Abstract
Expressed sequence tags (ESTs) currently encompass more entries in the public databases than any other form of sequence data. Thus, EST data sets provide a vast resource for gene identification and expression profiling. We have mapped the complete set of 176,915 publicly available Arabidopsis EST sequences onto the Arabidopsis genome using GeneSeqer, a spliced alignment program incorporating sequence similarity and splice site scoring. About 96% of the available ESTs could be properly aligned with a genomic locus, with the remaining ESTs deriving from organelle genomes and non-Arabidopsis sources or displaying insufficient sequence quality for alignment. The mapping provides verified sets of EST clusters for evaluation of EST clustering programs. Analysis of the spliced alignments suggests corrections to current gene structure annotation and provides examples of alternative and non-canonical pre-mRNA splicing. All results of this study were parsed into a database and are accessible via a flexible Web interface at http://www.plantgdb.org/AtGDB/.
The efforts of an international collaboration to obtain the complete genome sequence of the flowering plant Arabidopsis resulted in the release and annotation of 115.4 Mb of the genome (estimated at 125 Mb) in December of 2000 (Arabidopsis Genome Initiative, 2000). At that time, 25,498 protein-coding genes were identified in the five haploid chromosomes, but only 9% of these genes had been characterized experimentally, and only 69% could be functionally classified by similarity to proteins of known functions. In the interim, sequencing and annotation has progressed. The most current release of the Arabidopsis genome available at GenBank provides 117.3 Mb and 27,288 annotated protein-coding genes (see Data Sets in “Materials and Methods”). Annotation of the Arabidopsis genome and functional characterization of all the genes is an ongoing effort. Initial, high-throughput computational gene structure prediction has likely been successful in identifying most gene locations; however, these methods still suffer from limitations in predicting the precise gene structure for an entire gene, detection of intergenic regions, and identification of non-coding exon sequences (Pavy et al., 1999; Brendel and Zhu, 2002). Recent studies have concentrated on sequencing of full-length cDNAs to improve genome annotation (Haas et al., 2002; Seki et al., 2002).
Expressed sequence tags (ESTs) are single-pass sequencing reads of cDNA clones that have become a widely employed method for gene identification, expression profiling, and polymorphism analysis. Presently, more than 13.4 million EST entries have been deposited into the National Center for Biotechnology Information (NCBI) dbEST public database, including Arabidopsis with 176,915 ESTs and 21 other species with EST sets of more than 100,000 entries (http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html). In the absence of a whole-genome sequencing project for a particular species, clustering of ESTs into contigs that represent unique genes is one of the most promising strategies to glimpse the gene space of that organism. Challenges of EST clustering arise from poor average sequence quality, incomplete EST sampling, polymorphisms, alternative transcript isoforms, representation of highly similar transcripts from distinct members of multigene families, and cloning artifacts. Different strategies for EST clustering and the associated gene indexing databases have been reviewed by Bouck et al. (1999); for a recent method for EST clustering on parallel computers, see Kalyanaraman et al. (2003).
For Arabidopsis, up-to-date EST clusters are available in form of the UniGene clusters at NCBI (http://www.ncbi.nlm.nih.gov/UniGene/) and as a The Institute for Genome Research (TIGR) Gene Index (AtGI; http://www.tigr.org/tdb/tgi/agi/; Quackenbush et al., 2001). The current UniGene build (no. 28) comprises 27,248 clusters derived from 220,191 sequences (including 55,519 mRNAs). The current AtGI (release 9.0) comprises 38,462 clusters from 232,136 sequences. Whereas UniGene clusters are meant to represent all transcript isoforms derived from a gene locus, different transcript isoforms should split into distinct TIGR clusters. In either case, the clusters are constructed on the basis of mRNA sequence comparisons only. Of course, this is necessary for most species for which only limited genome sequence data are available. Here, we present results of Arabidopsis EST clustering based on direct spliced alignment of the ESTs onto the Arabidopsis genome. This approach has significant advantages when applicable (e.g. Kan et al., 2001; Yeh et al., 2001). First, accuracy should be greatly increased because cognate EST genomic locations can be easily identified for most ESTs, and clusters can be determined by proximity of EST locations on the genome scaffold. Second, the spliced alignments provide a rich data source to probe the extent and characteristics of alternative splicing, non-canonical splice sites, and other features of gene structure. We provide different sets of genome-confirmed EST clusters that can serve as standards for the comparison of programs and parameter settings for mRNA-based EST clustering. We discuss differences between current Arabidopsis gene structure annotation and EST-based gene annotation. All alignment results were imported into a relational database that is accessible via the Web at http://www.plantgdb.org/AtGDB/ and includes extensive tools for visualization and further analysis.
RESULTS
EST Spliced Alignments
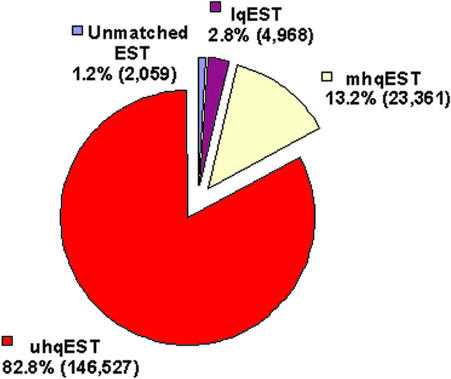
The Arabidopsis EST (ATest) data set employed in this study consists of 176,915 entries. As shown in Figure 1, only 2,059 EST sequences (1.2%) did not show any significant alignments with the genome. Further investigation based on BLASTN (Altschul et al., 1997) searches against the nonredundant nucleotide database at NCBI (E value < 1e-10) showed that about 40% (822) of those unmatched ESTs have no hits, about one-fifth (401) resulted from contamination (matching sequences from clone vectors, insects, fungi, etc.) or low-complexity sequences, and another 27% (557) came from the organelle genomes (mitochondrial and chloroplast). Surprisingly, most of the remaining sequences were found to have significant hits against sequences from Arabidopsis. Failure of these sequences to produce a valid spliced alignment could be attributed to either of two causes. First, the matching genomic sequences have not yet been assembled into the published Arabidopsis genome sequence. Thus, some ESTs clearly match with Arabidopsis bacterial artificial chromosomes (for example, EST gi:19837354 matches with bacterial artificial chromosome gi:18149207 derived from the centromere region of chromosome four) but do not match with the released Arabidopsis genome sequence. Second, with default parameters, GeneSeqer does not detect weak matches that may arise from poor sequence quality (for example, EST gi:9783909) or lowcomplexity regions (for example, EST gi:9787792). Such failed alignments are expected because no repeat masking or quality clipping was performed to preprocess the EST sequences before aligning them with the genome.
Figure 1.
Classification of Arabidopsis ESTs based on spliced alignment quality. Of a total of 176,915 ESTs, 2,059 ESTs have no significant hits in the Arabidopsis genome, 4,968 ESTs have only low-quality spliced alignments (lqEST), and the remaining 169,888 ESTs have hqSPAs. The latter category consists of 146,527 ESTs that match a unique (i.e. their cognate) locus in the genome (unique high-quality ESTs [uhqEST]) and 23,361 ESTs that have multiple hqESTs (mhqESTs), representing different loci of duplicated genes or multigene families.
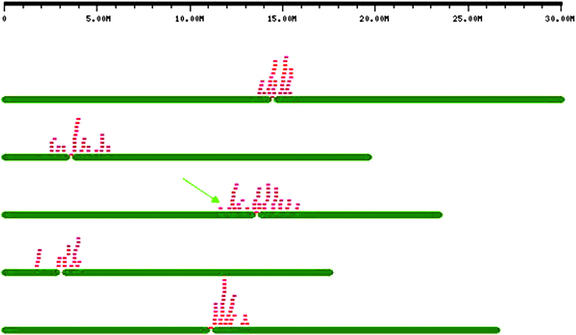
Of the ESTs, 96.0% have at least one high-quality spliced alignment (hqSPA; see “Materials and Methods”) with the Arabidopsis genome (such ESTs denoted as high-quality ESTs [hqESTs]), and about 13.2% have more than one hqSPA with the genome (such ESTs denoted as mhqESTs; see Fig. 1). The distribution of the number of hqSPAs per hqEST is shown in Table I. The majority of the ESTs have only one or two hqSPAs, but there are 38 ESTs with at least 10 hqSPAs. These ESTs were found to be associated with transposon families and other highly prolific genome elements. For example, EST gi:9787698 (with 170 hqSPAs) appears to be derived from an Arabidopsis putative retroelement polyprotein gene, clustered around all five centromeres of the Arabidopsis genome as shown in Figure 2.
