Abstract
Sulfate is a macronutrient required for cell growth and development. Arabidopsis has two high-affinity sulfate transporters (SULTR1;1 and SULTR1;2) that represent the sulfate uptake activities at the root surface. Sulfur limitation (–S) response relevant to the function of SULTR1;2 was elucidated in this study. We have isolated a novel T-DNA insertion allele defective in the SULTR1;2 sulfate transporter. This mutant, sel1-10, is allelic with the sel1 mutants identified previously in a screen for increased tolerance to selenate, a toxic analog of sulfate (Shibagaki et al., 2002). The abundance of SULTR1;1 mRNA was significantly increased in the sel1-10 mutant; however, this compensatory up-regulation of SULTR1;1 was not sufficient to restore the growth. The sulfate content of the mutant was 10% to 20% of the wild type, suggesting that induction of SULTR1;1 is not fully complementing the function of SULTR1;2 and that SULTR1;2 serves as the major facilitator for the acquisition of sulfate in Arabidopsis roots. Transcriptome analysis of approximately 8,000 Arabidopsis genes in the sel1-10 mutant suggested that dysfunction of the SULTR1;2 transporter can mimic general –S symptoms. Hierarchal clustering of sulfur responsive genes in the wild type and mutant indicated that sulfate uptake, reductive sulfur assimilation, and turnover of secondary sulfur metabolites are activated under –S. The profiles of –S-responsive genes further suggested induction of genes that may alleviate oxidative damage and generation of reactive oxygen species caused by shortage of glutathione.
Sulfur deficiency causes retarded and chlorotic growth of plants and significantly reduces crop productivities. Plants utilize sulfate for synthesis of various organic compounds through a complex metabolic network (Leustek and Saito, 1999; Leustek et al., 2000; Grossman and Takahashi, 2001). Once sulfate enters the cell, it is primarily assimilated through the reductive sulfur assimilation to form Cys and Met, which are the essential constituents of proteins. Cys is incorporated into glutathione (GSH), which serves for the redox control and conjugation of xenobiotics (Noctor and Foyer, 1998; Asada, 1999). Sulfur is further converted to a wide variation of secondary compounds essential for growth regulation and adaptation to environmental stresses through the sulfation and sulfonation pathways (Varin et al., 1997; Benning, 1998). The major targets of these secondary sulfur modification reactions are flavonoids, steroids, polysaccharides, and lipids. Upon sulfur limitation (–S), plants activate uptake of sulfate and primary sulfur assimilation for Cys synthesis. Interorgan transport of sulfate may be regulated for efficient distribution and utilization of the internal reserve of sulfate. In addition, the secondary sulfur metabolites can be remobilized to obtain sustainable amounts of sulfur source for primary assimilation. It is suggested that acclimation to –S stress includes complex regulation of sulfate transport and metabolisms, but the overall metabolic processes have remained poorly understood at the molecular level.
The studies on –S response of higher plants have been focused mostly on the regulation of sulfate transport and reductive assimilation. Cloning and characterization of proteins and enzymes for primary sulfur assimilation indicated that sulfate transporters and 5′-adenylylsulfate (APS) reductase are significantly regulated at the mRNA levels under –S conditions (Smith et al., 1995, 1997; Gutierrez-Marcos et al., 1996; Takahashi et al., 1997, 2000; Leustek et al., 2000; Shibagaki et al., 2002; Yoshimoto el al., 2002). These results suggested that activation of sulfate uptake and the metabolic control of sulfur flux in the reductive assimilation are the two important strategies taken for acclimation to –S. In addition to these primary responsive pathways, differential display demonstrated abundant accumulation of a putative NADPH oxidoreductase in –S-treated maize (Zea mays) seedlings (Petrucco et al., 1996). The function of this protein is postulated to be associated with restoration of NADPH or synthesis of antioxidants, but the exact role remains unclear. More recently, induction of nitrilase under –S suggested interaction of glucosinolate and auxin synthesis in Arabidopsis (Kutz et al., 2002). Under –S conditions, degradation of glucobrassicin, the precursor of indole-3-acetonitrile, is promoted, which necessitates synthesis of indole-3-acetic acid by increased accumulation of nitrilase.
