Abstract
In a recent bioinformatic analysis, we predicted the presence of multiple families of cell surface glycosylphosphatidylinositol (GPI)-anchored proteins (GAPs) in Arabidopsis (G.H.H. Borner, D.J. Sherrier, T.J. Stevens, I.T. Arkin, P. Dupree [2002] Plant Physiol 129: 486-499). A number of publications have since demonstrated the importance of predicted GAPs in diverse physiological processes including root development, cell wall integrity, and adhesion. However, direct experimental evidence for their GPI anchoring is mostly lacking. Here, we present the first, to our knowledge, large-scale proteomic identification of plant GAPs. Triton X-114 phase partitioning and sensitivity to phosphatidylinositol-specific phospholipase C were used to prepare GAP-rich fractions from Arabidopsis callus cells. Two-dimensional fluorescence difference gel electrophoresis and one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis demonstrated the existence of a large number of phospholipase C-sensitive Arabidopsis proteins. Using liquid chromatography-tandem mass spectrometry, 30 GAPs were identified, including six β-1,3 glucanases, five phytocyanins, four fasciclin-like arabinogalactan proteins, four receptor-like proteins, two Hedgehog-interacting-like proteins, two putative glycerophosphodiesterases, a lipid transfer-like protein, a COBRA-like protein, SKU5, and SKS1. These results validate our previous bioinformatic analysis of the Arabidopsis protein database. Using the confirmed GAPs from the proteomic analysis to train the search algorithm, as well as improved genomic annotation, an updated in silico screen yielded 64 new candidates, raising the total to 248 predicted GAPs in Arabidopsis.
Glycosylphosphatidylinositol (GPI)-anchored proteins (GAPs) are targeted to the plant cell surface and are likely to be involved in extracellular matrix remodeling and signaling (Sherrier et al., 1999; Borner et al., 2002). GPI anchoring is found in all eukaryotic organisms, and GAPs belonging to numerous protein families have been described (for recent review, see Ikezawa, 2002; Sharom and Lehto, 2002). Since the phosphatidylinositol moiety of the anchor is susceptible to cleavage by specific phospholipases (Griffith and Ryan, 1999), GAPs may exist in both a soluble and a membrane-associated form. In many organisms, GPI anchoring acts as a plasma membrane-targeting signal, and it has been implicated in polarized secretion (Brown et al., 2000), association with lipid microdomains (Ikonen, 2001), and recycling from the plasma membrane (Fivaz et al., 2002) in mammalian cells. Another intriguing feature of GAPs is their ability to transfer from one cell surface to the plasma membrane of an adjacent cell, a process known as “protein painting” (Premkumar et al., 2001). Whether the GPI anchor confers these interesting properties on the plant GAPs remains to be investigated.
A number of studies have highlighted the importance of GAPs at the plant cell surface. Arabinogalactan (AG) proteins (AGPs) have been implicated in signaling and differentiation (Showalter, 2001), and several AGPs are GPI anchored (Youl et al., 1998; Oxley and Bacic, 1999; Sherrier et al., 1999; Svetek et al., 1999; Schultz et al., 2000). However, current evidence suggests that plant GAPs are likely to have a wider range of functions. Arabidopsis expresses abundant and diverse GAPs in addition to the AGPs (Sherrier et al., 1999). Moreover, in a recent bioinformatic analysis, we identified over 200 potential GAPs in Arabidopsis that could be involved in cell signaling, adhesion, matrix remodeling, and pathogen response (Borner et al., 2002). In support of this view, several Arabidopsis mutants recently have revealed the involvement of likely GAPs in cell surface processes. For example, the COBRA protein appears to be a key factor in regulating cellulose deposition in root cells; evidence suggests that it is targeted in a polarized manner (Schindelman et al., 2001; Roudier et al., 2002). Similarly, deficiency in SOS5, a putative fasciclin-like adhesion protein, results in abnormal cell expansion (Shi et al., 2003). SKU5 (described as a BP-10 like GAP by Borner et al., 2002) is involved in directional root growth (Sedbrook et al., 2002), and PMR6, a pectate lyase-like protein, may play a role in interactions with pathogens (Vogel et al., 2002).
Despite the increasing amount of functional data, only two identified Arabidopsis proteins have been shown experimentally to be GPI anchored. The deglycosylated backbone of AtAGP10 is modified by C-terminal ethanolamine (Schultz et al., 2000), and SKU5 is sensitive to phosphatidylinositol-specific phospholipase C (Pi-PLC; Sedbrook et al., 2002). Direct evidence for the GPI anchoring of other predicted Arabidopsis GAPs, including COBRA, SOS5, and PMR6, is still lacking.
In this study, we have taken a proteomics approach to identify GAPs from Arabidopsis callus. The results provide strong support for the predictions of our previous bioinformatic analysis. We also update the genomic screen based on improved protein sequence data and a search algorithm trained on the confirmed GAPs.
RESULTS
In our previous genomic analysis of Arabidopsis, we identified more than 15 protein families comprising 210 putative GAPs (Borner et al., 2002). To validate these predictions, a proteomics approach was taken to identify bona fide GAPs.
Treatment with Pi-PLC in conjunction with Triton X-114 detergent phase partitioning is an established method to determine GPI anchoring of proteins (Sherrier et al., 1999; Hooper, 2001; Sedbrook et al., 2002). It relies on the partitioning of membrane proteins based on their hydrophobicities in a two-phase system. GAPs can be identified by a characteristic shift from the hydrophobic detergent-rich phase to the hydrophilic aqueous phase upon cleavage of the GPI-anchor with Pi-PLC. Using this method, GAPs were prepared from Arabidopsis callus membranes. To identify any abundant non-GAPs that contaminate the GAP-rich aqueous phase, a control experiment (without Pi-PLC) was performed.
