Abstract
Receptor-like kinases (RLKs) are a family of transmembrane proteins with versatile N-terminal extracellular domains and C-terminal intracellular kinases. They control a wide range of physiological responses in plants and belong to one of the largest gene families in the Arabidopsis genome with more than 600 members. Interestingly, this gene family constitutes 60% of all kinases in Arabidopsis and accounts for nearly all transmembrane kinases in Arabidopsis. Analysis of four fungal, six metazoan, and two Plasmodium sp. genomes indicates that the family was represented in all but fungal genomes, indicating an ancient origin for the family with a more recent expansion only in the plant lineages. The RLK/Pelle family can be divided into several subfamilies based on three independent criteria: the phylogeny based on kinase domain sequences, the extracellular domain identities, and intron locations and phases. A large number of receptor-like proteins (RLPs) resembling the extracellular domains of RLKs are also found in the Arabidopsis genome. However, not all RLK subfamilies have corresponding RLPs. Several RLK/Pelle subfamilies have undergone differential expansions. More than 33% of the RLK/Pelle members are found in tandem clusters, substantially higher than the genome average. In addition, 470 of the RLK/Pelle family members are located within the segmentally duplicated regions in the Arabidopsis genome and 268 of them have a close relative in the corresponding regions. Therefore, tandem duplications and segmental/whole-genome duplications represent two of the major mechanisms for the expansion of the RLK/Pelle family in Arabidopsis.
All living systems receive and process information at the cellular level through various classes of cell surface receptors. In plants, at least two different types of transmembrane kinases have been reported, including receptor-like Ser/Thr kinases (RLKs; for reviews, see Walker, 1994; Becraft, 1998; Torii, 2000; Shiu and Bleecker, 2001a) and receptor His kinases (for review, see Urao et al., 2001).
In Arabidopsis, RLKs belong to a large, monophyletic gene family (RLK/Pelle) with more than 610 members, including receptor kinases and non-receptor kinases (receptor-like cytoplasmic kinase [RLCK]; Shiu and Bleecker, 2001b). In addition to its large size, RLK family members vary greatly in their domain organization and the sequence identity in their extracellular domains (Shiu and Bleecker, 2001a). This diversity suggests that RLKs may function in the perception of a wide range of signals or stimuli. This notion is justified by the identification of putative protein and lipid ligands for RLKs, such as CLAVATA3 (CLV3) for CLAVATA1 (CLV1; Trotochaud et al., 2000; Brand et al., 2000), brassinosteroid for BRI1 (Wang et al., 2001), and SCR/SP11 for S-locus receptor kinase (SRK) (Kachroo et al., 2001; Takayama et al., 2001). On the basis of the presence/absence and identity of the extracellular domains and the kinase domain phylogeny, the RLK/Pelle family is subdivided into 45 subfamilies (Shiu and Bleecker, 2001b). The diversity of domain organization and large gene number in this family suggest that domain fusion contributed to the formation of novel receptor kinases, and subsequent gene duplications resulted in the expansion of the RLK/Pelle subfamilies in plants. On the basis of a preliminary analysis of four subfamilies on Arabidopsis chromosome 4, tandem duplication and whole-genome duplication and reshuffling are hypothesized to be two of the major mechanisms accounting for this expansion (Shiu and Bleecker, 2001b).
Within the Arabidopsis RLK gene family, more than 400 members have a domain configuration resembling transmembrane receptors, implying a major contribution of this class of proteins in the perception of cell surface signals in plants. Although relatively few RLKs have been studied in detail, recent studies implicate RLKs in a diverse range of processes. Some RLKs are involved in the control of plant growth and development under normal growth conditions, such as CLV1 in controlling meristem development (Clark et al., 1993), BRI1 in mediating brassinosteroid signaling (Li and Chory, 1997), and ERECTA in organ elongation (Torii et al., 1996). SRK from Brassica spp. is involved in self-incompatibility (Stein et al., 1996). Other RLKs involved in plant-microbe interactions and stress responses, such as rice (Oryza sativa) Xa21 in resistance to bacterial pathogen (Song et al., 1995), Arabidopsis FLS2 in flagellin perception (Gomez-Gomez and Boller, 2000), Lotus sp. SYMRK and Medicago sp. NORK in nodulation (Endre et al., 2002; Stracke et al., 2002), and tomato (Lycopersicon esculentum) SR160 in systemin signaling (Scheer and Ryan, 2002). On the other hand, more than 200 RLK/Pelle members do not have a receptor configuration. Very few of these RLCKs have known function. One of them, PBS1 of the RLCK VII subfamily is involved in disease resistance response (Swiderski and Innes, 2001). The large RLK gene family presents an intriguing subject for the analysis of the evolution of functional redundancy and divergence among duplicates. Because most RLK/Pelle members have no known function, it is unclear what the relative contribution is between redundancy and divergence in this gene family.
Genetic and biochemical analyses have contributed to the discovery of a number of proteins that interact with RLK/Pelle family members. Among them is an interesting class of proteins resembling the extracellular domains of several RLKs termed receptor-like proteins (RLPs; Jeong et al., 1999). The RLP CLV2, a putative transmembrane protein containing Leu-rich repeats (LRRs), has been shown to have a similar loss-of-function phenotype as its receptor partner, the LRR containing RLK CLV1 (Kayes and Clark, 1998). Furthermore, CLV2 interacts with CLV1 physically (Jeong et al., 1999). The maize (Zea mays) ortholog of CLV2, Fasciated Ear 2, also has similar function in meristem development, but its receptor counterpart is not known (Taguchi-Shiobara et al., 2001). The S-locus glycoprotein, resembling the extracellular part of SRK, is involved in self-incompatibility in Brassica spp., similar to its receptor counterpart (Cui et al., 2000). Another RLP, Too Many Mouths (TMM), is a transmembrane protein with LRRs involved in stomatal patterning (Nadeau and Sack, 2002). It is not known, however, whether TMM is functionally associated with an RLK. Given the diversity of extracellular domains in RLKs (Shiu and Bleecker, 2001a) and their potential functional association, it is of interest to investigate the diversity of RLPs and their relationship with RLK extracellular domains.
In this study, we present a detailed analysis of the RLK/Pelle family with the intent of further delineating the family and exploring the relationships between the RLK/Pelle members and all other kinases in Arabidopsis genome. We conducted a detailed analysis of kinases from available eukaryotic genomes to establish the relative abundance of this gene family in different eukaryotic lineages. To evaluate the importance of tandem and large-scale duplications in the expansion of the RLK family, the duplication pattern is examined for all RLK/Pelle subfamilies. Finally, a genome-wide survey is conducted to assess the relationships between RLKs and the structurally related RLPs.