Apparently, sulfur is globally spread in various secondary metabolic pathways, suggesting pleiotropic effects of –S stress on numerous bypassing pathways branching from the primary sulfur metabolism. To elucidate the global effects of sulfur nutrition in plants, we carried out a transcriptome analysis of approximately 8,000 Arabidopsis genes displayed on oligonucleotide chips. We applied a combinatorial transcriptome profiling of sulfur-responsive genes in the sel1 mutant and –S-treated wild-type plants. The sel1 mutant defective in SULTR1;2 sulfate transporter was originally isolated by selenate tolerance (Shibagaki et al., 2002), indicating the role of SULTR1;2 as a major facilitator for the uptake of sulfate and its toxic analog in Arabidopsis roots. The results of transcriptome analysis allowed us to evaluate a close correlation between the effects of genetic dysfunction of SULTR1;2 and –S stress. The present study provided a source of –S-responsive genes related to sulfate uptake, sulfur assimilation, remobilization of secondary sulfur metabolites, and mitigation of oxidative stress, indicating a global effect of sulfur nutrition deficiency on gene expression in higher plants.
RESULTS
Disruption of SULTR1;2 High-Affinity Sulfate Transporter Restricts Sulfate Uptake and Growth
The knockout mutant of the SULTR1;2 sulfate transporter in Arabidopsis was isolated by PCR screening of the pools of T-DNA insertion mutant collections of the University of Wisconsin Biotech Center (http://www.biotech.wisc.edu/Arabidopsis/; Krysan et al., 1999). The isolated mutant was a novel T-DNA insertion allele of the sel1 mutant (Shibagaki et al., 2002) and designated sel1-10 (Fig. 1A). Insertion of T-DNA was present 10 bp upstream of the junction of the ninth exon and intron of SULTR1;2 (Fig. 1A). Reverse transcription (RT)-PCR analysis indicated that expression of SULTR1;2 mRNA is completely eliminated in the mutant (Fig. 2A). The growth of the sel1-10 mutant was significantly retarded compared with the wild type, even though the plants were grown on GM medium (Valvekens et al., 1988) containing adequate sulfur (Fig. 1B). The sulfate content of the mutant was approximately 20% of the wild type both in leaves and roots (Fig. 2B), suggesting that SULTR1;2 transporter functions as a major component of the initial sulfate uptake system in Arabidopsis roots.
Figure 1.
High-affinity sulfate transporter, SULTR1;2, is disrupted by T-DNA insertion in the sel1-10 mutant. A, Location of the T-DNA insertion in SULTR1;2 Thick bars and lines indicate exons and introns, respectively. White bars indicate the 5′- and 3′-untranslated regions. B, Growth of the Ws wild type (upper) and sel1-10 mutant (lower). Plants were grown for 20 d on germination medium (GM) agar medium (Valvekens et al., 1988) in a growth chamber controlled at 22°C under 16-h-light/8-h-dark cycles.
Figure 2.
Determination mRNA and metabolite levels in the sel1-10 mutant. A, RT-PCR analysis of SULTR1;1 SULTR1;2 mRNA was carried out on 12-d-old plants as described in “Materials and Methods.” α-Tubulin (TUB) was used as internal control. B, The abundances of sulfate, Cys, Met, and GSH in the leaf and root tissues were determined by capillary electrophoresis and HPLC as described in “Materials and Methods.” Bars = sds (n= 3). Asterisks indicate statistically significant differences from the wild type (P< 0.05).