Two-Dimensional Difference Gel Electrophoresis (2D-DIGE)
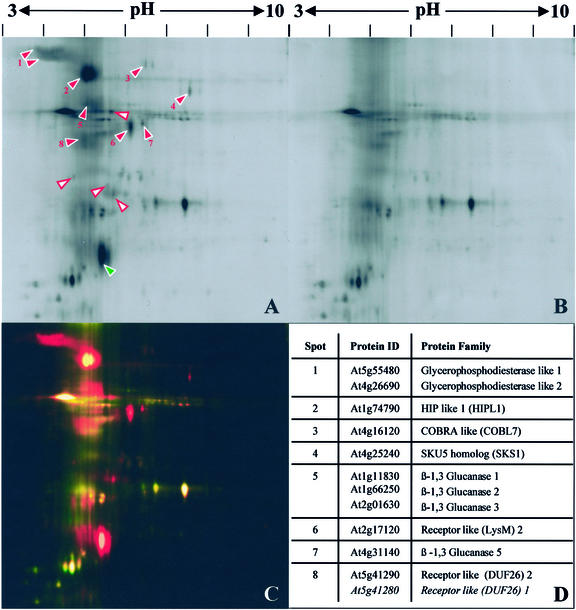
Preparations of GAPs were first analyzed by 2D-DIGE. The Pi-PLC-treated and control aqueous phases were labeled with different fluorescent CyDyes and pooled before isoelectric focusing (IEF; pH range 3–10) and SDS-PAGE in a single gel (Unlu et al., 1997). Proteins were detected with a dual-channel fluorescence scanner (Fig. 1, A and B). This revealed the presence of many proteins specifically enriched in the Pi-PLC-treated fraction and some background proteins present in both fractions. Because the samples were separated in a single gel, DIGE allowed unambiguous matching of spots and, thus, demonstrated a clear distinction between these two populations (Fig. 1C; Pi-PLC-sensitive proteins shown in red). The large number of Pi-PLC-sensitive proteins is in agreement with the results obtained by Sherrier et al. (1999).
Figure 1.
Identification of Arabidopsis GAPs sensitive to Pi-specific phospholipase C. Integral membrane proteins from callus were prepared by TX-114 phase partitioning and treated with Pi-PLC or buffer only (control). After repartitioning, proteins released into the aqueous phases were labeled with CyDyes. The samples were mixed and then separated by electrophoresis in a single two-dimensional gel. Gels were imaged with a fluorescence scanner. A to D, Broad pH range (3–10). A, Aqueous phase after Pi-PLC treatment. B, Control aqueous phase. C, False-color overlay of Pi-PLC-treated (red) and control (green) fractions. D, LC-MS/MS identifications of GAPs. Numbers correspond to red arrowheads in A. Some spots contained mixtures of related proteins. E to H, Narrow pH range (4–7). E, Aqueous phase after Pi-PLC treatment. F, Control aqueous phase. G, False-color overlay of Pi-PLC-treated (red) and control (green) fractions. H, LC-MS/MS identifications of GAPs. Numbers correspond to red arrowheads in E. Arrowheads in A and E indicate spots specifically enriched in the Pi-PLC-treated fractions. Red, numbered arrowheads correspond to identified GAPs (D and H). White arrowheads with red borders indicate unidentified Pi-PLC-sensitive proteins. Green arrowheads correspond to Pi-PLC-derived proteins.
Proteins clearly enriched in the Pi-PLC-treated fraction were excised, digested with trypsin, and subjected to liquid chromatography (LC)-tandem mass spectrometry (MS/MS). Eleven proteins were identified (Fig. 1D; Table I, Identification A). Analysis of their predicted protein sequences indicated that they all have the structural components necessary for GPI anchoring and that eight were predicted by Borner et al. (2002). Thus, the preparation and analysis method allowed identification of GAPs from Arabidopsis.
Table I.
Confirmed GPI-anchored proteins from Arabidopsis callus
Proteins were separated by two-dimensional difference gel electrophoresis (A, pH 3-10; B, pH 4-7) or one-dimensional SDS-PAGE (C, 12% [w/v] gel; D, 16% [w/v] gel). Proteins sensitive to Pi-PLC were identified by LC-MS/MS. Letters indicate experiments that yielded identification of a protein. Uppercase letters indicate identifications with greater than 95% confidence based on MASCOT scores (see “Materials and Methods”). The most likely cleavage sites (Udenfriend and Kodukula, 1995) are indicated (bold). The hydrophobic domain of each C-terminal signal peptide is underlined. Confirmations have been submitted to the Munich Information Center for Protein Sequences (MIPS).
| No. | Protein Family | MIPS No. | Identifications | C Terminus with Predicted Cleavage Site |
|---|---|---|---|---|
| 1 | Stellacyanin like | At5g20230 | C, D | TTPAGN AASSLGGATFLVAFVSAVVALF |
| 2 | Uclacyanin II | At2g44790 | C, D | PPPKAS GASKGVMSYVLVGVSMVLGYGLWM |
| 3 | Early nodulin like 1 | At2g25060 | C | APAPIS GSVRLGGCYVVLGLVLGLCAWF |
| 4 | Early nodulin like 2 | At4g27520 | D | PGQKKS SANGMTVMSITTVLSLVLTIFLSA |
| 5 | Early nodulin like 3 | At5g25090 | C | ASGGSA SSLTRQVGVLGFVGLLAIVLL |
| 6 | COBRA like 7 (COBL7) | At4g16120 | A, B, C, D | ILPMRS SQHRKHISVFLLALPVLALLILRA |
| 7 | glycerophosphodiesterase-like 2 (GPDL2) | At4g26690 | A, C, D | TPSTNA QAPSGQTRITLSLLLSVFAMVLASLLLL |
| 8 | glycerophosphodiesterase-like 1 (GPDL1) | At5g55480 | A, B, C, D | TPGPQS TGEKSPNGQTRVALSLLLSAFATVFASLLLL |
| 9 | Hedgehog interacting protein like 1 (HIPL1) | At1g74790 | A, B, C, D | SPSSSS SSCYKHINGFHGSLVVLFVSLSLILLGLLN |
| 10 | Hedgehog interacting protein like 2 (HIPL2) | At5g62630 | C | PQPLPS SARKLCFSVFLLLSLLMMFLTLLD |
| 11 | FLA7 | At2g04780 | D | KSSHKN SGQKLLLAPISMVISGLVALFL |
| 12 | FLA8 | At2g45470 | C, D | NSKSAN AAVGVSTPSLFTALVTIAAIAVSVSLCS |