RESULTS
The RLK/Pelle Family Constitutes 60% of All Kinases in Arabidopsis
It has been shown that Arabidopsis RLKs belong to a monophyletic gene family distinct from other families of eukaryotic protein kinases (Shiu and Bleecker, 2001). To determine the relative size of the RLK/Pelle gene family in the superfamily of eukaryotic protein kinases in Arabidopsis, a survey was conducted to retrieve all potential kinase sequences. In addition to known RLK/Pelle family members, 431 new kinase sequences were found (for a complete list, see Supplement A; supplemental data can be viewed at www.plantphysiol.org). To determine whether these sequences possess the kinase subdomain signatures, the predicted kinase domains were aligned with kinase family representatives (Shiu and Bleecker, 2001b). The 31 sequences that missed more than 40% of alignable regions or most of the conserved residues were removed from the kinase domain alignments. The remaining 400 genes have readily identifiable kinase subdomains.
In prior analyses, the RLK/Pelle family was delineated by first searching the Arabidopsis genome with known RLK sequences and then by phylogenetic analysis of candidate RLK sequences and kinase family representatives (Shiu and Bleecker, 2001b). To define the RLK/Pelle family members in the context of all kinases, the kinase domain alignment of the 400 kinase sequences and representative RLKs/RLCKs was used for phylogenetic reconstruction. Additional kinase family representatives, including sequences of previously defined RLK/Pelle subfamilies, were added to the alignment to facilitate the delineation of different kinase families. The result is shown in Figure 1 (see Supplement B for fully expanded phylogeny with gene names). The various kinases sequences can be readily divided into families based on the phylogeny. The RLK/Pelle representatives from Arabidopsis and human, as predicted, form a monophyletic group. In the 400 kinase sequences analyzed, 23 sequences are more closely related to the RLK/Pelle family than any other kinase families. Five of them fell into the same clade as RLK/Pelle representatives. The remaining 18 sequences form a distinct sister group to the RLK/Pelle family. Therefore, the size of the RLK/Pelle family is expanded to 625 representing 60% of the kinases in the Arabidopsis genome.
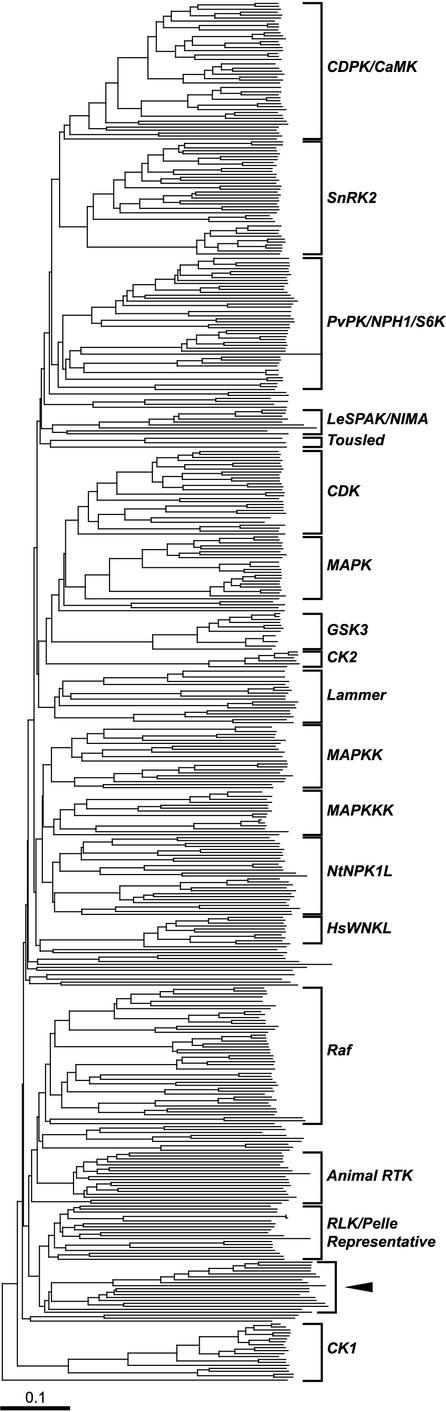
Figure 1.
Relationships among Arabidopsis protein kinases. The kinase sequences were identified from the Arabidopsis genome, and an alignment was generated using the kinase domain amino acid sequence. The phylogeny was inferred from the alignment using the neighbor-joining method with 100 bootstrap replicates (bootstrap values shown in Supplement B). The tentative kinase families were assigned based on nodes with more than 30% bootstrap support. The family names were given based on known sequences in each family. In the context of all kinases, representative RLK/Pelle members all fall into a monophyletic group as predicted. Interestingly, a group of kinases is found to form a sister group to the RLK/Pelle family (arrowhead a). In addition to RLKs in the RLK/Pelle family, the IRE1-like genes are the only other receptor kinases found in Arabidopsis (arrowhead b). The IRE1 family genes are not the closest relatives to RLK/Pelle members, suggesting that the receptor kinase configurations have arisen independently between these two kinase families. CDPK, Calcium-dependent protein kinase; CaMK, calmodulin-dependent kinase; SnRK2, SNF-related protein kinase 2; PvPK, bean protein kinase; NPH1, non-phototropic hypocotyl 1; S6K, ribosomal protein S6 kinase; LeSPAK, tomato self-pruning-associated kinase; NIMA, never in mitosis A; CDK, cyclin-dependent kinase; GSK3, glycogen synthase kinase 3; CK2, casein kinase 2; NtNPK1L, Nicotiana protein kinase 1 like; HsWNKL, human with no Lys kinase-like; RTK, receptor Tyr kinase; MAPK, mitogen-associated protein kinase; MAPKK, MAPK kinase; MAPKKK, MAPKK kinase.
The RLK/Pelle Family Is Much Larger in Arabidopsis Than in Non-Plant Eukaryote Genomes
The large Arabidopsis RLK/Pelle family was suggested to have undergone dramatic expansion in the land plant lineage (Shiu and Bleecker, 2001b). Because several eukaryote genomes representing a wider range of taxonomic groups are now available (Table I), it is of interest to determine the relative size of the RLK/Pelle family in these genomes to further assess the assertion of lineage-specific expansion in plants. To address this question, similar procedures for retrieving Arabidopsis kinases were used to identify kinases in the genomes of organisms listed in Table I. The RLK/Pelle family members were identified based on their relationships to the included RLK/Pelle representatives. The relationships of all non-Arabidopsis RLK/Pelle to Arabidopsis RLK/Pelle family were verified by building amino acid sequence alignments of the kinase domains for phylogenetic inference. The phylogeny is shown in Figure 2. Several Fugu sp. and Ciona sp. RLK/Pelle candidate sequences are outside of the RLK/Pelle family clade. As indicated in Table I, none of the four fungal genomes examined contained recognizable RLK/Pelle family members. However, the presence of family members in Plasmodium sp. and animal genomes suggests an origin for the RLK/Pelle family in a common ancestor of all genomes examined. The consistently low gene numbers in genomes other than Arabidopsis suggest that a dramatic expansion occurred in the plant lineage. On the other hand, the absence of family members in the fungal genomes examined indicates the RLK/Pelle family may be lost in fungal lineages after fungus-metazoan split.
Table I.