Table I.
Distribution of the no. of hqSPAs per hqEST
All hqESTs (Figure 1) were classified according to their no. of hqSPAs. The chromosomal distribution of the 170 hqSPAs of EST gi:9787698 is displayed in Figure 2.
| No. of hqSPAs | No. of ESTs |
|---|---|
| 1 | 146,527 |
| 2 | 16,116 |
| 3 | 3,697 |
| 4 | 2,235 |
| 5 | 945 |
| 6 | 196 |
| 7 | 68 |
| 8 | 46 |
| 9 | 20 |
| 10-19 | 22 |
| 20-99 | 14 |
| 164 | 1 |
| 170 | 1 |
| Total | 169,888 |
Figure 2.
Distribution of the 170 hqSPAs for EST gi:9787698 on the Arabidopsis genome. Each chromosome is represented by two dark-green bars, with the centromere marked by a space between the horizontal bars. Locations of the spliced alignments are shown by red bricks. Almost all hits are around the centromeres. Alignment scores suggest that the EST originates from the 12,075,567- to 12,075,806-bp region on chromosome three (marked by the green arrow). This EST shows high similarity with Arabidopsis gene At1g38360 (gi:18426880), a putative retroelement polyprotein gene. This display is shown as an example of the visualization tools at Arabidopsis Genome Database (AtGDB) that will dynamically generate similar graphics for any set of GenBank gi accessions or genes matched by common descriptions.
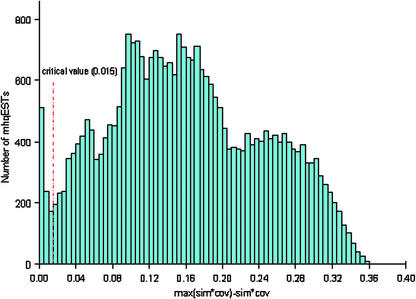
Overall, about 82.8% (146,527 entries) of the ATest data set are uhqESTs (see “Materials and Methods”), which align with a single locus in the genome. To properly position the remaining ESTs, which display multiple hqSPAs, we make the assumption that for each EST the alignment with maximal score (similarity score × coverage score) identifies the true cognate location of that EST. Such alignments are designated putative cognate spliced alignments (pcSPAs; see “Materials and Methods”). In this way, 172,137 pcSPAs were generated from 169,888 hqESTs and 206,833 hqSPAs. Because of virtual equalities among the scores of some hqSPAs for certain mhqESTs (Fig. 3), there are more pcSPAs than hqESTs.
Figure 3.
Distribution of the score differences between maximal and submaximal scoring hqSPAs for mhqESTs. Each hqSPA is scored by the product of similarity and coverage values (see “Materials and Methods”). Most of the score differences fall in the range 0.08 to 0.20. Based on the displayed distribution, a critical value 0.015 was set such that each hqSPA with a score difference smaller than 0.015 compared with the maximal scoring hqSPA for a given EST is designated pcSPA, representing the likely origin of this specific EST in the genome.
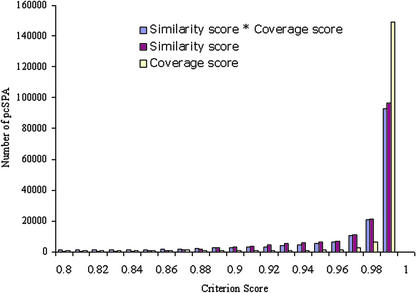
We should emphasize that our restriction on hqESTs largely eliminates typical problems of EST clustering and EST-based gene annotation, as caused by chimeric clones, for example. Thus, chimeric sequences would typically lead to alignments with coverage score below 0.8 because in any given genomic location, only one part of the sequence would match (or if the foreign sequence were only very short, it would not be used in the GeneSeqer spliced alignment, which optimizes the local alignment score). According to the aforementioned assumption, the similarity and coverage scores for each pcSPA correlate with our confidence in the prediction of cognate transcript origin for the hqEST in question. Higher alignment similarity and coverage scores denote greater confidence. The vast majority of pcSPAs have similarity and coverage scores in the 0.99 to 1.0 range (Fig. 4). This implies high confidence in the classification of these alignments as cognate. The designation of “putative” cognate is formally accurate, however, because the matched ESTs and genomic sequences were not isolated from the same plant. When considering the alignment of ESTs not derived from the Columbia ecotype on which the genomic sequences are based, cognate position implies the cognate origin of the most probable transcript ortholog to the aligned EST. According to dbEST annotation, about 98% of the Arabidopsis ESTs were derived from the Columbia ecotype. Three hundred of the 337 ESTs annotated as derived from ecotype Landsberg have pcSPAs with average similarity score 0.93 and average coverage score 0.94. Thus, the different Arabidopsis ecotypes appear to have such a high degree of sequence conservation that correct mapping of the ESTs onto the Columbia ecotype genome is unproblematic (see also Haas et al., 2002).
Figure 4.
Histogram showing the distribution of pcSPA similarity, coverage, and combined scores.
EST Clustering and Assembly
EST assembly refers to the problem of finding the correct orientation and order of EST sequences in a tiling path covering the cognate mRNA. Because EST sequences are typically generated by single-pass sequencing and, thus, contain a fair number of errors and ambiguous bases, this assembly can be difficult in the absence of genome sequence data. However, when the entire genome sequence is available, the spliced alignment of ESTs gives reliable assemblies and can be used for prediction of gene structure and alternative splicing (Kan et al., 2001; Yeh et al., 2001).
Because of the relative facility of EST sequencing, EST projects have outpaced genome sequencing projects for many species. EST clustering is typically the first analysis step in deriving a “unigene” set representing the transcriptome of the species. By clustering, EST sequences that share significant sequence similarity are partitioned into presumed gene-specific contigs, thus reducing the redundancy of the EST set. Such reduction is often dramatic, especially in the case of EST sets not derived from normalized libraries. Cluster-based reduction may be a practical necessity before EST assembly for large EST sets. Here, we are particularly interested in evaluating the utility of ESTs in gene identification. pc-SPAs, representing putative cognate gene locations, were clustered based on chromosome location. Each cluster contains ESTs from a single gene provided that the intergenic regions between neighboring genes are sufficiently long compared with the maximal allowed gap (negative overlap) set by the clustering parameters (see “Materials and Methods”). Because genome-based EST clustering does not depend on pair-wise EST sequence overlap, which is a necessary requirement for comparison-based assembly programs, small gaps in local genome coverage can be allowed, thereby joining partial gene annotations through a genome scaffolding scheme. In addition, high coverage as required for the pcSPAs excludes erroneous alignment of chimeric clones, which typically pose annoying problems for comparison-based assembly.
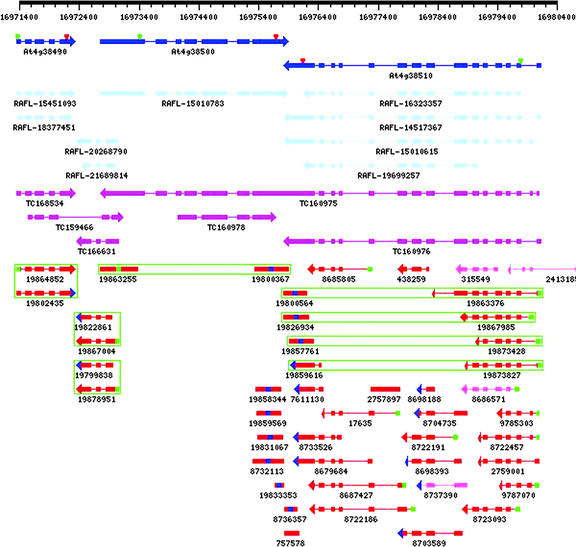
Figure 5 provides an example of the possibilities and difficulties of gene structure annotation by EST clustering. Full-length cDNA evidence indicates four genes in alternating directions in the displayed region of chromosome four. Current GenBank annotation misses the second gene, the 5′ end of which is overlapping the 5′ end of the third gene transcribed on the opposite strand. Genome-based EST clustering without using clone pair information would give the three clusters that are bounded by ESTs gi:19864852 and gi:19802435, gi:19822861 and gi:19863255, and gi:8732113 and gi:19863376, respectively. If clone pair information is used, the clusters resolve to four clusters that correctly identify the four genes. For comparison, Figure 5 also shows the alignment of TIGR Arabidopsis Gene Index tentative contigs (Quackenbush et al., 2001). Note the erroneous concatenation of ESTs in TC159466 and TC160975, resulting from clustering based on significant overlap only (but not coding strand identification).