| 13 | FLA10 | At3g60900 | C, D | NSNAKN AAFHVNAPALFTALVTIAATSLLL |
| 14 | FLA1 | At5g55730 | C, D | DATADD AGAVRIIGGAKAGLVVSLLCLFASSWLL |
| 15 | β-1,3 Glucanase 1 | At1g11830 | A | NSTEVA AGEATSRSLSRGFCVTIMILVTFSIL |
| 16 | β-1,3 Glucanase 2 | At1g66250 | A, C, D | ANSTTS SGIRSDLYYSRGIWSILTVMILNVANIL |
| 17 | β-1,3 Glucanase 3 | At2g01630 | A, c, D | ANSTTS GCIPKYYHHPHASFGDLTLLSLLLIIALVFL |
| 18 | β-1,3 Glucanase 4 | At3g13560 | C | PLGGNA NARIIFSYHLPILAPLALTLLQLLLQHDRLL |
| 19 | β-1,3 Glucanase 5 | At4g31140 | A, D | EDASEA SAMMPITRSTAVLLLLSICLYIVL |
| 20 | β-1,3 Glucanase 6 | At5g58090 | D | EPYYGG AAREHGFFFPLLMVAAIAVSIF |
| 21 | Lipid transfer protein like (LTPL) | At1g27950 | c, D | DKGGSA SAKDGHAVVALAVALMAVSFVLTLPRHVTLGM |
| 22 | SKU5 | At4g12420 | B, C, D | QKVSSS ASKSIGFTSLSMVVMALVMMMMLQH |
| 23 | SKU5 homolog (SKS1) | At4g25240 | A, D | KEQHHS AATSILNGHLKLMLLMVLLASVFRFC |
| 24 | Receptor like (Duf26) 1 | At5g41280 | a, b, D | PPPSRS GSFSIRGNNKILVGMILAVSVFAFLGL |
| 25 | Receptor like (Duf26) 2 | At5g41290 | A, B, D | PPPSRS GSFSHRGNNKLLGGMVLAVSVSVFAFLSLV |
| 26 | Receptor like (lysM) 1 | At1g21880 | D | GSISTA SASSVSYFFITFLISIASFSLALSS |
| 27 | Receptor like (lysM) 2 | At2g17120 | A, B, D | SPACPD SAGPDNYASTLSSSFNFVIVLIQCALLCLCLL |
| 28 | Auxin-induced protein AIR12 | At3g07390 | C, D | AGGPGN AGSLTRNVNFGVNLGILVLLGSIFIF |
| 29 | Unknown protein | At5g19230 | C | LTTTNS GAYAFGVNGLVSSSFLFLLFCFFMF |
| 30 | Unknown protein | At5g19250 | ca | SPASNS GAFAFGVNGLVSSSLMFLLFCFFMF |
Sequenced manually from MS/MS spectra to confirm identification.
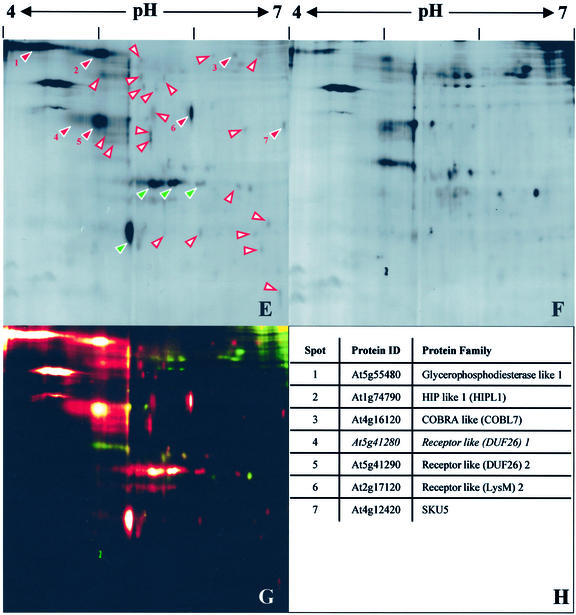
The most abundant Pi-PLC-sensitive proteins focused in a narrow pH range (from 4.5–5.5). To increase the resolution in this area of the gel, GAPs were analyzed by 2D-DIGE using a zoom strip (pH range 4–7) during IEF. The spot pattern obtained was comparable with the one observed with a broader pH-range but revealed numerous previously merged or undetectable proteins (Fig. 1, E–G). As before, Pi-PLC-sensitive proteins were excised, digested with trypsin, and subjected to LC-MS/MS. In addition to confirming several of the previous identifications, one further GAP (SKU5) was identified (Fig. 1H; Table I, Identification B). Several of the protein spots were of too low abundance to yield sufficient peptides for LC-MS/MS analysis. CyDye labeling is a highly sensitive detection method and allows visualization of very low protein quantities (Unlu et al., 1997).
Figure 1.
Identification of Arabidopsis GAPs sensitive to Pi-specific phospholipase C. Integral membrane proteins from callus were prepared by TX-114 phase partitioning and treated with Pi-PLC or buffer only (control). After repartitioning, proteins released into the aqueous phases were labeled with CyDyes. The samples were mixed and then separated by electrophoresis in a single two-dimensional gel. Gels were imaged with a fluorescence scanner. A to D, Broad pH range (3–10). A, Aqueous phase after Pi-PLC treatment. B, Control aqueous phase. C, False-color overlay of Pi-PLC-treated (red) and control (green) fractions. D, LC-MS/MS identifications of GAPs. Numbers correspond to red arrowheads in A. Some spots contained mixtures of related proteins. E to H, Narrow pH range (4–7). E, Aqueous phase after Pi-PLC treatment. F, Control aqueous phase. G, False-color overlay of Pi-PLC-treated (red) and control (green) fractions. H, LC-MS/MS identifications of GAPs. Numbers correspond to red arrowheads in E. Arrowheads in A and E indicate spots specifically enriched in the Pi-PLC-treated fractions. Red, numbered arrowheads correspond to identified GAPs (D and H). White arrowheads with red borders indicate unidentified Pi-PLC-sensitive proteins. Green arrowheads correspond to Pi-PLC-derived proteins.
One-dimensional SDS-PAGE
To identify further Pi-PLC-sensitive proteins that were not recovered from 2D gels, preparations of GAPs were analyzed by one-dimensional SDS-PAGE. Pi-PLC-treated and -untreated fractions were separated in parallel lanes in the same gel. Proteins were stained with Coomassie Blue (data not shown). Bands clearly enriched in the Pi-PLC-treated fraction were excised, digested with trypsin, and the peptides were subjected to LC-MS/MS. To identify proteins not visible with Coomassie Blue, both treated and control lanes were excised completely, cut into sections, digested with trypsin, and peptides analyzed by LC-MS/MS. Twenty-three Arabidopsis GAPs were found specifically enriched in the Pi-PLC-treated fraction (Table I, Identifications C and D). Only one protein (At4g20830) that has a sequence not predicted to confer GPI anchoring copartitioned with the GAPs.