Comparison of the size of the kinase superfamily in available eukaryotic genomes
| Organism | Predicted No. of Genesa | Size of the Kinase Superfamily | Size of the RLK/Pelle Family | Proteome | Kinase | Source Databaseb |
|---|---|---|---|---|---|---|
| % | ||||||
| Arabidopsis | 25,814 | 1,041 | 615 | 2.382 | 59.1 | MAtDB |
| Plasmodium falciparum | 5,334 | 94 | 1 | 0.019 | 1.1 | PlasmoDB |
| Plasmodium yoelii | 7,681 | 70 | 1 | 0.013 | 1.4 | PlasmoDB |
| Caenorhabditis elegans | 19,484 | 417 | 1 | 0.005 | 0.2 | WormBase |
| Fruitfly (Drosophila melanogaster) | 13,808 | 262 | 1 | 0.007 | 0.4 | BDGP |
| Anopheles gambiae | 15,088 | 216 | 1 | 0.007 | 0.5 | Ensemble |
| Ciona intestinalis | 15,852 | 316 | 6 | 0.038 | 1.9 | JGI |
| Fugu rubripes | 33,609 | 632 | 6 | 0.018 | 0.9 | Ensemble |
| Mus musculus | 22,444 | 495 | 3 | 0.013 | 0.6 | Ensemble |
| Human (Homo sapiens) | 22,980 | 472 | 4 | 0.017 | 0.8 | Ensemble |
| Brewer's yeast (Saccharomyces cerevisiae) | 6,449 | 113 | 0 | 0.000 | 0.0 | Pedant |
| Candida albicans | 6,164 | 95 | 0 | 0.000 | 0.0 | SGTC |
| Neurospora crassa | 10,082 | 104 | 0 | 0.000 | 0.0 | Pedant |
| Fission yeast (Schizosaccharomyces pombe) | 4,945 | 109 | 0 | 0.000 | 0.0 | Pedant |
Protein-coding genes only. b The URL for the source databases are: MAtDB, http://mips.gsf.de/proj/thal/; PlasmoDB, http://www.plasmodb.org/; WormBase, http://www.wormbase.org/; BDGP, http://www.fruitfly.org/; Ensemble, http://www.ensembl.org/; JGI, http://genome.jgi-psf.org/; Pedant, http://pedant.gsf.de/; SGTC, http://www-sequence.stanford.edu/.
Figure 2.
The RLK/Pelle family members from Arabidopsis and other eukaryotic genomes. The kinase domains of the Arabidopsis representatives listed in Table II are aligned with putative RLK/Pelle members from the genomes listed in Table I. The phylogeny inferred based on the alignments is shown on the left with branches leading to Arabidopsis sequences in green, to animal sequences in red, and to Plasmodium sp. in blue. The numbers on the base on the branch indicate bootstrap support out of 1,000 replicates. Branches with than 5% support are collapsed. All genomes except Arabidopsis have few RLK/Pelle members. The identifiers for Arabidopsis follow the convention: gene name, subfamily designation, and synonym (if available). For the other genomes, the identifier follows: species abbreviation, sequence name from the source database (see Table I), synonym (if available). AG, A. gambiae; DM, D. melanogaster; CE, C. elegans; CI, C. intestinalis; FR, F. rubripes; HS, Homo sapiens; MM, M. musculus.
Nearly All Putative Transmembrane Kinases in Arabidopsis Belong to the RLK/Pelle Family
Two-thirds of the RLK/Pelle family members are transmembrane receptor kinases. To determine whether other kinase families also contribute kinase domains to transmembrane receptors, the 424 non-RLK/Pelle kinase sequences were examined for the presence of putative signal sequences and transmembrane regions. We found that 15 of these sequences have putative signal sequences and three have putative transmembrane regions N-terminal to the kinase domains (see Supplement A). Two sequences, At5g24360 and At4g12020, were found to possess both signal peptides and transmembrane regions with C-terminal kinases. Further examination of At4g12020 indicates that it represents a misannotation between two genes. On the other hand, At5g24360 is closely related to the IRE1 ER membrane protein found in most eukaryotes (Urano et al., 2000). At least two other IRE1-like genes can be found in Arabidopsis based on kinase domain similarity (At2g17520 and At3g11870). To see whether IRE1-like genes are more closely related to RLKs than to other kinases, a phylogenetic analysis was conducted using representatives of different kinase families from both Arabidopsis and human. Arabidopsis IRE1-like genes and human IRE1 form a well-supported clade with a high bootstrap value (Fig. 3). However, they are not the closest relatives to RLKs, Rafs, or animal Tyr kinases. This conclusion is also supported by the phylogenetic analysis of Arabidopsis kinases (Fig. 1, arrow b). These findings suggest that the receptor configurations of RLKs and IRE1-like genes may have arisen independently, and IRE1-like genes represent the only other receptor Ser/Thr kinases identified in Arabidopsis genome.
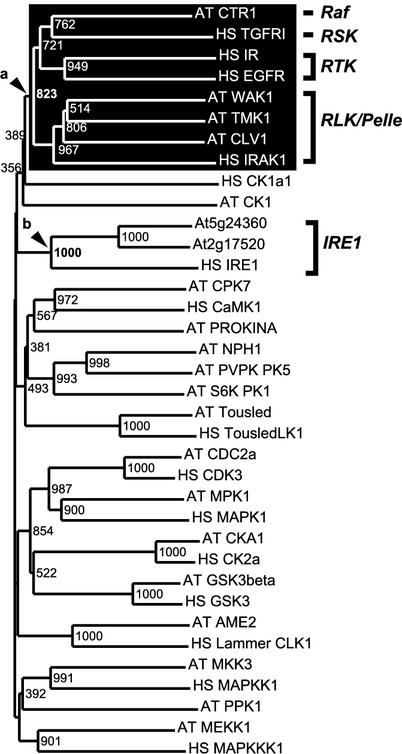
Figure 3.
The relationships between IRE1-like genes and RLK/Pelle family kinases. The kinase domain amino acid sequences of IRE1-like genes and kinase family representatives from Arabidopsis and human were used for generating the neighboring-joining tree with 1,000 bootstrap replicates. The RLK/Pelle family forms a close relationship to Raf kinases and human receptor Tyr kinases (arrowhead b). The Arabidopsis IRE1-like genes are closely related to human IRE1 with high bootstrap support (arrowhead b). The identifiers follow the convention: species abbreviation followed by gene name. RSK, Metazoan receptor Ser/Thr kinase. RTK, Animal receptor Tyr kinase.
The RLK/Pelle Family in Arabidopsis Can Be Classified into Multiple Subfamilies
RLKs in the RLK/Pelle family contain a variety of extracellular domains, and RLKs with similar extracellular domains have similar kinase domains (Fig. 4A). This relationship provides a phylogenetic basis for classifying the RLK/Pelle family. However, a few interesting exceptions to this generalization are present. For example, the S-domain, LRK10-like, and CrRLK-like RLKs can be found in more than two different clades (Fig. 4A, arrows). To further investigate the validity and correctness of the phylogeny generated based on kinase domain sequences, the exon-intron organization of all RLK/Pelle family members was examined based on the gene models released by the MIPS Arabidopsis Database (MAtDB). We first determined the number of predicted exons in each gene and superimposed the information onto the kinase phylogeny (Fig. 4B). Numbers of exons correlate well with both the kinase phylogeny and the identity of extracellular domains.