Figure 5.
Visual assessment of EST clustering and gene characteristics for a region of the Arabidopsis genome. In the display, which is available for all genomic regions at http://www.plantgdb.org/AtGDB/, pcSPAs originating from EST spliced alignments are shown in red and non-pcSPAs in pink. For multi-exon alignments, the arrow indicates the direction of transcription, inferred from the implied splice site patterns (Usuka et al., 2000). Multi-exon 5′ ESTs are marked by green color at their 5′ terminus, and multi-exon 3′ ESTs are marked by blue color at their 3′ terminus. Single-exon ESTs have corresponding 5′/3′ labels at the center of their representations. Pairs of 5′ and 3′ ESTs from the same clone are grouped by green boxes. PcSPAs originating from cDNA spliced alignments are shown in light blue, and non-pcSPAs are shown in gray. Dark-blue gene structures represent the current GenBank gene annotations for this region. The 5′ and 3′ boundaries of the corresponding coding regions are indicated by green and red triangles, respectively. Note that the current annotation misses the gene represented by clone pair ESTs gi:19867004 and gi:19822861 and gi:19878951 and gi:19799838. The purple structures represent the spliced alignments of TIGR Arabidopsis Gene Index tentative contigs. The figure also shows an alternatively spliced internal mini-exon. This exon of 16 nucleotides occurs in the 5′-UTR of At4g38510, an H+-transporting ATPase (EC 3.6.1.35). The transcript isoform including this intron is supported by ESTs gi:9785303 and gi:8722457. In the same region, EST gi:9787070 supports a different internal exon of 73 nucleotides, and EST gi:19867985 (equal to RAFL-15010615) indicates an alternative transcription start. Note that all sequence records at AtGDB are identified by their unique GenBank gi identifiers. The Riken Arabidopsis full-length (RAFL) cDNAs (Seki et al., 2002) thus indicated as RAFL-15451093, RAFL-18377451, RAFL-20268790, RAFL-21689814, RAFL-15010783, RAFL-14517367, RAFL-16323357, RAFL-15010615, and RAFL-19699257 correspond to clones RAFL05-11-M12, U16016, RAFL06-81-F18, U11966, RAFL03-01-G10, RAFL04-09-A19, U12748, RAFL07-17-H08, and U12937, respectively.
Choosing various clustering parameters from 50-bp overlap to 100-bp gap was shown to alter the number of clusters by less than 12% (Table II). The following results are based on the 27,611 clusters obtained by allowing a maximal gap of 60 bp (other criteria give similar results; data not shown). About one-half of the clusters contain only one or two pcSPAs (Table III). Large clusters correspond to highly expressed genes (e.g. Fernandes et al., 2002), including Rubisco, PSII type I chlorophyll a/b-binding protein, seed storage protein, and ribosomal proteins (for descriptions of all clusters with at least 100 pcSPAs, see http://www.plantgdb.org/AtGDB/prj/ZSB03PP/virtualNorthern.html). More than 64.5% (17,609) of the annotated genes have at least one pcSPA within their annotated boundaries, with an average of about seven ESTs supporting each of these annotations (range: 1–1,014). Of the 27,288 annotated gene-coding regions, 22.5% (6,141) are fully covered by an EST cluster, and for 44.8% (12,226) of the annotated genes, clone pair-joined clusters confirm the annotated extent of the coding region.
Table II.
Effect of clustering criterion on the no. of EST clusters
ESTs were clustered based on their genomic locations, derived from pcSPAs. Several clustering parameters were tested, ranging from requiring a minimum of 50-bp overlap between clustered ESTs to a maximal gap of 100 bp between clustered EST ends.
| Clustering Criterion | No. of Clusters |
|---|---|
| =50-bp overlap | 30,154 |
| =0-bp overlap | 28,883 |
| =50-bp gap | 27,787 |
| =60-bp gap | 27,611 |
| =100-bp gap | 26,956 |
Table III.
Distribution of EST cluster size
Cluster size is given in no. of ESTs. The displayed nos. are based on the clusters derived with the criterion of a 60-bp maximal gap (Table II).
| Cluster Size | No. of Clusters |
|---|---|
| 1 | 9,488 |
| 2 | 4,977 |
| 3-4 | 4,927 |
| 5-8 | 3,971 |
| 9-16 | 2,378 |
| 17-32 | 1,132 |
| 33-64 | 472 |
| 65-128 | 185 |
| 129-256 | 60 |
| 257-512 | 113 |
| 513-1024 | 8 |
| Total | 27,611 |
Gene Identification by ESTs
As described in the next section, some of our spliced alignment results contradict particular gene models in the most recent Arabidopsis genome annotation. To safeguard against possible errors in our employed methods, we exploited a set of 5,000 nonredundant full-length cDNAs derived in a Ceres/TIGR collaboration (Haas et al., 2002; ATcdna; see “Materials and Methods”) for benchmarking. In particular, we sought to determine, first, whether the cDNA spliced alignments were consistent with the genome annotation and, second, how the EST spliced alignments and assemblies compared with the cDNA spliced alignments. It should be noted that the Ceres/TIGR full-length cDNAs were derived from the Wassilewskija and Landsberg erecta, rather than Columbia, Arabidopsis ecotypes; however, Haas et al. (2002) reported more than 99% average identity between the three ecotypes, confirmed by our spliced alignment results.
The results showed that 4,999 of the cDNAs have at least one hqSPA. The only unmatched cDNA (gi:21405014, Ceres identification no. CT23693) matches mitochondrial DNA. Generally, the pcSPA of a full-length cDNA is regarded to be the most decisive experimental evidence to define gene structures. Therefore, the cDNA-derived pcSPAs provide a reliable set to assess EST-based gene prediction. Overall, the 4,999 cDNAs have 4,691 uhqSPAs, 308 mhqSPAs, and 5,013 pcSPAs. Surprisingly, 1,100 (21.9%) of the pcSPAs are embedded in longer EST clusters (see http://www.plantgdb.org/AtGDB/prj/ZSB03PP/extendedCoverage.html). This discrepancy may result from alternative transcription initiation and termination sites or systematic biases in the cDNA cloning process (Haas et al., 2002). Alternative transcription initiation and termination sites may also reflect polymorphisms among different Arabidopsis ecotypes. Of the pcSPAs, 91.0% (4,563) are at least partially covered with ESTs, with an average of 10 EST-derived hqSPAs supporting each (partially) covered gene (range: 1–652). On the intron level, 81.8% (13,980) of 17,091 introns (including low-quality introns) deduced from pcSPAs of full-length cDNAs are supported by EST alignments. The majority of the annotated introns are consistent with the high-quality introns derived from cDNA spliced alignments as we expected, but there are still 28 annotated introns that are contradicted, associated with 23 distinct annotated genes (see http://www.plantgdb.org/AtGDB/prj/ZSB03PP/geneAnnotationVScdna.html).
Because the cDNA-covered gene set is not representative of the entire Arabidopsis gene set (highly expressed genes have a greater chance to be cloned and sequenced both as ESTs and full-length cDNAs), the 91% fraction of pcSPAs from full-length cDNAs covered also by the EST-derived pcSPAs is an upper bound of the estimated fraction of genes identified by ESTs. The comparison confirms that both ESTs and cDNAs were accurately mapped to the genome with our method and that these approaches provide both alternative and complementary paths to gene discovery.
AtGDB
Spliced alignments of ATest and ATcdna and the recent annotation of the Arabidopsis genome were parsed and imported into an MySQL relational database, which was named AtGDB. An elaborate Web interface was designed for the database to allow users to browse the genome and query the database by sequence similarity, identifiers, or description (http://www.plantgdb.org/AtGDB/). In general, the Web interface is composed of three parts: the genomic context view, the query view, and the sequence view. The genomic context view allows users to browse a specific genomic region in the context of multiple annotation resources. The region graphic displays these multiple sources of alignment information relative to one another. Each is colored with respect to its specific annotation source (see Fig. 5). The query view allows users to view and interact with the results of a user query. Stored EST/cDNA alignments and annotated transcripts each have an individual page, the sequence view, which glues together sequence data, analysis tools, and related external links. This Web interface efficiently presents the database entries on the fly and facilitates data access and utilization as described below.