Summary of the Proteomic Analysis
The proteomic analysis identified 30 proteins in total that were sensitive to Pi-PLC and have sequence features expected of GAPs, indicating that they are GPI anchored (Table I). Furthermore, 21 of them had been predicted in the genomic analysis (Borner et al., 2002). Eight of the novel GAPs were not correctly annotated in the protein database used by Borner et al. (2002); therefore, it was not possible to predict them at that time. The ninth has a charge in the C-terminal signal peptide (At1g27950). However, all the novel GAPs belong to the families previously predicted.
Updated Genomic Analysis
Because the first genomic prediction was very effective but limited by apparently incomplete or incorrect annotation of the genome in August 2001, the MIPS Arabidopsis data set (November 30, 2002) was analyzed according to Borner et al. (2002). Of the 26,618 proteins in the database, 239 GAPs were predicted, including 55 new candidates. One hundred eighty-four proteins described in the previous analysis were confirmed, whereas 26 were no longer predicted to become GPI anchored in their present annotation.
Candidate GAPs were sorted into families based on sequence similarity to known proteins. A single new protein family, consisting of three pectate lyases not correctly annotated in the August 2001 database, was added to the previously described list. Unknown and hypothetical proteins were also grouped on the basis of sequence similarity. The results are summarized in Table II.
Table II.
Predicted and confirmed GPI-anchored proteins in Arabidopsis
Confirmed GPI-anchored proteins are marked by superscript nos. (Table I). Bold indicates the presence of probable AG glycomodules. Entries in parentheses are candidates that have weak predictions for N- or C-terminal peptides. Families of homologous unknown/hypothetical proteins are separated by semicolons. Predictions have been submitted to MIPS.
| Protein Family | MIPS Protein Entry Codes | Total |
|---|---|---|
| Classical AGPs | At1g35230, At1g68725, At2g14890, At2g22470, At2g23130, At2g47930, At3g01700, At3g06360a, At4g09030, At4g16985, At4g37450, At4g40091, At5g10430, At5g14380, At5g18690, At5g64310, At5g65390 | 17 |
| AG peptides | At1g55330, (At2g46330), At3g01730, At3g13520, At3g20865, At3g57690, (At3g61640), At4g26320, At5g11740, At5g40730, At5g53250, At5g56540 | 12 |
| Extensin related | At1g02405, At1g23040, At1g70990, At3g06750, At4g16140, At5g11990, At5g49280 | 7 |
| Phytocyanins (28) | ||
| Stellacyanin like | At2g26720, At2g31050, At5g202301, At5g26330 | 4 |
| Uclacyanin like | At1g22480, At1g72230, At2g32300, At2g447902, At3g27200, At3g60270, At3g60280, At5g07475 | 8 |
| Early nodulin like | At1g08500, At1g48940, At1g79800, At2g23990, At2g250603, At3g01070, At3g18590, At3g20570, At4g275204, At4g28365, At4g30590, At4g31840, At4g32490, At5g15350a, At5g250905, At5g53870 | 16 |
| COBRA family (10) | ||
| COBRA | At1g09790, At3g02210, At3g29810, At5g15630, At5g60920 | 5 |
| COBRA like | At3g16860, At3g20580, (At4g161206), (At4g27110), At5g49270 | 5 |
| GPDL | At1g66970, At3g20520, At4g266907, At5g554808, At5g58050, At5g58170 | 6 |
| HIPL | At1g747909, At5g39970, At5g6263010 | 3 |
| Fasciclin-like AGPs (FLAs) | At1g03870, At2g0478011, At2g20520, At2g24450, At2g4547012, At3g12660, At3g46550, At3g6090013, At4g12730, At4g31370, At5g03170, At5g44130, At5g5573014, At5g60490 | 14 |
| Fasciclin like | At1g30800, At4g12950 | 2 |
| β-1,3 Glucanases | At1g1183015, (At1g18650), At1g26450, At1g32860a, At1g64760, At1g6625016, At1g69295, At1g77780, At2g0163017, At2g19440, At2g26600, At3g04010, At3g1356018a, At3g15800, (At3g24330), At3g58100, At4g29360, At4g3114019, At5g08000, (At5g18220), At5g20870, At5g42100, At5g42720, At5g56590, At5g5809020, At5g58480, At5g61130, At5g64790 | 28 |
| Polygalacturonase | At3g15720 | 1 |
| Pectate Lyases | At3g53190, At3g54920, At5g04310 | 3 |
| Proteases (16) | ||
| Aspartyl- | At1g05840, At1g08210, At1g65240, At2g17760, At3g02740, At3g51330, At3g51350, At4g35880, At5g10080, At5g36260 | 10 |
| Metallo- | At1g24140, At1g59970, At1g70170, (At2g45040), (At4g16640) | 5 |
| Cys- | At3g43960 | 1 |
| LTPL | At1g05450, At1g18280, At1g2795021a, At1g36150, (At1g55260), At1g62790, At1g73890, At2g13830, At2g27130, At2g44290, At2g44300, At2g48130, At2g48140, At3g22600, At3g22611, At3g43720, At3g58550, At4g08670, At4g12360, At4g14805, At4g14815, At4g22630, At4g22640, At5g09370, At5g13900, At5g64080 | 26 |
| SKU5 family | At4g1242022, At4g2524023, At5g51480 | 3 |
| Receptor like (16) | ||
| RLK3 like (DUF26) | At1g63550, At1g63580, At5g4128024, At5g4129025, At5g41300 | 5 |
| PRK5 like | At1g20030, At4g36010,At4g38660 | 3 |
| Lectin like | At1g07460 | 1 |
| LysM domains | At1g2188026, At1g77630, At2g1712027 | 3 |
| Other | At1g10375 | 1 |
| Cf-2/Cf-5 like | At1g80080, At2g42800, At4g28560 | 3 |
| GPI-Anchored peptides (GAPEPs) | At3g01940, (At3g01950), At5g14110, (At5g40960), At5g40970a, At5g40980, (At5g50660), At5g63500 | 8 |
| Other | At5g07190, At5g62200, At5g62210; At3g0739028; At1g24520, At4g15460 | 6 |
| Unknown/hypothetical | At1g54860, At3g06035, At5g1923029, At5g1925030; At1g07135, At1g09175, At3g04640, At3g55790; At1g29980, At2g34510, At5g14150; At2g20700, At4g28280, At5g56170; At3g18050, At4g28100; At3g27410, At5g40620, (At1g23050), At1g70985; At5g26290a, At5g26300a; At3g24518, At5g35890; At1g21090; At1g56320; At1g61900; At1g64640; At2g28410; (At2g29660); At3g26110; At3g44100a; At3g58890; At3g61980; At4g14746; At4g28085; At4g38140; At5g08210; At5g14190; At5g16670; At5g22430; At5g67131 | 42 |
| Total | 248 |
Sequences found in the extended genomic analysis.