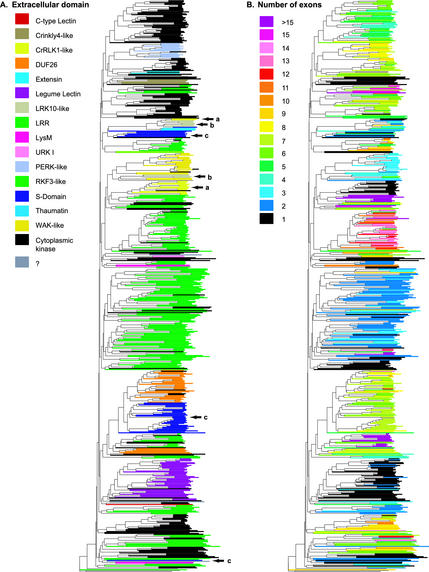
Figure 4.
The relationships between the kinase phylogeny, the extracellular domains, and the number of exons. The phylogenetic trees in (A) and (B) were generated from an alignment of 610 RLK/Pelle family members previously published (Shiu and Bleecker, 2001). A, Branches of the phylogenetic tree were colored according to the identity of the extracellular domains as predicted by SMART and Pfam databases. RLKs with similar extracellular domains are generally clustered together. RLCKs are clustered into several groups also. Exceptions are the CrRLK1-like genes (arrow a), the LRK10-like genes (arrow b), and the S-domain RLKs (arrow c). For detailed domain organization for all genes, see Supplement C. B, Branches of the phylogenetic tree were colored according to the number of exons based on the predicted gene model of RLK/Pelle members. Related RLK/Pelle members as defined by the kinase phylogeny or extracellular domain identities, in general, have identical or similar numbers of exons. For the locations, placements, and phases of introns, see Supplement C.
To investigate whether the introns in related RLKs are homologous, we generated protein sequence alignments for each subfamily and examined the intron locations and phases (the location of the splice junction within a codon). We found that there are at least 43 distinct intron insertion sites in the kinase domains of all RLKs (data not shown). In most cases, members of the subfamilies defined according to the kinase phylogeny and the identity of extracellular domains have conserved intron placements and phases within the kinase domains and within the full-length genes (for the graphical representation of domain prediction, intron information, and classification, see Supplement C). On the basis of the kinase phylogeny, the extracellular domains, and/or the intron information, the RLK/Pelle family is subdivided into 46 different subfamilies (Table II). At1g66980 shared high similarity with LRKs10-like 2 subfamily members but has an unrelated extracellular domain resembling glycerophosphoryl diester phosphodiesterase. It is therefore classified into a single-member subfamily not present in previously published classification schemes (Shiu and Bleecker, 2001b).
Table II.
Total numbers and extent of tandem clustering in different RLK/Pelle subfamilies —, not determined.
| Subfamily | Representative | No. of Subfamily Members | Genes in Cluster |
|---|---|---|---|
| % | |||
| C-Lectin | At1g52310 | 1 | N.A.a |
| CR4-like | At2g39180 | 8 | 0 |
| CRPK1-like1 | At3g51550 | 15 | 13 |
| CRPK1-like2 | At5g39030 | 2 | 100 |
| DUF26 | RLK3 | 45 | 84 |
| Extensin-like | At5g56890 | 5 | 0 |
| L-Lectin | LecRK1 | 45 | 65 |
| LRK10-like1 | At1g18390 | 5 | 40 |
| LRK10-like2 | At1g66930 | 7 | 100 |
| GDPDb | At1g66980 | 1 | N.A. |
| LRR I | At3g46420 | 49 | 62 |
| LRR II | BAK1 | 14 | 14 |
| LRR III | RKL1 | 47 | 0 |
| LRR IV | At2g45340 | 3 | 0 |
| LRR V | At1g53730 | 9 | 0 |
| LRR VI | At5g07150 | 11 | 0 |
| LRR VII | At3g28040 | 10 | 0 |
| LRR VIII-1 | At5g01950 | 8 | 38 |
| LRR VIII-2 | At3g14840 | 14 | 73 |
| LRR IX | TMK1 | 4 | 0 |
| LRR X | BRI1 | 16 | 0 |
| LRR XI | CLV1 | 28 | 7 |
| LRR XII | FLS2 | 10 | 40 |
| LRR XIII | ER | 7 | 0 |
| LRR XIV | At2g16250 | 3 | 0 |
| LysM | At3g21630 | 5 | 0 |
| PERK | PERK1 | 19 | 21 |
| RKF3-like | RKF3 | 2 | 0 |
| RLCK I | At2g40270 | 3 | 0 |
| RLCK II | At4g35230 | 7 | 29 |
| RLCK III | At1g67470 | 5 | 60 |
| RLCK IV | At5g58940 | 3 | 0 |
| TAK(RLCK V) | TAK1 | 11 | 0 |
| RLCK VI | At5g57670 | 14 | 0 |
| RLCK VII | PBS1 | 46 | 9 |
| RLCK VIII | At3g59350 | 11 | 36 |
| RLCK IX | At1g78940 | 21 | 10 |
| RLCK X | At1g80640 | 4 | 0 |
| RLCK XI | At4g25390 | 4 | 0 |
| RLCK XII | At1g01740 | 17 | 29 |
| S-domain1 | ARK1 | 32 | 91 |
| S-domain2 | RLK4 | 7 | 0 |
| S-domain3 | At2g41890 | 1 | N.A. |
| Thaumatin | PR5K | 3 | 33 |
| URK I | At1g49730 | 2 | 0 |
| WAK-like | WAK1 | 25 | 68 |
| Not classified | — | 16 | N.A. |
| Annotated pseudogene | — | 7 | N.A. |
N.A., Not applicable. b The sole member of this subfamily is originally classified as LRK10-like2 based on its high similarity on the kinase domain.
It should be noted that, although the three molecular characters examined are largely in agreement with one another, there are a noticeable number of exceptions. Where the multiple sequence alignments indicate missing or extra sequences between members of a subfamily, the junctions of such anomalies in many cases coincide with the predicted intron junctions.
Proteins That Resemble the Extracellular Domains of Several RLK Subfamilies Are Present in the Arabidopsis Genome
RLPs, proteins that resemble the extracellular domains of RLKs, have been shown to play roles in RLK signaling (Jeong et al., 1999). Given the diversity and abundance of RLKs in Arabidopsis, it is not known what types of RLKs have corresponding RLPs. In addition, the elucidation of RLPs may shed light on the potential origin of novel RLKs. It is therefore of interest to determine the diversity and abundance of RLPs in the Arabidopsis genome. Using the extracellular domains of RLKs as queries, candidate RLPs were obtained from the Arabidopsis genome through similarity search with BLAST. After eliminating known RLK sequences and sequences similar to nucleotide binding site-LRR putative R genes, the 178 candidate RLP sequences were combined with the representative extracellular domain sequences. These sequences were clustered with the unweighted pair group method with arithmetic mean (UPGMA) algorithm using transformed E values as the distance measure. The resulted cluster diagram reflects the similarity between sequences over the alignable regions where a shorter bifurcating branch implies a higher sequence similarity (Fig. 5).