Applications
After mapping the ESTs to the genome, we not only acquired the genomic loci each EST originated from but also confirmation of other annotation resources by comparison with the EST spliced alignments. Here, we explored several applications listed below. However, we should emphasize that we cannot describe in-depth analysis of these data within the scope of this manuscript and rather wish to point out possibilities of further studies based on the rich data source provided by the comprehensive EST mapping.
Consistency of Gene Structure Annotation
The annotation of the Arabidopsis genome referred to in this study was published on August 20, 2002, and represents the most current genome annotation released by the Arabidopsis Genome Initiative. Because much of the annotation is still computationally produced without human expert scrutiny, EST evidence may not always have been incorporated into the gene models. To estimate the extent of this problem, we compared annotated intron positions with predicted intron sequences based on our EST spliced alignments. As a result, 58,120 of the 115,949 annotated introns were confirmed. Another 1,272 annotated introns are inconsistent with high-quality predicted introns inferred from the spliced alignments. These introns occur in 977 distinct gene models or about 3.4% of the annotated genes (data available at http://www.plantgdb.org/AtGDB/prj/ZSB03PP/geneAnnotationVSest.html). Although these discrepancies may be caused by alternative transcript isoforms, erroneous gene prediction seems a more parsimonious explanation in the absence of other evidence.
In addition to suggesting corrections to current gene annotations, the EST spliced alignments also identify novel gene locations. Thus, of the 27,611 EST contigs assembled on the basis of proximity in their genomic locations, 129 occur in regions without any annotated gene models and contain open reading frames (ORFs) longer than 100 residues that show no significant hits with annotated Arabidopsis proteins using BLASTP (threshold 1e-10). Eighty-two of these show no hits at the same threshold when compared against the NCBI nonredundant protein database, and the remaining 47 EST contigs show at least one hit (data available at http://www.plantgdb.org/AtGDB/prj/ZSB03PP/novelGenes.html). For example, ESTs gi:19863912, gi:9786135, and gi:8721866 form a cluster that supports an ORF of 108 residues between genes At4g02400 and At4g02410; the existence of a gene in that region is also supported by full-length cDNAs gi:14596167 and gi:20148266. In other cases, the novel ORFs may correspond to upstream or downstream exons of incompletely annotated genes. The display at AtGDB allows users to provide updated annotation upon more in-depth analysis of individual cases.
5′- and 3′-Untranslated Regions (UTRs) in mRNAs
Most annotated gene models correspond to the coding portions of exons only. Although attempts have been made recently to predict the UTR portions of mRNAs by genome sequence inspection (Davuluri et al., 2000, 2001; Tabaska et al., 2001), this has proven to be a difficult endeavor. If UTRs are annotated, the annotations are derived mostly from full-length cDNAs. ESTs provide a more accessible resource to gain UTR information, provided accurate EST assembly and mapping onto the genome is possible.
The gene density in the Arabidopsis genome is high, with about one gene every 5 kb. Therefore, intergenic regions are typically very short, which may make accurate UTR assignments difficult. We cataloged high-quality predicted introns that mapped into annotated intergenic regions into potential 5′-UTR or 3′-UTR introns, depending on whether the constituent hqSPAs extend from the flanking coding region into the upstream or downstream region, respectively (note that in some cases the additional exons may extend an annotated ORF; thus, the derived set of potential UTR introns is a superset of EST-confirmed UTR introns). In this way, 2,282 potential 5′-UTR introns in 2,023 annotated genes (including 199 genes with multiple potential 5′-UTR introns; all data displayed at http://www.plantgdb.org/AtGDB/prj/ZSB03PP/upstreamUTRintrons.html) and 570 potential 3′-UTR introns in 487 annotated genes (including 47 genes with multiple potential 3′-UTR introns; all data displayed at http://www.plantgdb.org/AtGDB/prj/ZSB03PP/downstreamUTRintrons.html) were identified. Seventy-two genes have both potential 5′-UTR and potential 3′-UTR introns. Thus, at least 9% of Arabidopsis genes may have introns in their UTRs. Our listing of these features at AtGDB should provide a valuable resource to study possible roles for these introns in the regulation of gene expression and to develop models for UTR prediction (see also dbUTR; Pesole et al., 2002).
Non-Canonical Splice Sites
Almost all introns contain the canonical GT-AG splice site junctions, but other varieties also exist. It was estimated that about 1% of Arabidopsis introns are non-canonical GC-AG introns (Brown et al., 1996), slightly higher than the proportion identified in mammals (Burset et al., 2000, 2001). In all other respects, GC-AG introns seem to be analogous to the canonical GT-AG introns, and they are processed in the same splicing pathway (U2-type spliceosome). AT-AC introns, with consistently low frequency in diverse eukaryotic taxa, are another well-studied type of non-canonical introns, which are typically spliced by a distinct U12-type spliceosome (Wu and Krainer, 1996; Wu et al., 1996; Burge et al., 1998).
In this study, 738 introns (1.7% of the 43,165 high-quality predicted introns derived from EST alignments) were found to have non-canonical splice sites (Table IV). GC-AG introns represent the large majority of non-canonical introns (453 cases, or about 1.0% of all high-quality predicted introns). AT-AC introns comprise the second largest category (25 cases). Many of the non-canonical introns have short direct repeats spanning the donor and acceptor sites. In these cases, the exact intron position cannot be unambiguously determined by spliced alignment; thus, some of the classifications in Table IV may prove incorrect. The complete listing of apparent noncanonical introns (http://www.plantgdb.org/AtGDB/prj/ZSB03PP/ncSpliceSites.html) should facilitate experimental investigation of splicing in the absence of the standard splice site features.
Table IV.
Non-canonical introns (NN represents any dinucleotide)
The intron types were assigned by the terminal intron dinucleotides based on high-quality spliced alignments.
| Type | No. |
|---|---|
| GC-AG | 453 |
| NN-AG (not including GC-AG and GT-AG) | 99 |
| GT-NN (not including GT-AG) | 80 |
| AT-AC | 25 |
| GC-NN (not including GC-AG) | 14 |
| Others (26 patterns, each with less than six hits) | 67 |
| Total | 738 |
The 453 GC-AG introns (http://www.plantgdb.org/AtGDB/prj/ZSB03PP/non_canonical/gc_ag.html) have the consensus donor sequence (non-U)AG/GCAAGU (donor site boldfaced) exactly as reported before for other data sets (Burset et al., 2000). These introns exhibit a similar distribution of predicted splice site scores as do GT-AG introns (Brendel and Kleffe, 1998; data not shown). This suggests that the mechanism of splicing of GC-AG introns may be the same as that of GT-AG introns but involve more highly conserved sequence features apart from the GC dinucleotide.
Dietrich et al. (1997) reported that U12-type introns are more likely to be determined by the conserved motifs around the donor site and the branch site than by the dinucleotide-termini of the intron. Consistently, some AT-AC introns are spliced by the U2-type spliceosome, whereas some GT-AG introns are spliced by the U12-type spliceosome. In this study, all but two of the 25 AT-AC introns (http://www.plantgdb.org/AtGDB/prj/ZSB03PP/non_canonical/at_ac.html) exhibit both the ATATCCTY donor site motif and the TCCTTRAY branch site element (Wu and Krainer, 1996; Burge et al., 1998). The two exceptions (derived from the uhqSPAs of ESTs gi:931334 and gi:19874656) may not be typical U12-type AT-AC introns and could also be classified as non-canonical TT-CC and TC-CA introns, respectively. In addition, one AT-AA intron and 17 GT-AG introns were identified as likely U12-type introns based on a more detailed motif search (see “Materials and Methods”). All 41 likely U12-introns and related information are listed at http://www.plantgdb.org/AtGDB/prj/ZSB03PP/u12Introns.html.
Mutual comparison of the genes containing the putative U12-type introns shows that some of them may correspond to duplications within gene families. For example, the genes At1g56280, At5g26990, At3g06760, At3g05700, and At4g02200 all encode a drought-induced-19 like protein. A detailed study shows that all five genes have a U12-type intron between coding exons three and four (At4g02200 has a U12-dependent GT-AG intron, whereas the other four genes have a U12-dependent AT-AC intron). Similarly, the genes At3g53520 (Fig. 6A) and At3g62830, which encode a dTDP-Glc 4-6-dehydratase-like protein, also both have a U12-dependent AT-AC intron in the same location. Inspection of a homologous rice gene shows that the U12-type intron location is not only conserved among the Arabidopsis paralogs but also across the monocot/dicot divide (Fig. 6B). This observation is consistent with the conjecture of the early origin of U12-class introns (Wu et al., 1996; Burge et al., 1998; Wu and Krainer, 1999).