The new set of predictions included 28 of the 30 GAPs confirmed by proteomic methods. The two missing proteins, At3g13560 (β-1,3 glucanase) and At1g27950 (LTPL), have all the structural hallmarks of GAPs but are unusual in that they have very hydrophilic or charged residues in their hydrophobic C termini.
Extended Genomic Analysis
To investigate the possible occurrence of further such GAPs in Arabidopsis, we modified the C-terminal hydrophobic segment requirements of our search algorithm, GPT (GPI anchoring prediction tool). The algorithm was permitted to ignore two hydrophilic or charged residues while the minimum summed hydrophobicity was significantly increased. We trained this new version of GPT on the set of 30 confirmed Arabidopsis GAPs shown in Table I.
Analysis of the MIPS protein database resulted in the identification of nine novel candidate GAPs. In addition to the two unusual confirmed GAPs from the proteomic analysis, these included a classical AGP, a phytocyanin, a further β-1,3 glucanase, and a GAPEP, all strong candidates for GPI anchoring. The new sequences were added to Table II (footnote a), resulting in a total of 248 predicted or confirmed Arabidopsis GAPs. The GAP predictions and confirmations are available at MIPS.
One of the most unexpected predictions of Borner et al. (2002) was that 42% of GAPs have (hydroxy) Pro-rich AG-type glycosylation modules (Kieliszewski, 2001; Zhao et al., 2002). In the updated analysis, we still predict that 100 of 248 (40%) GAPs have putative AG glycomodules, of which only 29 are classical AGPs or AG peptides (Table II).
DISCUSSION
In this study, evidence was obtained for the GPI anchoring of 30 proteins from Arabidopsis callus, using sensitivity to Pi-PLC as the diagnostic tool and LC-MS/MS for protein identification. In addition, novel sequence features of confirmed GAPs and new gene predictions were exploited for an improved genomic analysis.
Correct annotation of the N- and C-terminal signal peptides is essential for the GAP prediction algorithm. Although the total number of annotated Arabidopsis proteins has changed little since the first publication by the Arabidopsis Genome Initiative (2000), gene predictions have improved substantially. Thus, the updated genomic analysis predicted 248 GAPs in Arabidopsis, including 184 of our previous predictions (Borner et al., 2002). Despite this significant increase, only one new protein family of three pectate lyases was found. These proteins were not correctly annotated in the database at the time of the first genomic screen. Thus, although further improvements of the Arabidopsis genome annotation will allow us to uncover more putative GAPs, it is likely that nearly all will have sequence similarity to proteins described here.
Several recent studies have revealed interesting functions for predicted GAPs, but experimental evidence for GPI anchoring of these proteins was mostly lacking. COBRA, a predicted GAP, is proposed to be involved in cellulose deposition (Schindelman et al., 2001) and is related to 10 other proteins in Arabidopsis (Borner et al., 2002; Roudier et al., 2002). With the exception of the truncated COBL5, they have all been predicted to be GAPs (Borner et al., 2002; Roudier et al., 2002). We provide evidence here for the GPI anchoring of a COBRA family protein, COBL7. Interestingly, this protein appears to have an unusual omega cleavage site in its C-terminal signal peptide. Because the other predicted GAPs in the family have more conventional C-terminal signal peptides, it is very likely that they are also GPI anchored. Similarly, we show here for the first time, to our knowledge, that the fasciclin-like putative adhesion proteins FLA1, FLA7, FLA8, and FLA10 are Pi-PLC sensitive. This strongly suggests that their close homologs are also GPI anchored, including SOS5 (Shi et al., 2003), previously described as FLA4 (Gaspar et al., 2001). The proteomic analysis further demonstrated the GPI anchoring of SKU5's closest homolog SKS1 and confirmed the recently reported Pi-PLC sensitivity of SKU5 (Sedbrook et al., 2002).
The proteomic analysis also demonstrated the occurrence of GPI anchoring in several families of largely uncharacterized proteins. The largest group of confirmed GAPs are six predicted β-1,3 glucanases. The glucanases are also the largest family of predicted GAPs, with 28 members. GPI anchoring has been predicted for numerous phytocyanins (Nersissian et al., 1998; Borner et al., 2002). It is particularly noteworthy that this study showed GPI anchoring of members of all three subfamilies of phytocyanins with predicted GAPs, the early nodulin-like phytocyanins, stellacyanins, and uclacyanins. Similarly, a large group of predicted GAPs related to lipid transfer proteins now has a confirmed GPI-anchored member. Further families without previously confirmed GAPs include the Hedgehog-interacting protein-like (HIPL) proteins and the glycerophosphodiesterase-like (GPDL) proteins (Borner et al., 2002). Two of the major GAPs seen in the DIGE gels were identified as HIPL1 (At1g74790, Fig. 1Fig. 1, Spot 2) and GPDL1 (At5g55480, Fig. 1Fig. 1, Spot 1) and correspond to the abundant GAPs AtGPIP1 and AtGPIP2 of Sherrier et al. (1999). An extracellular glycerophosphodiesterase activity has recently been shown in Arabidopsis cell cultures, but GPI anchoring was not investigated (van der Rest et al., 2002). Finally, this study confirmed one of the most intriguing predictions of Borner et al. (2002), namely the existence of GPI-anchored receptor-like proteins in Arabidopsis. GPI anchoring of four putative receptors was demonstrated, including two proteins with LysM binding domains, which may recognize bacterial peptidoglycan (Bateman and Bycroft, 2000). A curious observation was the copartitioning with the confirmed GAPs of a putative oxidoreductase (At4g20830). Its sequence, lacking a C-terminal signal peptide, is incompatible with GPI anchoring. A possible interpretation is that this protein is tightly bound to a GAP that is sensitive to Pi-PLC.