Figure 5.
Similarity clustering and domain organization of RLPs from Arabidopsis. On the basis of pair wise comparison of putative RLPs and the extracellular domain sequences of representative RLKs with BLAST, a distance matrix is generated with transformed E values and used for clustering with the UPGMA algorithm implemented in MEGA2. The clustering diagram is a representation of relatedness between sequences. More closely related sequence pairs are joined together with branches of shorter length. The domain organizations as predicted by SMART/Pfam are shown on the left of the clustering diagram. The identifiers for the representative sequences follow the convention: gene name, subfamily, and synonym (if available). The intracellular domains of all representative RLKs are not shown. The classification of RLPs is shown right to the domain organizations. Red arrows indicate sequences representing misannotated RLKs. Branches are colored in three RLP classes: green, TMM-like; red, LRR II-like; and blue, LRR-I-like.
The candidate RLPs can be subdivided into several clusters based on sequence similarity alone. After superimposing the domain prediction on the clusters, the boundary between clusters becomes clearer. Five of these 178 candidate RLPs however are located right next to a downstream kinase on the chromosome (Fig. 5, red arrows). In all cases, closely related sequences resemble the fusions between these five proteins and their downstream kinases. Therefore, they likely represent the N-terminal ends of misannotated RLKs.
More than one-half of the putative RLPs contain various numbers of LRRs. However, most of these LRR-containing RLPs are more similar to one another than they are to LRR containing RLKs. The exceptions are the subfamilies LRR I (Fig. 5, blue branches) and LRR II (Fig. 5, red branches). A large number of LRR RLPs are more closely related to the tomato resistance gene Cf (data not shown). Several other RLK subfamilies also have corresponding RLPs, including Lysin motif, DUF26, CrRLK1-like, glycerophosphoryl diester phosphodiesterase-like, LRK10-like, thaumatin, legume lectin, and S-locus receptor kinase subfamilies. More than 50% of these RLPs are found in tandem repeats (based on the criteria outlined in “Materials and Methods”). Moreover, there are 21 RLPs found in close proximity to a related RLK. Some of these RLK-RLP tandems are located within large clusters of RLKs, suggesting that they may be derived from or may give rise to RLKs with similar extracellular domains (ECDs).
One-Third of the RLK/Pelle Family Members in Arabidopsis Are Found in Tandem Clusters
The RLK family represents approximately 60% of kinases and 2.5% of all predicted protein-coding genes in Arabidopsis. The large number of genes in this family raises several questions concerning its expansion. By examining the chromosomal distribution of four RLK/Pelle subfamilies on chromosome 4, it was found that tandem duplication and large-scale segmental duplication/reshuffling may represent the major mechanisms contributing to the expansion of the RLK/Pelle family in Arabidopsis (Shiu and Bleecker, 2001b). To expand the analysis to all members of this gene family and to evaluate the relative importance of these two mechanisms in the whole Arabidopsis genome, we examined the chromosome locations in conjunction with the phylogenetic relationship of all RLK/Pelle family members and the chromosome duplication patterns hypothesized by the Arabidopsis Genome Initiative (2000). On the basis of the criteria that tandem duplicated genes are located within 10 predicted open reading frames or within 30 kb of each other and that only genes of the same RLK subfamilies are considered, we found that there were 50 clusters of RLKs with 2 to 19 genes. A total of 210 genes were found in tandem repeats, representing 33.6% of all RLKs (Table III; see also Supplement D for the location and identity of duplicated genes). Subfamilies vary greatly in the extent of tandem duplication. In the 20 subfamilies with more than 10 members, DUF26, l-lectin, LRR I, LRR VIII-2, S-domain I, and WAK-like subfamilies all have more than 60% of their members in tandem repeats. These findings indicate that tandem duplication represents one of the major mechanisms of RLK expansion in Arabidopsis.
Table III.
Distribution of RLK family genes in duplicated regionsa
| Outside of Duplicated Regions | Within Duplicated Regions
|
||
|---|---|---|---|
| With Match | Without Match | ||
| Total | 155 | 268 | 202 |
| Singular | 123 | 125 | 172 |
| Tandem repeats | 32 (10)b | 143 (19)b | 30 (11)b |
According to Arabidopsis Genome Initiative (2000), see Supplement D. Data does not include RLCK XII. b The numbers in the brackets are the number of distinct tandem clusters.
Large-Scale Duplications Represent Another Major Mechanism for the RLK/Pelle Expansion
To determine the contribution of the whole-genome duplication and reshuffling that has been postulated (Arabidopsis Genome Initiative, 2000), we compared the chromosome duplication pattern with the location of RLKs/RLCKs and their phylogeny. The results are shown in Table III (see also Supplement D for a list matching the genes in the duplicated regions). Among the 625 RLKs, 470 were found in the hypothesized duplicated/reshuffled regions, whereas 155 were located outside. Within the duplicated regions, 125 were singular genes (genes not in tandem repeats) with a close relative found in the corresponding duplicated regions. In addition, 143 genes in 19 clusters were found to have one or more close relatives in the corresponding duplicated regions. Therefore, 43% (263) of the Arabidopsis RLK/Pelle members can be accounted for by the large-scale duplication pattern. However, the remaining 202 RLK/Pelle sequences within the duplicated regions do not have a corresponding relative, suggesting the involvement of gene loss or more localized duplications. Nonetheless, these results indicate that large-scale segmental duplications, in conjunction with tandem duplications, are partly responsible for the expansion of this gene family in Arabidopsis.
DISCUSSION
The Predominance of the RLK/Pelle Family in Arabidopsis
RLKs in plants have been found to play important roles in a multitude of different processes. Given the complexity of cellular communication in multicellular organisms and the size of the RLK family, it is widely speculated that this family may represent one of the major receptor systems for intercellular signaling pathways in plants. Another receptor kinase family found in Arabidopsis is the receptor His kinases with six members including ethylene receptor ETR1 (Bleecker and Kende, 2000) and cytokinin receptor CRE1 (Inoue et al., 2001). In a prior analysis, we found that RLKs belong to a large gene family with more than 600 members. However, only a few RLK homologs (based on kinase domain sequences) can be found for human, fly, and worm (Shiu and Bleecker, 2001b). With an extended analysis of completed eukaryotic genomes, this gene family still has very low gene number in all animals, although two rounds of genome duplications seem to have occurred in the vertebrates (Fig. 2). The presence of only one gene in each Plasmodium sp. genome suggests that the ancestral gene number for this gene family is likely to be small before the divergence of plants, animals, fungi, and protists. Because RLKs are absent in all fungal genomes examined, a subsequent gene loss may have occurred after the split between animal and fungi and before the divergence of the ascomycetes genomes analyzed. It should be noted that the bootstrap support for the monophyly was merely 15% (Fig. 2). However, it has been demonstrated that plant RLK/RLCKs and metazoan Pelle kinases are monophyletic (Shiu and Bleecker, 2001b). In addition, the metazoan sequences in the RLK/Pelle family all have the same domain configuration (data not shown). The low bootstrap support for the RLK/Pelle family is most likely due to the sequence divergence among the subfamily representatives included. On the basis of an analysis of the whole kinase complement in Arabidopsis, we show that the RLK/Pelle gene family makes up 60% of kinases and is the predominant form of putative receptor kinases in the Arabidopsis genome. Within the kinase superfamily, IRE1-like genes are the only other type of receptor Ser/Thr kinases found (Figs. 1 and 3). IRE1 is an endoplasmic reticulum-resident protein mediating the unfolded protein response (Urano et al., 2000). The closest relatives of RLKs are Raf-type kinases, but not IRE1-like genes. Their relationships argue that the receptor configurations found in these two receptor families have arisen independently.