Figure 6.
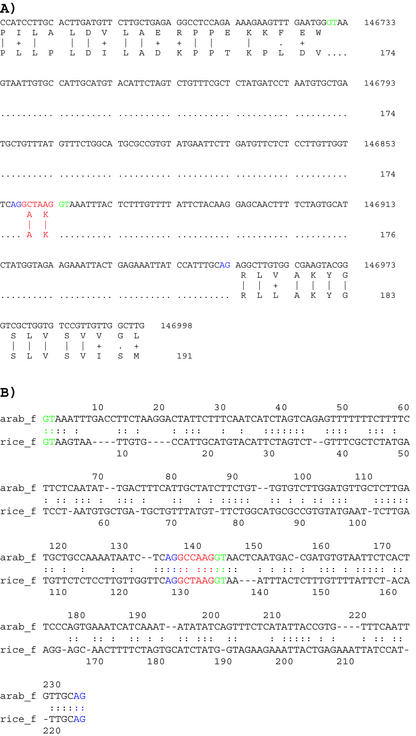
Spliced alignment of Arabidopsis EST gi:5839990 with: A, the Arabidopsis At3g53520 gene encoding a dTDP-Glc 4-6-dehydratase-like protein; and B, a rice (Oryza sativa) genomic sequence (accession no. AP003271). The two alignments reveal conserved gene structure between Arabidopsis and rice, including a conserved AT-AC intron. C, Pair-wise alignment of the orthologous AT-AC intron sequences. The conserved donor site (ATATCCTY) and branch site motifs (TCCTTRAY) are highlighted in red color.
The analysis of U12-type introns gives an example of how to utilize the EST data and AtGDB resource, and it also exposes several annotation problems. For instance, of the 23 AT-AC U12-type introns, only four AT-AC introns are explicitly annotated (At3g53520, At5g22650, At5g26990, and At5g27380). One AT-AC U12-type intron in gene At3g62830 is incorrectly annotated as a CA-TA intron, even with the presence of six cognate full-length cDNAs. In addition, gene structures predicted by AB INITIO methods will typically never include non-canonical introns (for example, the gene At1g76170). Furthermore, EST data can provide a check on the accuracy of the genome sequence (Brendel and Zhu, 2002). For example, 24 ESTs supporting the AT-AC U12-type intron in the drought-induced-19-like gene At1g56280 clearly suggest that one adenosine should be inserted after the 20,673,745-bp position in chromosome one of the current genome assembly. This inference is also supported by two cognate full-length cDNAs.
Alternative Splicing
Current research suggests that approximately 40% to 60% of human genes are alternatively spliced (Black, 2000; Brett et al., 2002; Modrek and Lee, 2002). Identification of alternative splicing is generally based on cDNA or EST evidence (Kan et al., 2001; Coward et al., 2002; Huang et al., 2002; Modrek and Lee, 2002). Based on strict criteria and manual inspection (see “Materials and Methods”), we identified 327 cases of alternative splicing among Arabidopsis genes and categorized them into five groups: (a) alternative donor sites (102 cases), (b) alternative acceptor sites (190 cases), (c) alternative introns that are shifted in position at both sites (three cases), (d) exon skipping (21 cases), and (e) composite alternative splicing (different combinations of several alternative splicing events, 11 cases). All cases and the EST evidence are displayed at http://www.plantgdb.org/AtGDB/prj/ZSB03PP/alternativeSplicing/. Intron retention may in part result from inconsequential inefficient splicing or inclusion of incompletely spliced transcripts in EST libraries; thus, evidence for intron retention is not discussed further here (338 cases; see (http://www.plantgdb.org/AtGDB/prj/ZSB03PP/alternativeSplicing/intron_retention.html). Based on the EST evidence, we calculated a lower bound for the fraction of alternatively spliced genes as 1.2% (327 of 27,288). Although EST sampling and coverage remains limited, alternative splicing would seem to be much less pervasive than observed in mammalian systems. For example, if we assume that 5% (or 20%) of the transcripts of an alternatively spliced gene represent the alternative isoform, then the average of seven ESTs per gene result in a 30% (or 79%) detection rate of this gene as alternatively spliced. Thus, limited EST sampling alone should not account for the low estimate of the fraction of alternatively spliced genes.
Mini-Exons and Mini-Introns
Currently, there are two nonexclusive models regarding the mechanisms of splicing: Intron definition purports interactions of splice site recognition factors across the intron, whereas exon definition suggests interactions of splicing factors at the acceptor and donor sites from consecutive introns across the interspersed exon (Berget, 1995). The latter model provides a conceptual framework for the molecular recognition of the very long introns occurring in some mammalian genes, whereas the former model may be the simplest model for recognition of terminal and short introns. For either model, the existence of very short introns and exons raises difficult questions about the steric accommodation of multiple splicing factors.
Based on EST evidence, we did not find any introns less than 50 bp. According to the GenBank annotation, there are 46 introns ranging from 1 to 10 bp, but it seems likely that these are annotation mistakes. One 27-bp intron was annotated in the gene At3g53740, which is supported by full-length cDNA CT267357 (gi:21405387). However, 33 pcSPAs uniformly support a continuous exon in that position. It is possible that this region is polymorphic between Columbia and other ecotypes and that the cognate origin of CT267357 includes a standard-sized intron.
Conversely, 128 nonterminal mini-exons are supported by EST evidence. These exons range in size from 5 to 25 bp, with 13 of them no longer than 10 nucleotides in length (http://www.plantgdb.org/AtGDB/prj/ZSB03PP/miniexons.html). In a few cases, these mini-exons may occur in regions of increased alternative splicing activity. An example of this is given in Figure 5. However, most mini-exons appear to be constitutively spliced, as confirmed by the consistent alignments of several ESTs. For example, a six-nucleotide exon in At5g14030 is unanimously confirmed by 12 EST spliced alignments and conserved in an apparent rice homologous gene (Figs. 7 and 8). Due to steric constraint imposed by their size, we find it difficult to explain the accurate splicing of mini-exons by exon definition, and intron-definition and/or facilitation of splicing by splicing enhancers may be a more plausible splice site selection model in this case. Interestingly, most mini-exons are characterized by high splice prediction scores in the flanking exon-intron junctions (data not shown), suggesting that the associated spliceosome and mechanism of splicing involved in resolving mini-exons may be highly similar to that of normal exons.
Figure 7.
Visualization of an annotated, normally expressed internal mini-exon. The exon of six nucleotides found in the 3′-coding region of At5g14030 (encoding an unknown protein) is supported by 12 different EST spliced alignments. Strikingly, this miniature exon is also conserved in what appears to be a rice homolog of this gene (see Fig. 8). Symbols are as in Figure 5. The three cDNAs identified by GenBank gi as CT-21404330, RAFL-14517445, RAFL-22136543 correspond to Ceres/TIGR full-length cDNA 16313 and RAFL clones RAFL02-05-J08 and U12778, respectively.
Figure 8.
Evolutionary conservation of a miniexon. A, Spliced alignment of the translated ORF (bottom lines) originating from the EST cluster shown in Figure 7 with a rice genomic clone (GenBank accession no. AP003727); the alignment was made with the GeneSeqer program (Usuka and Brendel, 2000). B, Alignment of the Arabidopsis mini-exon and its flanking introns with a homologous region of the rice genome. The mini-exon is highlighted in red characters, the intron donor sites in green, and the intron acceptor sites in blue.