In summary, the proteomic analysis identified GAPs belonging to 11 functional categories (Table I). These confirmed GAPs are representatives of most of the major families and subfamilies of predicted GAPs (Table II). This increases confidence in the predictions that the related proteins are GPI anchored. Previously, only the classical AGP and SKU5 families had one experimentally confirmed member each (Schultz et al., 2000; Sedbrook et al., 2002). Now, a substantial proportion (148 of 248) of the predicted GAPs belong to families with at least one confirmed GPI-anchored member.
GAPs from a few of the predicted families were not detected in the proteomic analysis. There are several possible reasons. The classical AGPs, AG peptides, and extensin-like proteins are probably heavily glycosylated and yield few or no tryptic peptides for LC-MS/MS analysis. Similarly, the GAPEPs are also a class of proteins without any confirmed members. They are very small proteins in their predicted mature forms (only 1–2 kD) and, thus, would not be detected in the gels used.
The remaining unconfirmed families include proteases, pectate lyases including PMR6 (Vogel et al., 2002), subfamilies of the receptor-like proteins, and several groups of unknown and hypothetical proteins. Possibly, they were not expressed in callus tissue, or their expression levels were too low for detection on the gels or for sequencing by mass spectrometry. We will prepare GAPs from other tissues to investigate the differences in expression. However, some GAPs may be resistant to cleavage by Pi-PLC (Ikezawa, 2002) and, therefore, will not be detected in a PLC/phase partitioning approach.
Three of the 30 confirmed GAPs have unexpected sequence features. Two have charges in the hydrophobic stretch of the C terminus, as has been reported in a few yeast GAPs (Hamada et al., 1999). Furthermore, the most likely omega cleavage site at the C terminus of COBL7 is the triplet “SSQ.” This omega site has not been described in a GAP before to our knowledge. Nevertheless, COBL7 was identified in multiple experiments (Table I). Clearly, sequence requirements for Arabidopsis transamidase(s) appear to be different from those of metazoan and protozoan homologs. Thus, it will be important to develop plant-specific versions of algorithms such as Big-PI (Eisenhaber et al., 1999), to predict plant omega cleavage sites.
The analysis of the recent protein database identified a novel classical AGP (AtAGP28, At4g16985) and two novel AG peptides (AtAGP29, At3g01730; AtAGP40, At3g20865) in addition to those previously described (Borner et al., 2002; Schultz et al., 2002). Thus, in Arabidopsis, classical AGPs and AG peptides constitute 29 of 248 predicted GAPs (12%). Nevertheless, as found in Borner et al. (2002), over 40% of the predicted GAPs have putative AG glycomodules. Proteins that contain both AG glycomodules and non-glycosylated modules, such as the GPDLs, FLAs, phytocyanins, and LTPLs, are much more numerous than the classical AGPs and AG peptides in Arabidopsis.
With this study, the Arabidopsis proteome of GAPs is one of the best characterized of any organism. The identification of so many diverse GAPs is likely to facilitate investigations into trafficking of lipid rafts, polarized targeting, and many other aspects of plant cell surface processes.
MATERIALS AND METHODS
Genomics
Database Analysis
The annotated Arabidopsis protein database was retrieved from MIPS http://mips.gsf.de/proj/thal/db/index.html. Where necessary, gene entries from MIPS were re-annotated using GenBank ESTs or cDNA information found in MIPS.
Bioinformatic analysis of the Arabidopsis protein database was carried out as described by Borner et al. (2002). In brief, our own algorithm GPT was used to generate a list of candidate GAPs with putative N- and C-terminal signal peptides. This list was subsequently refined using SignalP version 2.0 (Nielsen et al., 1997), TMHMM (Sonnhammer et al., 1998), and the rules established by Udenfriend and Kodukula (1995). Thus, the method will not identify any GAP with a signal anchor, such as DAMP1 of mammalian cells (Kupzig et al., 2002).
A modified version of GPT was used to screen for GAPs with charged C termini. The confirmed GAPs from our proteomic analysis were used as the training set. The most stringent settings that identified >90% of the training sequences, including the two GAPs with charged C termini (At3g13560 and At1g27950), were determined to the following values: signal length at C terminus 12 amino acids and hydrophobicity –25 kJ mol–1 (GES scale; Engelman et al., 1986), ignoring up to two internal hydrophilic or charged residues.
Auxiliary Programs
Sequence alignments were performed using ClustalW (Thompson et al., 1994) on the European Bioinformatics server (http://www.ebi.ac.uk/clustalw). BLAST searches (Altschul et al., 1990, 1997) for homologous sequences from all organisms and searches for conserved domains using Reverse Position-Specific BLAST (Altschul et al., 1997) were performed on the National Centre for Biotechnology server (http://www.ncbi.nlm.nih.gov/BLAST). BLAST searches of the Arabidopsis genome were performed on The Arabidopsis Information Resource server (http://www.Arabidopsis.org/Blast).
Proteomics
Plant Cell Culture
Liquid cultures of Arabidopsis Columbia callus were established and maintained as described (Prime et al., 2000). Cells were grown in large culture flasks for 3 weeks before being harvested for biochemical fractionation.
Biochemical Fractionation and Preparation of GAPs
To prepare total cell membranes, callus tissue was resuspended in two volumes of cold homogenization buffer (12% [w/v] Suc, 1 mm EDTA, and 100 mm Tris-HCl [pH 8.0]). Three pulses of 15 s at 5,700 rpm with a polytron (Kinematica, Littav, Switzerland) were used to homogenize the tissue at 4°C. The homogenate was centrifuged three times for 10 min at 1,600g to remove cell debris. The membrane suspension was then pelleted onto a 1.8 m Suc cushion at 100,000g for 35 min. The membrane fraction was harvested and diluted 5-fold in cold TNE (25 mm Tris-HCl, 150 mm NaCl, and 5 mm EDTA [pH 7.5]). After centrifugation at 100,000g for 2 h, membrane pellets were resuspended in cold TNE and homogenized in a 1-mL Dounce glass homogenizer. Membrane preparations were frozen in liquid nitrogen and stored in aliquots at –80°C.