Molecular Diversity of the RLKs and the Presence of RLPs in the Arabidopsis Genome
On the basis of the combined analysis of the kinase phylogeny and extracellular domains, the RLK/Pelle family was subdivided into 46 different subfamilies. This classification is in general supported by the predicted gene structures of all RLKs/RLCKs. It should be noted that the kinase phylogeny alone is, in some cases, insufficient for delineating subfamilies due to weak bootstrap support. Additional criteria, such as domain organization and gene structures, provide additional evidence to support or refute the relationships inferred strictly from phylogeny. Interestingly, the same extracellular domains are found to form RLKs with kinase domains of distinct evolutionary origin within the RLK/Pelle family. In the case of S-domain, fusions with three different RLK/Pelle subfamilies are evident (S.H. Shiu, unpublished data). If these domains form fusion with other proteins with equal frequency and efficiency, the multiple independent fusion events observed between these domains and the kinases from the RLK gene family but not any other kinase families or non-kinase proteins would indicate that the fusions formed with the kinase of the RLK/Pelle were retained preferentially. In the analysis of the human genome, it is found that molecular complexity and diversity of proteins can be accomplished through fusion between unrelated protein domains (Lander et al., 2001; Venter et al., 2001). This process is not a metazoan-specific process but is likely to be a general theme in all living organisms. In plants, the RLK/Pelle family represents a good example for domain reuse in different molecular contexts.
In addition to the diversity of RLKs, a large number of RLPs resembling the RLK extracellular domains are present in the Arabidopsis genome. For the RLPs with known function, both CLV2 and SLG have been implicated to function in the signaling pathway involving RLKs, CLV1, and SRK, respectively (Jeong et al., 1999; Cui et al., 2000). Interestingly, both CLV2 and CLV1 extracellular domain contain multiple LRRs. SLG is related to the extracellular domains of SRK (which is similar to the S-domain subfamilies). It seems to suggest that some RLKs may require a signaling partner, secreted or associated with membrane, and that partner may be related to the extracellular domains of RLKs in question. In this analysis, several distinct groups of RLPs were found to be related to RLKs. These RLPs may be functionally related to their RLK counterparts. After excluding potentially misannotated sequences, there were 173 RLPs which fell into 20 similarity clusters (Fig. 5). Interestingly, 21 of these RLPs were found to be within 10 predicted genes to related RLKs. In all 3′ regions examined, no sequence resembles known kinases at either the protein or the nucleotide level. However, it is not known whether such association has any functional significance. Several of these RLPs are located in large clusters of RLKs and may have arisen through unequal crossovers.
Xa21D, an RLP from rice illustrates the potential roles of RLPs found in RLK clusters. Xa21D presumably is derived from a duplication event that gave rise to itself and Xa21, a rice RLK conferring resistance to bacterial pathogen (Wang et al., 1998). Subsequent transposon insertion in Xa21D resulted in a truncated protein with only the extracellular domain. Surprisingly, Xa21D still confers partial disease resistance. This finding suggests that RLPs recently derived from RLKs may still have similar function to its RLK progenitor. It is therefore possible that RLKs and RLPs in a tandem cluster have overlapping functions. On the other hand, the RLPs may be created fortuitously and become pseudogenes quickly. A comparison of synonymous and non-synonymous substitution rates in these genes may help resolve this issue. It should be noted that several subfamilies of RLKs do not have obvious RLP homologs in the Arabidopsis genome. Because the search was conducted with an E value cutoff of 1 × 10–10, related but divergent sequences may be excluded. However, the absence may also indicate differences in the signaling mechanisms of these RLKs from those with closely related RLPs.
Expansion of the RLK/Pelle Family
A large size difference exists between the numbers of RLK/Pelle family members in different eukaryotes, and there is little question that this family has undergone expansion in land plants based on the over-representation of RLK/Pelle expressed sequence tags (ESTs) in various land plant lineages (Shiu and Bleecker, 2001b) and a preliminary analysis of the draft versions of rice genomes (S.-H. Shiu, unpublished data). It is known that both tandem duplications and segmental duplications, or even whole-genome duplications have occurred in Arabidopsis (Arabidopsis Genome Initiative, 2000; Vision et al., 2000). In this study, the locations of all RLKs/RLCKs were examined to determine the relative importance of tandem duplication and segmental duplication. We found that 33% of RLK/Pelle family members are found in 50 clusters. The percentage of tandem repeats in this gene family is 2-fold higher than the average of tandem duplicated genes in the whole Arabidopsis genome (Arabidopsis Genome Initiative, 2000). It should be noted that we estimated tandem duplication in a conservative fashion by considering only closely related genes of the same subfamily instead of using similarity score cutoff alone. The differences between the percentage of RLK/Pelle members in tandem repeats and that of the genome average would be even more pronounced if the same criteria were used.
From the regions that underwent segmental duplication as defined by the Arabidopsis Genome Initiative (2000), we found that 470 RLK/Pelle family members are located within the duplicated regions. Among them, 268 genes have at least one related gene of the same subfamily in the corresponding duplicated regions. These genes are most likely remnants of large-scale duplication events. However, 202 RLKs/RLCKs in these regions do not have a corresponding paralog. It is possible that the corresponding paralogs had been mutated beyond recognition and lost over time. It is also possible that some of these genes were derived from relatively more localized scale duplication events not examined, such as suggested by Vision et al. (2000). Another possibility is transposition facilitated by transposon or retrotransposon activity, although no convincing case of such activity has been found in this gene family (data not shown). A total of 155 RLK genes are located outside of the duplicated regions. Some of these genes may lie inside the more localized duplicated regions. In addition, the original detection scheme may be sensitive to differential gene loss and may have missed regions that were duplicated but have become too degenerate to be detected with the approaches used. It has been shown that more duplicated blocks are recovered through comparisons of all segments with similar but not identical gene contents (Simillion et al., 2002). A detailed analysis based on the delineated area would likely reduce the number of genes lying outside of the duplicated regions. In any case, a much higher proportion of tandem duplicates is found in the regions of proposed large-scale duplications, indicating potential interactions between these two mechanisms. It is possible that, after large-scale duplication events, these duplicates were directly involved in the generation of tandem repeats.