DISCUSSION
ESTs have become the most popular method for gene discovery in eukaryotic species without a whole-genome sequencing project and a key technology for genome annotation when genome sequence data are available. We are particularly interested in systematic, functional, and phylogenetic comparisons of the gene repertoires of plants. Currently, a near-complete genome has been assembled for only Arabidopsis and rice. In contrast, some of the largest species-specific EST collections are from plants, including wheat (Triticum aestivum; more than 415,000), barley (Hordeum vulgare; more than 310,000), soybean (Glycine max; more than 305,000), maize (Zea mays; more than 195,000), and Medicago truncatula (more than 180,000; http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html). Kalyanaraman et al. (2003) present a novel algorithm and software program (PaCE) to cluster large sets of ESTs into contigs that represent distinct gene fragments and its application to 22 plant species EST sets. Our motivation for the mapping of Arabidopsis ESTs onto the Arabidopsis genome was in part derived from the need for a confirmed standard of proven EST clusters against which to gauge the success of EST clustering programs that do not incorporate genome sequence data. Here, we derived a number of different standards from uniquely mapped Arabidopsis ESTs depending on the minimal overlap required between different EST spliced alignments. All spliced alignments are displayed at a novel Web resource, http://www.plantgdb.org/AtGDB/, which was specifically designed to view and explore all Arabidopsis gene structure annotation and evidence.
In comparison with other indexing methods such as UniGene or the TIGR Gene Indices that work entirely on the mRNA level, genome location-based clustering not only has the advantage of accuracy but also allows using low-quality ESTs more effectively. For example, EST gi:8332684 has a uhqSPA with a similarity score marginally higher than 0.8, but the GeneSeqer spliced alignment still accurately reveals the exon-intron boundaries of the gene At1g20620 (catalase 3). This EST is clustered with hundreds of other cognate ESTs located in the same region. However, although labeled as weakly similar to At1g20620, it is clustered as a singleton in the TIGR Arabidopsis Gene Index.
Surprisingly, the complete EST mapping revealed a large number of discrepancies between the current gene structure annotation and assignments of exons and introns indicated by the spliced alignment. Previously, Haas et al. (2002) reported that 1,591 Arabidopsis genes were incorrectly annotated at the time of their comparison with the 5,000 full-length Ceres/TIGR cDNAs, and an additional 240 putative novel genes were identified by the same set of cDNAs. This suggested that full-length cDNA data should greatly improve genome annotation efforts. The most recent release of Arabidopsis genome annotation from TIGR, used in this study, does incorporate full-length cDNA spliced alignments, thereby reducing the number of contradictory annotations compared with prior annotations. However, there are still about 1,000 genes inaccurately annotated according to our analysis. Furthermore, GeneSeqer alignments using the same full-length cDNA data set as Haas et al. (2002) indicates that for 23 matching genes the current annotation remains erroneous (http://www.plantgdb.org/AtGDB/prj/ZSB03PP/geneAnnotationVScdna.html), suggesting that even with full-length cDNAs, gene identification is still not trivial. Interestingly, about 20% of the gene locations of this representative set of full-length cDNAs are embedded in longer EST alignments (http://www.plantgdb.org/AtGDB/prj/ZSB03PP/extendedCoverage.html). Haas et al. (2002) also reported length differences between the Ceres/TIGR full-length cDNAs (ATcdna set, from ecotypes Wassilewskija and Landsberg erecta) and RIKEN full-length cDNAs (ecotype Columbia; Seki et al., 2002) that may reflect alternative transcription initiation and termination sites, possibly polymorphic among different ecotypes. However, these differences are minor and do not in any other way obscure gene structure prediction. In particular, our comparison with EST spliced alignments show that, except for a few cases, the cDNA confirmed introns are identically predicted by the EST alignments, about 98% of which are with ESTs from the Columbia ecotype. The current sampling of ESTs from different ecotypes is insufficient to assess differences in gene expression or splicing patterns between the ecotypes.
In addition to providing the standards for EST clustering and data for refining basic gene structure annotation, the spliced alignments also provide a rich resource for more in-depth analysis of pre-mRNA processing, including assessment of the extent of alternative splicing and use of non-canonical splices sites. Based on very stringent spliced alignment criteria, we established alternative splicing (excluding possible intron retention) for only about 1.5% of the Arabidopsis genes. The majority of alternative splicing occurs at either the donor site or the acceptor site of an intron but not on both ends simultaneously (292 of 327 cases). We also observe that most alternative splice sites are within 50 bp of the common splice site (220 of 292). Specifically, in 134 cases, the distances between the alternative splice site and the common splice site are less than 10 bp. Such transcript isoforms with minor difference may be easily over-looked in conventional EST clustering and transcript assembly. For example, the gene At1g02500 has two alternative isoforms with a difference of only 3 bp in the location of the acceptor site of its sole intron. Each of the isoforms has at least six ESTs to support its unique gene structure. However, all of these ESTs are assembled into one index in the TIGR Arabidopsis Gene Index (Cluster ID:TC149272).
Most certainly, these estimates of the occurrence of alternative splicing are very conservative. First, these estimates were based on only very good spliced alignments that leave no doubt as to the origin of the respective ESTs. Second, the ATest collection is still very small compared with the human collection, for example, which is about 30 times larger. However, we still estimate the occurrence of alternative splicing in Arabidopsis much lower than the reported 40% to 60% of human genes (Black, 2000; Brett et al., 2002; Modrek and Lee, 2002).
Currently, most gene identification efforts rely heavily on ab initio gene prediction programs (Pavy et al., 1999). However, few ab initio gene identification programs successfully make alternative splicing predictions, consider non-canonical splice sites, or allow other exceptional cases. For example, a special situation where the start codon (ATG) of a gene is interrupted by an intron would confuse almost every ab initio gene prediction algorithm currently available. Similarly, mini-exons (EST confirmed examples of which are displayed at http://www.plantgdb.org/AtGDB/prj/ZSB03PP/miniexons.html) will generally be neglected due to their small coding potential, especially if the length of the mini-exon is a multiple of three. Thus, it would seem imperative that spliced alignment be a key technology of genome annotation. The GeneSeqer program (Usuka et al., 2000) is very convenient for that purpose.
To facilitate refined genome annotation and further study of pre-mRNA processing based on the spliced alignment data, all of our results were stored in a MySQL database and are visually presented on a special Web site, AtGDB (http://www.plantgdb.org/AtGDB/). Several established and comprehensive Arabidopsis databases are already available to date, such as TAIR (http://www.arabidopsis.org/), Munich Information Center for Protein Sequences (http://mips.gsf.de/proj/thal/), and the TIGR Arabidopsis Database (http://www.tigr.org/tdb/e2k1/ath1/). All displays in AtGDB are linked to the corresponding entries in those databases. AtGDB adds a convenient sequence-centered view of the genome. Users of AtGDB can easily find the distribution of target sequences in the genome, see their related annotations, and exact genomic coordinates (based upon the most recent release of Arabidopsis genome annotation) of ESTs and cDNAs. Analytical tools are linked to the displays to allow further analysis with additional data, for example spliced alignment with ESTs from sources other than Arabidopsis. We hope that this analysis and the new Web tools will contribute to more complete and accurate genome annotation.
MATERIALS AND METHODS
Data Sets
The five chromosome sequences of Arabidopsis were obtained from GenBank (http://www.ncbi.nih.gov/entrez/query.fcgi?db=Nucleotide) as accessions NC_003070 (chromosome I, dated August 20, 2002, 30,028,691 bp), NC_003071 (chromosome II, dated August 20, 2002, 19,646,746 bp), NC_003074 (chromosome III, dated August 20, 2002, 23,467,821 bp), NC_003075 (chromosome IV, dated August 20, 2002, 17,550,036 bp), and NC_003076 (chromosome V, dated August 20, 2002, 26,583,670 bp). Arabidopsis ESTs were downloaded from the dbEST database (http://www.ncbi.nlm.nih.gov/dbEST/). Our analysis was based on 176,915 EST records available October 25, 2002 (data set label: ATest). According to the GenBank records, 111,155 non-RIKEN ESTs were derived from the Columbia ecotype. An additional 61,481 ESTs are from RIKEN, and these ESTs also were from Columbia (Seki et al., 2002). Only 337 ESTs are indicated as ecotype Landsberg, and no ecotype information is given for the remaining about 4,000 ESTs. A set of 27,288 putative Arabidopsis proteins was obtained from TIGR (ftp://ftp.tigr.org/pub/data/a_thaliana/ath1/SEQUENCES/ATH1.pep), which represented the latest annotation of the Arabidopsis genome made by TIGR (data set label: ATpep, version: July 25, 2002). Full-length cDNAs (5,017) sequenced by Ceres, Inc. were downloaded from the TIGR ftp site (ftp://ftp.tigr.org/pub/data/a_thaliana/ath1/ceres/Ceres.arab.cdna). Only the subset of 5,000 sequences deposited in GenBank (Entrez search: Arabidopsis [ORGN] AND FLI_CDNA [KYWD] AND Haas [AUTH]) were used in this study (data set label: ATcdna, version: March 2, 2001). These cDNAs were derived from the Wassilewskija and Landsberg erecta ecotypes (Haas et al., 2002).