For the preparation of GAPs, 10 to 15 mg of total membrane protein was processed for a single experiment using a protocol adapted from Sherrier et al. (1999). Membranes were resuspended in 2% (v/v) Triton X-114/TNE at 37°C. After removal of insoluble material by centrifugation at 4°C, phase separation was induced by raising the temperature to 37°C. The detergent phase was washed three times by repartitioning with Tris-buffered saline (10 mm Tris and 150 mm NaCl [pH 7.4 at 37°C]), split into two equal fractions, and diluted 20-fold in Tris-buffered saline. Pi-PLC (Sigma, St. Louis) was added to one sample to a final concentration of 1.5 units mL–1. Both samples were incubated at 37°C for 1 h and 30 min as described by Sherrier et al. (1999). The aqueous phases were separated and washed twice with fresh Triton X-114. Gelatin (Sigma) or bovine serum albumin (New England Biolabs, Beverly, MA) was added to both samples to increase total protein concentration. After ultrafiltration (Centricon, molecular weight cutoff = 10,000, Millipore, Bedford, MA) to reduce the volume, proteins were precipitated with 80% (w/v) acetone at –20°C overnight. Proteins were air dried and resuspended in AUT sample buffer (10 mm Tris/HCl [pH 8.5], 7 m urea, 2 m thiourea, and 2% [w/v] amino-sulfobetaine detergent [ASB14]) for two-dimensional electrophoresis and in SDS sample buffer for one-dimensional electrophoresis.
Fluorescent Labeling for DIGE
Non-saturating labeling of samples was performed using NHS esters of Cy3 or Cy5 (CyDye DIGE fluors, Amersham Biosciences, Amersham, UK). Dye (200 pmol) was added to 30 μg of sample in AUT sample buffer to give a typical total volume of 10 to 15 μL and left on ice in the dark for 30 min. The reaction was quenched by the addition of 10 nmol Lys and further incubation for 10 min. The sample was then prepared for IEF by the addition of an equal volume of sample buffer (20 mg mL–1 dithiothreitol, 2% appropriate immobilized pH gradient (IPG) buffer [Amersham Biosciences], 7 m urea, 2 m thiourea, and 2% [w/v] ASB14).
Electrophoretic Analyses
Two-dimensional electrophoresis was carried using 13-cm pH 3 to 10 or pH 4 to 7 IPG strips in conjunction with an IPGPhor (Amersham Biosciences). IPG strips were rehydrated for 12 h with rehydration buffer (2 mg mL–1 dithiothreitol, 1% [w/v] appropriate IPG buffer, 7 m urea, 2 m thiourea, and 2% [w/v] ASB14) to which the sample was added to give a final volume of 250 μL. Focusing was carried out for a total of 41,700 V h. Separation in the second dimension was carried out using a 12% (w/v) SDS-PAGE gel (Hoefer SE600, Amersham Biosciences,). Gels were scanned with the appropriate excitation and emission wavelengths for Cy3 and Cy5 using a 2920–2DMasterImager (Amersham Biosciences). Images were exported as TIF Files. False coloration and contrast enhancement of scans were performed with Adobe Photoshop (Adobe Systems, Mountain View, CA).
One-dimensional SDS-PAGE was performed using a standard protocol (Sambrook et al., 1990) except that all solutions were filtered and polymerized for at least 12 h. Pi-PLC-treated and control fractions were separated next to each other on a large (20-cm) 12% (w/v) polyacrylamide gel for optimum resolution. Bands significantly enriched in the treated fraction were excised and analyzed by LC-MS/MS. Alternatively, Pi-PLC-treated and control fractions were separated on a 16% (w/v) acrylamide minigel (4 cm). Both lanes were excised completely, cut into regular sections, and analyzed by LC-MS/MS. The total composition of both fractions was compared.
Mass Spectrometry
For analysis by mass spectrometry, proteins were stained with Coomassie G250 and excised manually. Proteins within the gel-excised spots were first reduced, carboxyamide methylated, and then digested to peptides using trypsin on a MassPrepStation (Micromass, Manchester, UK). The resulting peptides were applied to LC-MS/MS. The liquid chromatographic separation was achieved with a PepMap C18, 180-μm i.d., 15-cm column (LC Packings, Amsterdam). The mass spectrometer was a QTof (Micromass). Fragmentation data was used to search the National Center for Biotechnology Information Arabidopsis database using the MASCOT search engine (http://www.matrixscience.com). Probability-based MASCOT scores were used to evaluate identifications. Only matches with P < 0.05 for random occurrence were considered significant (further explanation of MASCOT scores can be found at http://www.matrixscience.com). Manual sequence assignment was assisted using the peptide sequencing feature of BioLynx (Micromass).
Acknowledgments
We thank Matthias Mann, Bernhard Kuester, and Len Packman for help in the initial stages of the proteomics project. We also thank Azam Razzaq for help in DIGE gel preparation and Svenja Hester and Julie Howard for assistance with mass spectrometry. The LC-MS/MS data are available from the Cambridge Centre for Proteomics on request.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.021170.