Retention of Duplicates
Gene duplication occurs at a high rate in eukaryotes, but the vast majority of gene duplicates were lost within a few million years (Lynch and Conery, 2000). Most gene families in living organisms are small, and the large gene families such as the RLK/Pelle family are exceptions rather than the norm (S.-H. Shiu and K. Mayer, unpublished data). Therefore, it is of interest to consider how natural selection appears to have favored retention of duplicated genes in the RLK/Pelle family in plants. The distribution of this gene family in the genomes examined supports the notion that the RLK/Pelle family maintained a low gene number before the divergence of plants, animals, and Plasmodium sp. If each tandem duplicated region is regarded as a single locus, there will be a total of 465 RLK “loci” because 210 of the 625 RLK/Pelle members are found in 50 clusters. Assuming the presence of four ancestral RLK genes before the split between plants and animals and that all duplicated genes were retained, then seven rounds of whole-genome duplication could account for the number of RLK loci in the Arabidopsis genome. Vision et al. (2000) proposed that at least four large-scale duplication events had occurred 100 to 200 million years ago at approximately the time of angiosperm diversification. This notion is also supported by a similar study accounting for potential gene loss (Simillion et al., 2002). Although these duplication events do not necessarily encompass the whole genome, they account for some of the required duplication events leading to the expansion of the RLK gene family in Arabidopsis. Because the inference of large-scale duplication events involved evolutionary rate estimates, it is possible that older events are not detected due to the saturation of substitutions. Given the evidence from EST analysis of plant lineages that the RLK family had already undergone significant expansion early in the evolution of land plants (Shiu and Bleecker, 2001b), the time scale involved will have provided many opportunities for additional large scale duplication events that are not recognized.
One important assumption in estimating the number of whole-genome duplications required is that most, if not all, duplicated RLK genes were retained. Because the rate for duplication and loss are both high in general, it is likely that the observed duplication events may reflect only a subset of the duplications that have occurred. Nonetheless, the RLK/Pelle family members seem to be retained at a much higher rate compared with the average gene or gene families in Arabidopsis. For any pair of gene duplicates, they may first experience relaxation of selection because a loss-of-function mutation on one copy would not have deleterious consequences. Some duplicates may become pseudogenes within several million years. This then raises the question whether many members in this gene family are pseudogenes or are descendants of very recent duplication events. The average protein sequence similarity for each RLK/Pelle subfamily ranges from 30% to 90%. This indicates that some subfamilies are quite divergent and argues against the notion that they were duplicated recently. The subfamilies with high similarity are mostly genes located within tandem clusters that may indicate their recent origin or a propensity for gene conversion. Five genes in addition to the 625 RLK/Pelle members are annotated as pseudogenes. There are potentially more pseudogenes because some members of this family are truncated when compared with their close relatives. But the fact that members that show anomaly in their domain organizations account for less than 3% of the whole gene family argues against the notion that pseudogenes contribute much to the size of the RLK family. In addition, nearly 50% of genes in this family have one or more ESTs. More detailed expression analyses will likely show that more RLK/Pelle members are expressed.
Another explanation for the large gene number in this gene family is that the duplicates were quickly established to take on a subset of the original functions or novel functions. Considering the functions of RLKs in plant development, RLK/Pelle duplicates might have been retained because they conferred additional controls for tissue differentiations to provide control system necessary for the development of more complex multicellular traits. Alternatively, several RLK/Pelle members have been implicated in disease resistance. During the period of relaxed selection on gene duplicates, the regions responsible for binding specificity may diversify and result in genes with new specificity. Subsequently, these genes were retained for their contribution in recognizing new components of the pathogens or symbiotic organisms. Continued studies on the functions of individual RLKs in plants, coupled with comparative genomic studies, will certainly provide insights into the evolutionary history of this interesting family of plant receptors.
MATERIALS AND METHODS
Sequence Retrieval and Annotation
For a list and the accession numbers of the 610 RLK family members analyzed, see Shiu and Bleecker (2001). The eukaryotic Ser/Thr/Tyr kinases in Arabidopsis were identified with the procedures detailed below. Following Hanks and Hunter (1995) and Hardie (1999), we used 52 plant and animal sequences from different eukaryotic Ser/Thr/Tyr kinase families to conduct batch BLAST searches (Altschul et al., 1997) against the July 4, 2001, release of the Arabidopsis genome from MAtDB (http://mips.gsf.de/proj/thal/) with an E-value cutoff of 1. In a preliminary analysis, we found that an E-value cutoff of 0.1 allowed us to recover all selected kinase family representatives. Therefore, a more relaxed E value of 1 was chosen in an attempt to include all potential kinases. A total of 1,235 sequences retrieved based on this criteria were regarded as “potential” kinases. Structural domains of all sequences were defined according to the SMART (Schultz et al., 2000) and Pfam (Sonnhammer et al., 1998) databases. One thousand and thirty-one sequences had kinase domains detected by either of these two databases with the default parameters. After eliminating sequences in the RLK family, 431 of these “candidate” kinases were included in further analysis as described in the next section. To determine whether these candidates were putative transmembrane proteins, the database outputs from SMART were examined. Only sequences with both signal sequences and transmembrane regions were regarded as putative receptor kinases. The Arabidopsis kinase complement defined was updated according to the November 12, 2002, release of the Arabidopsis proteome from MAtDB. Similar procedures were used to retrieve kinase sequences from other genomes listed in Table I.
Alignment and Phylogenetic Inference
The kinase domain sequences of the candidates defined in the previous section were compiled and aligned using ClustalX (Higgins et al., 1996). All sequences were aligned against sequence profiles of representative eukaryotic kinases (Shiu and Bleecker, 2001b). The weighing matrix used was BLOSUM62 with the penalty of gap opening 10 and gap extension 0.2. The alignments generated were manually adjusted according to the subdomain signatures of eukaryotic kinases (Hanks and Hunter, 1995). A candidate kinase was regarded as “true” kinases if more than 60% of the defined kinase domains was present and were “alignable.” The alignable criterion is based on a detailed examination to determine whether the target sequences contained most, if not all, of the kinase subdomain signatures. Phylogeny of the aligned sequences was generated with the neighbor-joining method (Saitou and Nei, 1987), and bootstrap analyses were conducted with 100 replicates, for each organism listed in Table I. Aminoglycoside kinase (APH(3′) III) from Staphylococcus sp. (P00554) and the Arabidopsis homolog of RIO1 family kinases (S61006) were used as outgroups. These two genes are divergent members of the eukaryotic kinase superfamily and are hypothesized to be ancestral eukaryotic kinases (Hon et al., 1997; Leonard et al., 1998). The RLK/Pelle family is defined as the clade that contains RLK/Pelle representative sequences and forms sister group relationships to Raf kinases and receptor Tyr kinases.
Domain Organization and Gene Structure of the RLK Gene Family Members
The gene models used in this analysis were based on the released information available from MAtDB July 4, 2001, sequence release and updated according to the November 12, 2002, release. The gene models for all RLK family members were extracted and used to calculate intron location and insertion phase (the position of intron insertion relative to the bases in a codon). For each RLK subfamily defined by Shiu and Bleecker (2001b), a full-length alignment was generated. After manual adjustment of alignments, the intron locations were mapped onto the alignments to determine if the intron sites were homologous between members of the same subfamilies. All output files of SMART queries using full-length RLKs/RLCKs were parsed to obtain information on domain identity and coordinates of domain junctions. The domain organization and intron information of all RLK family members were consolidated and the graphical display was generated (see Supplement C).