EST Mapping by Spliced Alignment
Alignment of cDNAs or ESTs to a genomic template is known as spliced alignment because the alignment must correctly reflect the removal of introns from the pre-mRNA copy of the genomic template. Several programs and services are available for this task, including PROCRUSTES (Gelfand et al., 1996), NAP (Huang et al., 1997), SIM4 (Florea et al., 1998), est_genome (Mott, 1997), Spidey (Wheelan et al., 2001), and GeneSeqer (Usuka et al., 2000; Usuka and Brendel, 2000). The alignments discussed here were derived with the GeneSeqer program. The program involves preprocessing of the cDNA/EST set to generate a suffix array of these sequences, subsequent fast matching of cDNAs/ESTs to the genome based on significant blocks of sequence identity, and spliced alignment by dynamic programming based on predicted splice site probabilities and sequence similarity scores. Using default parameter settings, the entire mapping of ATest was achieved in about 120 h on a 1-GHz Pentium Pro III processor CPU.
Selection of hqEST Alignments
The default GeneSeqer parameters are set to allow detection of gene structure through alignment of ESTs from non-cognate ESTs derived from a homologous gene elsewhere in the genome (or even ESTs from a homologous locus in a related species). For some of the questions studied here, it was necessary to restrict the data to only the cognate alignments. Because of allelic variation and sequencing errors, even cognate alignments will not necessarily display 100% sequence matching; however, the overall alignment quality generally should be much higher than for heterologous alignments. For a given EST, GeneSeqer assesses alignment quality by two parameters: a similarity score, defined as the ratio of the observed alignment score over the maximum possible alignment score obtained in the absence of any substitutions and insertions or deletions; and a coverage score, defined as the fraction of the EST nucleotides involved in the displayed alignment (because the GeneSeqer spliced alignment is local, any poorly matching N- or C-terminal EST regions are culled from the displayed alignment). Here, we define hqEST spliced alignments (hqSPAs) as alignments that give similarity and coverage scores both of at least 0.8. ESTs with at least one hqSPA are defined as hqEST. An hqEST is further categorized according to the number of hqSPAs derived from the given EST. It is called a uhqEST if the EST matches a unique locus in the genome, and it is called an mhqEST if the EST matches multiple sites in the genome (presumably corresponding to duplicated genes). The corresponding spliced alignments are referred to as uhqSPAs and mhqSPAs.
The major task of spliced alignment discussed in this paper was to identify cognate positions for each entry of ATest. Because the EST set was not masked or filtered to remove contaminations, low-complexity regions, or repeats, and because high-sensitivity/low-specificity default GeneSeqer parameters were applied for the spliced alignment, we limited most of our derived results to hqSPAs and hqESTs. The product of similarity and coverage scores was utilized as a measure to identify the pcSPAs, based on the assumption that the pcSPA should have the best score among hqSPAs for each specific hqEST. Due to recent gene duplications, possible genome assembly errors, or other uncertain reasons, some hqESTs may have several hqSPAs with identical or near-identical score in different locations of the genome. Thus, the pcSPA for each hqEST is not necessarily unique. The distribution of score differences among multiple hqSPAs for an EST is shown in Figure 3. Based on this distribution, all hqSPAs with scores strictly within 0.015 of the maximal score for that EST were labeled as pcSPA. With default parameters, a GeneSeqer-reported similarity score of s corresponds to 0.5 × (1 + s) × 100% sequence identity (for an alignment without gaps). Thus, two alternative full-length alignments of an EST will be distinguished as cognate and non-cognate if the weaker match has on average one additional mismatch to the genomic sequence per 100 nucleotides compared with the better match. The average nucleotide difference between the duplicated genes identified by hqESTs was calculated as 11.4% ± 4.6%. Therefore, the given criterion would safely distinguish duplicated genes except for very recent duplications that result in such minor sequence differences that they are indistinguishable from EST sequencing error rates.
EST Clustering and Assembly
hqESTs were mapped to the Arabidopsis genome based on pcSPAs as described in the previous section. The mapped hqESTs were clustered according to genome coordinates derived from their pcSPAs requiring a defined minimal overlap length or a maximal coverage gap size. Precisely, let est1 map to region [a,b] and est2 to region [c,d], where a ≤ c on the same chromosome; then est1 and est2 are clustered if c ≤ b + G + 1, where G is the clustering parameter. G could be negative (overlap required) or positive (specifying the maximal allowed gap). For ESTs giving multiple exonspliced alignments, the overlap rule is superceded by the requirement for consistency of strand orientation as indicated by GeneSeqer. Thus, ESTs from overlapping genes in opposite transcriptional directions can be separated into different clusters (compare with Fig. 5). In addition, ESTs from the same plasmid (clone pairs) were used to join clusters independent of their local map coordinates. Different sets of clusters based on alignment and clustering parameters are available at http://www.plantgdb.org/AtGDB/prj/ZSB03PP/ESTclustering.html. ESTs of each cluster were further assembled by the built-in function of GeneSeqer to generate alternative gene structures and predicted peptide sequences (PPSs) derived from long ORFs in the alternative gene structures. The PPSs were searched against ATpep via BLASTP to locate putative novel genes as described below.
Quality Control
The set of full-length cDNAs was aligned to the genome similarly to the EST alignments (the GeneSeqer option –x 30 –y 50 was used, which probes for potential gene locations by about 50-base identities in the suffix array, thus quickly identifying cognate loci). These alignments served as quality control in two ways. First, the results test the integrity of our analysis method. Because these full-length cDNAs were used previously to improve the Arabidopsis genome annotation (Haas et al., 2002), the cDNA spliced alignments from GeneSeqer are expected to be consistent with the genome annotation. Second, we can check whether the pcSPAs are consistent with the cDNA alignments in regions of overlap. The 5,017 full-length cDNAs can be regarded as a random sample of the total gene set of Arabidopsis. Comparing the coverage of ESTs relative to these cDNAs tests the limits of EST projects.
Database and Web Interface
The raw output of GeneSeqer occupied a total of 1.6 billion bytes of disc space. The output was parsed and imported into an MySQL relational database management system (http://www.mysql.com) for further analysis. The database is accessible via the Web at http://www.plantgdb.org/AtGDB/. Supplementary data for the results of this study are available at http://www.plantgdb.org/AtGDB/prj/ZSB03PP/.
High-Quality Predicted Introns
GeneSeqer gives two scores to each splice site, a prediction score and a local similarity score. The prediction score is between 0 and 1.0, based on a statistical model for the probability of the site to function as a splice site. Non-canonical splice sites receive 0 as a prediction score. The local similarity score measures sequence matching in the 40- to 50-bp flanking exon regions derived from the spliced alignment. This score is also normalized to 1.0 for complete identity. For exons shorter than 40 bp, the local similarity scores of the flanking splice sites are both set to 0. In this study, high-quality predicted introns were selected as predicted introns with: (a) splice site prediction scores for the donor and acceptor sites both higher than 0, i.e. the intron should be a canonical intron; and (b) local similarity scores for the donor and acceptor sites both higher than 0.95 (implying that the flanking exons should be no less than 40 bp and that at most, one mismatch is allowed in the 40–50-bp flanking exon region alignment).
Identification of U12 Introns
The 5′ site motif ATCC in positions +3 to +6 is highly conserved in U12 introns (Wu and Krainer, 1996; Sharp and Burge, 1997; Burge et al., 1998), where the numbering +1 to +6 denotes the first six nucleotides of the intron starting at the 5′ splice site. On the basis of this observation, we selected one AT-AA and 153 GT-AG introns as potential U12-class introns among all the EST-confirmed introns (in addition to the 23 U12-dependent AT-AC introns discussed in the text). To further classify these sites, we used a procedure similar to those described by Burge et al. (1998) and Levine and Durbin (2001). First, MEME (Bailey and Elkan, 1994) was used to define motifs for the donor and branch sites of the 23 manually verified U12-class AT-AC introns. These motifs were then used to query the additional 154 candidates via the MAST application (E-value threshold set to 1.0; Bailey and Gribskov, 1998), and 18 introns with motif E-values less than 1.0 for both motifs were characterized as likely U12-introns.
