubMed] [Google Scholar]
- Arkin A, Ross J, McAdams HH (1998) Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics 149: 1633–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes SA, Nishizawa NK, Quaggio RB, Whitelam GC, Chua N-H (1996) Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell 8: 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Huala E (1999) Blue-light photoreceptors in higher plants. Annu Rev Cell Dev Biol 15: 33–62 [DOI] [PubMed] [Google Scholar]
- Cashmore AR (1997) The cryptochrome family of photoreceptors. Plant Cell Environ 20: 764–767 [Google Scholar]
- Cashmore AR, Jarillo JA, Wu Y-J, Liu D (1999) Cryptochromes: blue light receptors for plants and animals. Science 284: 760–765 [DOI] [PubMed] [Google Scholar]
- Chon HP, Briggs WR (1966) Effect of red light on the phototropic sensitivity of corn coleoptiles. Plant Physiol 41: 1217–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschigano KT, Melo-Oliveira R, Lim J, Coruzzi GM (1998) Arabidopsis gls mutants and distinct Fd-GOGAT genes: implications for photorespiration and primary nitrogen assimilation. Plant Cell 10: 741–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh C-H, Minokawa T, Amore G, Hinman V, Arenas-Mena C et al. (2002) A genomic regulatory network for development. Science 295: 1669–1678 [DOI] [PubMed] [Google Scholar]
- D'haeseleer P, Liang S, Somogyi R (2000) Genetic network inference: from co-expression clustering to reverse engineering. Bioinformatics 16: 707–726 [DOI] [PubMed] [Google Scholar]
- Dijkwel PP, Huijser C, Weisbeek PJ, Chua N-H, Smeekens S (1997) Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell 9: 583–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J (1997) Light control of plant development. Annu Rev Cell Dev Biol 13: 203–229 [DOI] [PubMed] [Google Scholar]
- Genoud T, Metraux JP (1999) Crosstalk in plant cell signaling: structure and function of the genetic network. Trends Plant Sci 12: 503–507 [DOI] [PubMed] [Google Scholar]
- Genoud T, Trevino Santa Cruz MB, Metraux JP (2001) Numeric simulation of plant signaling networks. Plant Physiol 126: 1430–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss PJE, Peccoud J (1998) Quantitative modeling of stochastic systems in molecular biology by using petri nets. Proc Natl Acad Sci USA 95: 6750–7655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Mockler T, Duong H, Lin C (2001) SUB1, an Arabidopsis Ca2+-binding protein involved in cryptochrome and phytochrome coaction. Science 291: 487–490 [DOI] [PubMed] [Google Scholar]
- Hsieh M-H (1999) Metabolic regulation of asparagine synthetase genes and characterization of a putative nitrogen sensor (PII) in Arabidopsis. PhD thesis. New York University, New York
- Huijser C, Kortstee A, Pego J, Weisbeek P, Wisman E, Smeekens S (2000) The Arabidopsis sucrose uncoupled-6 gene is identical to abscisic acid insensitive-4: involvement of abscisic acid in sugar responses. Plant J 23: 577–585 [DOI] [PubMed] [Google Scholar]
- Halford NG, Purcell PC, Hardle DG (1999) Is hexokinase really a sugar sensor in plants? Trends Plant Sci 4: 117–119 [DOI] [PubMed] [Google Scholar]
- Jang J-C, Leon P, Zhou L, Sheen J (1997) Hexokinase as a sugar sensor in higher plants. Plant Cell 9: 5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Christopher DA, Mullet JE (1993) Direct evidence for selective modulation of psbA, rpoA, rbcL and 16S RNA stability during barley chloroplast development. Plant Mol Biol 22: 447–463 [DOI] [PubMed] [Google Scholar]
- Koch KE (1996) Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 509–540 [DOI] [PubMed] [Google Scholar]
- Lam HM, Coschigano K, Oliveira IC, Melo-Oliveira R, Coruzzi GM (1996) The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47: 569–593 [DOI] [PubMed] [Google Scholar]