Survey of Arabidopsis Proteins with Similarity to the Extracellular Domain of RLKs
Thirty-five representative RLKs were chosen from all RLK subfamilies with at least 50% members that contain predicted transmembrane regions in similar positions. The ECD was defined as the region between the end of signal sequence (or the first residue if signal sequence was absent) and the beginning of transmembrane regions. These putative ECDs were used to conduct BLAST search against the Arabidopsis protein set. With a cutoff threshold of 1 × 10–10, after eliminating known RLK sequences and sequences that were more similar to nucleotide binding site-LRR putative R genes, candidate RLPs were obtained. Molecular features adjacent to these candidate RLPs were examined to determine whether they likely represented RLKs that split into two parts. In addition, the 3′ regions were examined for kinases not annotated. The putative RLPs are defined as the sequences that do not represent split genes and have no unannotated kinase sequences in the 3′ direction with the same orientation. For the classification of putative RLPs, the sequences of the ECDs of RLK representatives and the putative RLPs were used to conduct BLAST search against the database formatted with the same sequence set. The E values of pair wise comparisons were transformed to represent distance measures and were used to generate similarity clusters with the UPGMA algorithm implemented in the program MEGA2 (Kumar et al., 2001).
Physical Map Location and Criteria for Determining Duplicated RLK Family Members
The locations of RLKs in the form of coordinates on the Arabidopsis chromosomes assembled from BAC sequences were obtained from MAtDB. Adjacent genes were regarded as tandem repeats if they were within 10 predicted genes apart or within 30 kb of each other and if they belonged to the same subfamily. On the basis of the postulated large-scale duplication and reshuffling of the Arabidopsis genome (Arabidopsis Genome Initiative, 2000), the coordinates of the genes flanking the duplicated regions (kindly provided by Dr. Heiko Schoof from MAtDB) were used to determine whether any given RLKs/RLCKs were located in the duplicated regions (see Supplement D). Comparisons of all RLK family genes in each pair of duplicated regions were then conducted to determine whether any RLK sequences was likely products from the duplication events. RLK sequences were regarded as products of the duplication events if one or more relatives from the same subfamily could be found in the corresponding regions.
Supplementary Material
Acknowledgments
We thank Dr. Wen-Hsiung Li for suggestions and comments, the MAtDB for providing sequence information and detailed annotation information, and Dr. Heiko Schoof for providing the Arabidopsis whole-genome duplication data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.021964.
The work was supported by a National Institutes of Health National Research Service Award (grant no. 1F32GM066554–01 to S.-H.S.) and by the Department of Energy (grant no. DE–FG02–91ER20029 to A.B.B.).
The online version of this article contains Web-only data. The supplemental material is available at http://www.plantphysiol.org.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Becraft PW (1998) Receptor kinases in plant development. Trends Plant Sci 3: 384–388 [Google Scholar]
- Bleecker AB, Kende H (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16: 1–18 [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM (1993) CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119: 397–418 [DOI] [PubMed] [Google Scholar]
- Cui Y, Bi YM, Brugiere N, Arnoldo M, Rothstein SJ (2000) The S locus glycoprotein and the S receptor kinase are sufficient for self-pollen rejection in Brassica. Proc Natl Acad Sci USA 97: 3713–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kalo P, Kiss GB (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Hanks SK, Hunter T (1995) The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FAESB J 9: 576–596 [PubMed] [Google Scholar]
- Hardie DG (1999) Plant protein serine/threonine kinases: classification and functions. Annu Rev Plant Physiol Plant Mol Biol 50: 97–131 [DOI] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol 266: 383–402 [DOI] [PubMed] [Google Scholar]
- Hon W-C, McKay GA, Thompson PR, Sweet RM, Yang DS, Wright GD, Berghuis AM (1997) Structure of an enzyme required for aminoglucoside antibiotic resistance reveals homology to eukaryotic protein kinases. Cell 89: 887–895 [DOI] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE (1999) The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo A, Schopfer CR, Nasrallah ME, Nasrallah JB (2001) Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 293: 1824–1826 [DOI] [PubMed] [Google Scholar]
- Kayes JM, Clark SE (1998) CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125: 3843–3851 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobson IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W et al. (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Leonard CJ, Aravind L, Koonin EV (1998) Novel families of putative protein kinases in bacteria and archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome Res 8: 1038–1047 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. 90: 5. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD (2002) Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Scheer JM, Ryan CAJ (2002) The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc Natl Acad Sci USA 99: 9585–9590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P (2000) SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acid Res 28: 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001a) Plant receptor-like kinase gene family: diversity, function, and signaling. Sci Signal Transduction Knowledge Environ 113: RE22. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001b) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y (2002) The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 13627–13632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH et al. (1995) A receptor kinase-like protein encoded by the rice disease resistance gene Xa21. Science 270: 1804–1806 [DOI] [PubMed] [Google Scholar]
- Sonnhammer ELL, Eddy SR, Birney E, Bateman A, Durbin R (1998) Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acid Res 26: 320–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JC, Dixit R, Nasrallah ME, Nasrallah JB (1996) SRK, the stigma-specific S locus receptor kinase of Brassica, is targeted to the plasma membrane in transgenic tobacco. Plant Cell 8: 429–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K et al. (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Swiderski MR, Innes RW (2001) The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J 26: 101–112 [DOI] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D (2001) The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev 15: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Shimosato H, Shiba H, Funato M, Che FS, Watanabe M, Iwano M, Isogai A (2001) Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 413: 534–538 [DOI] [PubMed] [Google Scholar]
- Torii KU (2000) Receptor kinase activation and signal transduction in plants: an emerging picture. Curr Opin Plant Biol 3: 361–367 [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud AE, Jeong S, Clark SE (2000) CLAVATA3, a multimeric ligand for the CLAVATA1 receptor-kinase. Science 289: 613–617 [DOI] [PubMed] [Google Scholar]
- Urano F, Bertolotti A, Ron D (2000) IRE1 and efferent signaling from the endoplasmic reticulum. J Cell Sci 113: 3697–3702 [DOI] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Shinozaki K (2001) Plant histidine kinases: an emerging picture of two-component signal transduction in hormone and environmental responses. Sci Signal Transduction Knowledge Environ 113: RE18. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA et al. (2001) The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD (2000) The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117 [DOI] [PubMed] [Google Scholar]
- Walker JC (1994) Structure and function of the receptor-like protein kinases of higher plants. Plant Mol Biol 26: 1599–1609 [DOI] [PubMed] [Google Scholar]
- Wang GL, Ruan DL, Song WY, Sideris S, Chen L, Pi LY, Zhang S, Zhang Z, Fauquet C, Gaut BS et al. (1998) Xa21D encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subject to adaptive evolution. Plant Cell 10: 765–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Seto H, Fujioka S, Yoshida S, Chory J (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.