Analysis of EST Spliced Alignments
The mapped ESTs provide a rich data set for studying many aspects of genome and gene structure. Here, we have explored the following issues.
Consistency of Gene Structure Annotation
EST spliced alignments reveal partial or full gene structures; thus, they are helpful to check and refine ab initio gene predictions (Brendel and Zhu, 2002). Distinct introns derived from all EST alignments were utilized to identify what fraction of annotated introns is supported by EST evidence. Only high-quality predicted introns were used to identify annotated introns that are not supported but contradicted by EST evidence. Even in the well-annotated Arabidopsis genome, there may still be some genes that are not yet described. We defined putative (partial) novel genes as EST-derived conceptual transcripts with an ORF longer than 300 bp (PPSs with more than 100 amino acid residues) but displaying no significant similarity to proteins in ATpep (threshold 1e-10 using BLASTP) and have no overlap with annotated genes.
5′- and 3′-UTRs in mRNAs
We used the EST evidence to identify UTR exons and introns.
Non-Canonical Splice Sites
Non-canonical splice sites obtain a prediction score of 0 in the GeneSeqer spliced alignments. Therefore, it is very simple to identify potential noncanonical introns. To exclude questionable spliced alignments and remove redundancy, only distinct introns with flanking exons of at least 40 bp and local similarity score greater than 0.95 were selected for further analysis and categorization according to the observed intron borders.
Alternative Splicing
All high-quality introns were mutually compared with find overlapped but nonidentical introns, indicating different types of alternative splicing (except intron retention, cases of which were identified separately).
Mini-Exons and Mini-Introns
Mini-exons were selected from hqSPAs containing at least one exon of at most 25 bp, with 100% alignment identity over the entire exon region and canonical splice sites as boundaries. Similar criteria were also applied to seek mini-introns not exceeding 50 bp in length.
Acknowledgments
The authors would like to thank Peter T. Vedell for early contributions to this work and Jessica A. Schlueter for critical reading of the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.018101.
This work was supported in part by the National Science Foundation (grant no. DBI–0110254 to V.B.).
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Bailey TL, Gribskov M (1998) Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14: 48–54 [DOI] [PubMed] [Google Scholar]
- Berget SM (1995) Exon recognition in vertebrate splicing. J Biol Chem 270: 2411–2414 [DOI] [PubMed] [Google Scholar]
- Black DL (2000) Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell 103: 367–370 [DOI] [PubMed] [Google Scholar]
- Bouck J, Yu W, Gibbs R, Worley K (1999) Comparison of gene indexing databases. Trends Genet 15: 159–162 [DOI] [PubMed] [Google Scholar]
- Brendel V, Kleffe J (1998) Prediction of locally optimal splice sites in plant pre-mRNA with applications to gene identification in Arabidopsis thaliana genomic DNA. Nucleic Acids Res 26: 4748–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel V, Zhu W (2002) Computational modeling of gene structure in Arabidopsis thaliana. Plant Mol Biol 48: 49–58 [PubMed] [Google Scholar]
- Brett D, Pospisil H, Valcarcel J, Reich J, Bork P (2002) Alternative splicing and genome complexity. Nat Genet 30: 29–30 [DOI] [PubMed] [Google Scholar]
- Brown JW, Smith P, Simpson CG (1996) Arabidopsis consensus intron sequences. Plant Mol Biol 32: 531–535 [DOI] [PubMed] [Google Scholar]
- Burge CB, Padgett RA, Sharp PA (1998) Evolutionary fates and origins of U12-type introns. Mol Cell 2: 773–785 [DOI] [PubMed] [Google Scholar]
- Burset M, Seledtsov IA, Solovyev VV (2000) Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res 28: 4364–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burset M, Seledtsov IA, Solovyev VV (2001) SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res 29: 255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward E, Haas SA, Vingron M (2002) SpliceNest: visualizing gene structure and alternative splicing based on EST clusters. Trends Genet 18: 53–55 [Google Scholar]
- Davuluri RV, Grosse I, Zhang MQ (2001) Computational identification of promoters and first exons in the human genome. Nat Genet 29: 412–417 [DOI] [PubMed] [Google Scholar]
- Davuluri RV, Suzuki Y, Sugano S, Zhang MQ (2000) CART classification of human 5′ UTR sequences. Genome Res 10: 1807–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich RC, Incorvaia R, Padgett RA (1997) Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol Cell 1: 151–160 [DOI] [PubMed] [Google Scholar]
- Fernandes J, Brendel V, Gai X, Lal S, Chandler VL, Elumalai RP, Galbraith DW, Pierson EA, Walbot V (2002) Comparison of RNA expression profiles based on maize expressed sequence tag frequency analysis and micro-array hybridization. Plant Physiol 128: 896–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W (1998) A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res 8: 967–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand MS, Mironov AA, Pevzner PA (1996) Gene recognition via spliced sequence alignment. Proc Natl Acad Sci USA 93: 9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Volfovsky N, Town CD, Troukhan M, Alexandrov N, Feldmann KA, Flavell RB, White O, Salzberg SL (2002) Full-length messenger RNA sequences greatly improve genome annotation. Genome Biol 3: research 0029.1–0029.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Adams MD, Zhou H, Kerlavage AR (1997) A tool for analyzing and annotating genomic sequences. Genomics 46: 37–45 [DOI] [PubMed] [Google Scholar]
- Huang YH, Chen YT, Lai JJ, Yang ST, Yang UC (2002) PALS db: Putative Alternative Splicing database. Nucleic Acids Res 30: 186–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyanaraman A, Kothari S, Brendel V, Aluru S (2003) Efficient clustering of large EST data sets on parallel computers. Nucleic Acids Res 31: in press [DOI] [PMC free article] [PubMed]
- Kan Z, Rouchka EC, Gish WR, States DJ (2001) Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res 11: 889–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A, Durbin R (2001) A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res 29: 4006–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek B, Lee C (2002) A genomic view of alternative splicing. Nat Genet 30: 13–19 [DOI] [PubMed] [Google Scholar]
- Mott R (1997) EST_GENOME: a program to align spliced DNA sequences to unspliced genomic DNA. Comput Appl Biosci 13: 477–478 [DOI] [PubMed] [Google Scholar]
- Pavy N, Rombauts S, Déhais P, Mathé C, Ramana DVV, Leroy P, Rouzé P (1999) Bioinformatics 15: 887–899 [DOI] [PubMed] [Google Scholar]
- Pesole G, Liuni S, Grillo G, Licciulli F, Mignone F, Gissi C, Saccone C (2002) UTRdb and UTRsite: specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Update 2002. Nucleic Acids Res 30: 335–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res 29: 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y et al. (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Sharp PA, Burge CB (1997) Classification of introns: U2-type or U12-type. Cell 91: 875–879 [DOI] [PubMed] [Google Scholar]
- Tabaska JE, Davuluri RV, Zhang MQ (2001) Identifying the 3′-terminal exon in human DNA. Bioinformatics 17: 602–607 [DOI] [PubMed] [Google Scholar]
- Usuka J, Brendel V (2000) Gene structure prediction by spliced alignment of genomic DNA with protein sequences: increased accuracy by differential splice site scoring. J Mol Biol 297: 1075–1085 [DOI] [PubMed] [Google Scholar]
- Usuka J, Zhu W, Brendel V (2000) Optimal spliced alignment of homologous cDNA to a genomic DNA template. Bioinformatics 16: 203–211 [DOI] [PubMed] [Google Scholar]
- Wheelan SJ, Church DM, Ostell JM (2001) Spidey: a tool for mRNA-togenomic alignments. Genome Res 11: 1952–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Gaubier-Comella P, Delseny M, Grellet F, Van Montagu M, Rouzé R (1996) Non-canonical introns are at least 10(9) years old. Nat Genet 14: 383–384 [DOI] [PubMed] [Google Scholar]
- Wu Q, Krainer AR (1996) U1-mediated exon definition interactions between AT-AC and GT-AG introns. Science 274: 1005–1008 [DOI] [PubMed] [Google Scholar]
- Wu Q, Krainer AR (1999) AT-AC pre-mRNA splicing mechanisms and conservation of minor introns in voltage-gated ion channel genes. Mol Cell Biol 19: 3225–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh RF, Lim LP, Burge CB (2001) Computational inference of homologous gene structures in the human genome. Genome Res 11: 803–816 [DOI] [PMC free article] [PubMed] [Google Scholar]