This work was supported by the Biotechnology and Biological Sciences Research Council (research studentship to G.H.H.B.), by the Biotechnology and Biological Sciences Research Council Investigating Gene Function Initiative (GARNet), and by the Studienstiftung des Deutschen Volkes (scholarship to G.H.H.B.).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bateman A, Bycroft M (2000) The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J Mol Biol 299: 1113–1119 [DOI] [PubMed] [Google Scholar]
- Borner GHH, Sherrier DJ, Stevens TJ, Arkin IT, Dupree P (2002) Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: a genomic analysis. Plant Physiol 129: 486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown O, Cowen RL, Preston CM, Castro MG, Lowenstein PR (2000) Subcellular post-transcriptional targeting: delivery of an intracellular protein to the extracellular leaflet of the plasma membrane using a glycosyl-phosphatidylinositol (GPI) membrane anchor in neurons and polarised epithelial cells. Gene Ther 7: 1947–1953 [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Bork P, Eisenhaber F (1999) Prediction of potential GPI-modification sites in proprotein sequences. J Mol Biol 292: 741–758 [DOI] [PubMed] [Google Scholar]
- Engelman DM, Steitz TA, Goldman A (1986) Identifying nonpolar transbilayer helices in amino-acid-sequences of membrane-proteins. Annu Rev Biophys Biophys Chem 15: 321–353 [DOI] [PubMed] [Google Scholar]
- Fivaz M, Vilbois F, Thurnheer S, Pasquali C, Abrami L, Bickel PE, Parton RG, van der Goot FG (2002) Differential sorting and fate of endocytosed GPI-anchored proteins. EMBO J 21: 3989–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar Y, Johnson KL, McKenna JA, Bacic A, Schultz CJ (2001) The complex structures of arabinogalactan-proteins and the journey towards understanding function Plant Mol Biol 47: 161–176 [PubMed] [Google Scholar]
- Griffith OH, Ryan M (1999) Bacterial phosphatidylinositol-specific phospholipase C: structure, function, and interaction with lipids. Biochim Biophys Acta 1441: 237–254 [DOI] [PubMed] [Google Scholar]
- Hamada K, Terashima H, Arisawa M, Yabuki N, Kitada K (1999) Amino acid residues in the omega-minus region participate in cellular localization of yeast glycosylphosphatidylinositol-attached proteins. J Bacteriol 181: 3886–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper NM (2001) Determination of glycosyl-phosphatidylinositol membrane protein anchorage. Proteomics 1: 748–755 [DOI] [PubMed] [Google Scholar]
- Ikezawa H (2002) Glycosylphosphatidylinositol (GPI)-anchored proteins. Biol Pharmacol Bull 25: 409–417 [DOI] [PubMed] [Google Scholar]
- Ikonen E (2001) Roles of lipid rafts in membrane transport. Curr Opin Cell Biol 13: 470–477 [DOI] [PubMed] [Google Scholar]
- Kieliszewski MJ (2001) The latest hype on Hyp-O-glycosylation codes. Phytochemistry 57: 319–323 [DOI] [PubMed] [Google Scholar]
- Kupzig S, Korolchuk VI, Rollason R, Sugden A, Banting G (2002) DAMP-1 is a lipid raft-associated protein with a novel topology. 55th Harden Conference: “Dynamics of Membrane Traffic.” Ambleside, UK, p S18
- Nersissian AM, Immoos C, Hill MG, Hart PJ, Williams G, Herrmann RG, Valentine JS (1998) Uclacyanins, stellacyanins, and plantacyanins are distinct subfamilies of phytocyanins: plant-specific mononuclear blue copper proteins. Protein Sci 7: 1915–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JE, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10: 1–6 [DOI] [PubMed] [Google Scholar]
- Oxley D, Bacic A (1999) Structure of the glycosylphosphatidylinositol anchor of an arabinogalactan protein from Pyrus communis suspension-cultured cells. Proc Natl Acad Sci USA 96: 14246–14251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar DR, Fukuoka Y, Sevlever D, Brunschwig E, Rosenberry TL, Tykocinski ML, Medof ME (2001) Properties of exogenously added GPI-anchored proteins following their incorporation into cells. J Cell Biochem 82: 234–245 [DOI] [PubMed] [Google Scholar]
- Prime TA, Sherrier DJ, Mahon P, Packman LC, Dupree P (2000) A proteomic analysis of organelles from Arabidopsis thaliana. Electrophoresis 21: 3488–3499 [DOI] [PubMed] [Google Scholar]
- Roudier F, Schindelman G, DeSalle R, Benfey PN (2002) The COBRA family of putative GPI-anchored proteins in Arabidopsis: a new fellowship in expansion. Plant Physiol 130: 538–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1990) Molecular Cloning: A Laboratory Manual, Ed 2, Vol 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN (2001) COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev 15: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Johnson KL, Currie G, Bacic A (2000) The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell 12: 1751–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Rumsewicz MP, Johnson KL, Jones BJ, Gaspar YM, Bacic A (2002) Using genomic resources to guide research directions: the arabinogalactan protein gene family as a test case. Plant Physiol 129: 1448–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook JC, Carroll KL, Hung KF, Masson PH, Somerville CR (2002) The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharom FJ, Lehto MT (2002) Glycosylphosphatidylinositol-anchored proteins: structure, function, and cleavage by phosphatidylinositol-specific phospholipase C. Biochem Cell Biol 80: 535–549 [DOI] [PubMed] [Google Scholar]
- Sherrier DJ, Prime TA, Dupree P (1999) Glycosylphosphatidylinositol-anchored cell-surface proteins from Arabidopsis. Electrophoresis 20: 2027–2035 [DOI] [PubMed] [Google Scholar]
- Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK (2003) The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15: 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM (2001) Arabinogalactan-proteins: structure, expression and function. Cell Mol Life Sci 58: 1399–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E, von Heijne G, Krogh A (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. In J Glasgow, T Littlejohn, F Major, R Lathrop, D Sankoff, C Sensen, eds, Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, pp 175–182 [PubMed]
- Svetek J, Yadav MP, Nothnagel EA (1999) Presence of a glycosylphosphatidylinositol lipid anchor on rose arabinogalactan proteins. J Biol Chem 274: 14724–14733 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udenfriend S, Kodukula K (1995) How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu Rev Biochem 64: 563–591 [DOI] [PubMed] [Google Scholar]
- Unlu M, Morgan ME, Minden JS (1997) Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 18: 2071–2077 [DOI] [PubMed] [Google Scholar]
- van der Rest B, Boisson AM, Gout E, Bligny R, Douce R (2002) Glycerophospho-choline metabolism in higher plant cells: evidence of a new glyceryl-phosphodiester phosphodiesterase. Plant Physiol 130: 244–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Schiff C, Somerville SC (2002) PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14: 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youl JJ, Bacic A, Oxley D (1998) Arabinogalactan-proteins from Nicotiana alata and Pyrus communis contain glycosylphosphatidylinositol membrane anchors. Proc Natl Acad Sci USA 95: 7921–7926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZD, Tan L, Showalter AM, Lamport DT, Kieliszewski MJ (2002) Tomato LeAGP-1 arabinogalactan-protein purified from transgenic tobacco corroborates the Hyp contiguity hypothesis. Plant J 31: 431–444 [DOI] [PubMed] [Google Scholar]