- Lam HM, Coschigano K, Schultz C, Melo-Oliveira R, Tjaden G, Oliveira I, Ngai N, Hsieh MH, Coruzzi GM (1995) Use of Arabidopsis mutants and genes to study amide amino acid biosynthesis. Plant Cell 7: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H-M, Hsieh M-H, Coruzzi G (1998) Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana. Plant J 16: 345–353 [DOI] [PubMed] [Google Scholar]
- Lam H-M, Peng S-Y, Coruzzi G (1994) Metabolic regulation of the gene encoding glutamine-dependent asparagine synthetase in Arabidopsis thaliana. Plant Physiol 106: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C (2000) Plant blue-light receptors. Trends Plant Sci 5: 337–342 [DOI] [PubMed] [Google Scholar]
- Liscum E, Briggs WR (1995) Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell 7: 473–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancinelli AL (1994) The physiology of phytochrome action. In RE Kendrick, GHM Kronenberg, eds, Photomorphogenesis in Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 211–269
- Melo-Oliveira R, Olivseira IC, Coruzzi G (1996) Arabidopsis mutant analysis and gene regulation define a nonredundant role for glutamate dehydrogenase in nitrogen assimilation. Proc Natl Acad Sci USA 93: 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr H (1994) Coaction between pigment systems. In RE Kendrick, GHM Kronenberg, eds, Photomorphogenesis in Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 353–372
- Moller SG, Ingles PJ, Whitelam GC (2002) The cell biology of phytochrome signaling. New Physiol 154: 553–590 [DOI] [PubMed] [Google Scholar]
- Nagy F, Schafer E (2002) Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu Rev Plant Physiol Plant Mol Biol 53: 329–355 [DOI] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J (2000) Light: an indicator of time and place. Genes Dev 14: 257–271 [PubMed] [Google Scholar]
- Nelson VP, Nagel TH (1995) Logic circuit analysis and design. In VP Nelson, TH Nagel, JD Irwin, eds, Logic Circuit and Design. Prentice Hall Press, Englewood Cliffs, NJ, pp 175–211
- Ngai N, Coruzzi G (1998) Dissecting light repression of the asparagine synthetase gene (AS1) in Arabidopsis. In FL Schiavo, RL Last, G Morelli, NV Raikhel, eds, Cellular Integration of Signaling Pathways in Plant Development, NATO ASI Series. Springer-Verlag, Berlin, pp 147–157
- Oliveira IC, Coruzzi GM (1999) Carbon and amino acids reciprocally modulate the expression of glutamine synthetase in Arabidopsis. Plant Physiol 121: 301–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell Suppl 2002: S185–S205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shasha DE, Kouranov AY, Lejay LV, Chou MF, Coruzzi GM (2001) Using combinatorial design to study regulation by multiple input signals. A tool for parsimony in the post-genomics era. Plant Physiol 127: 1590–1594 [PMC free article] [PubMed] [Google Scholar]
- Sheen J, Zhou L, Jang J-C (1999) Sugars as signaling molecules. Curr Opin Plant Biol 2: 410–418 [DOI] [PubMed] [Google Scholar]
- Short T (1999) Overexpression of Arabidopsis phytochrome B inhibits phytochrome A function in the presence of sucrose. Plant Physiol 119: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S (2000) Sugar-induced signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol 51: 49–81 [DOI] [PubMed] [Google Scholar]
- Smith H (1994) Sensing the light environment: the functions of the phytochrome family. In RE Kendrick, GHM Kronenberg, eds, Photomorphogenesis in Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 377–416
- Weaver DC, Workman CT, Stormo GD (1999) Modeling regulatory networks with weight matrices. Pac Symp Biocomputing 4: 29–40 [DOI] [PubMed] [Google Scholar]
- Yuh CH, Bolouri H, Davidson EH (1998) Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science 279: 1896–1902 [DOI] [PubMed] [Google Scholar]