In Arabidopsis roots, SULTR1;2 is colocalized with the –S-inducible isoform of sulfate transporter, SULTR1;1, in the epidermis, cortex, and root hairs (Takahashi et al., 2000; Shibagaki et al., 2001; Yoshimoto et al., 2002). The level of SULTR1;1 mRNA was significantly increased in the sel1-10 mutant (Fig. 2A). The F2 plants of the sel1-10/Wassilewskija (Ws) backcross segregated into 1:3 ratio (16:44) for homozygous insertion of T-DNA in SULTR1;2, and the homozygous plants showed significant induction of SULTR1;1 mRNA, indicating that up-regulation of SULTR1;1 mRNA was caused by a single T-DNA insertion in SULTR1;2. The increase of SULTR1;1 mRNA correlated with substantial decrease of the sulfate and GSH contents (Fig. 2B). Decrease of sulfate and GSH contents indicate reduction of sulfate uptake and inefficient synthesis of downstream metabolites in the mutant (Fig. 2B). These results suggest that induction of SULTR1;1 mRNA is closely associated with the shortage of internal pool of sulfur. Furthermore, the results suggested that induced expression of SULTR1;1 mRNA may work compensatory for the acquisition of sulfate but is not fully restoring the sulfate uptake capacity of SULTR1;2 at the root surface.
–S Transcriptome Experiments
The transcript levels of approximately 8,000 Arabidopsis genes were examined using Arabidopsis Genome GeneChip array (Affymetrix, Santa Clara, CA; http://www.affymetrix.com/index.affx). Details of the experimental designs and procedures of chip hybridization are summarized as a web supplemental file (see Supplemental Table I at http://www.plantphysiol.org) compliant with the MIAME checklist format (http://www.mged.org/Workgroups/MIAME/miame.html).
Table I.
Experimental conditions of GeneChip experiments
Sulfate was extracted from roots and quantified by capillary electrophoresis (n = 3). The Columbia ecotype was used for the transfer experiments, HL (high sulfur to low sulfur), LL (low sulfur to low sulfur), HH (high sulfur to high sulfur), and LH (low sulfur to high sulfur). Ws is the background ecotype of the sel1-10 mutant (SULTR/i2 knockout, KO).
| Label | Conditions | SO42- Contents |
|---|---|---|
| nmol mg-1 fresh wt | ||
| HL (wild type) | 1.5 mM Sulfate, 13 d → 0 mM sulfate, 24 h | 2.55 ± 0.18 |
| LL (wild type) | 0.05 mM Sulfate, 13 d → 0.05 mM sulfate, 24 h | 1.18 ± 0.55 |
| KO (sel1-10 mutant) | 1.7 mM Sulfate, 12 d | 1.50 ± 0.15 |
| Ws (wild type) | 1.7 mM Sulfate, 12 d | 7.47 ± 0.36 |
| HH (wild type) | 1.5 mM Sulfate, 13 d → 1.5 mM sulfate, 24 h | 5.11 ± 0.49 |
| LH (wild type) | 0.05 mM Sulfate, 13 d → 1.5 mM sulfate, 24 h | 5.18 ± 1.52 |
Six different sources of plant materials representing the –S or +S conditions were prepared (Table I). For the HL experiment, 13-d-old plants grown on agar medium (Inaba et al., 1994) containing 1.5 mm sulfate were transferred to agar medium containing no sulfate to examine the effect of –S stress in 24 h. For LL experiment, plants grown on agar medium containing 0.05 mm sulfate were transferred to the same low-sulfate media to examine the effect of continuous –S stress. HH represents the control for HL and LL. LH is the experiment for determination of the effect of sulfur repletion (+S) in 24 h. LL was used as a control of LH. The sel1-10 mutant and the wild-type plants were grown on GM medium (Valvekens et al., 1988) containing 1.7 mm sulfate for 12 d and analyzed in parallel. These sets of experiments were validated with the abundances of sulfate in roots. The sulfate contents in the sel1-10 mutant and the –S-treated plants (HL and LL) were significantly lower than those of the wild type or the +S-treated plants (HH and LH; Table I), showing strong correlations between the –S treatment and the mutant. We used Col ecotype for the –S and +S transfer experiments and Ws ecotype for evaluation of sel1-10 mutant. Combinatorial analysis using these two ecotypes enabled us to select –S-responsive genes that are regulated by sulfur nutrition irrespective of the differences in the ecotypes.
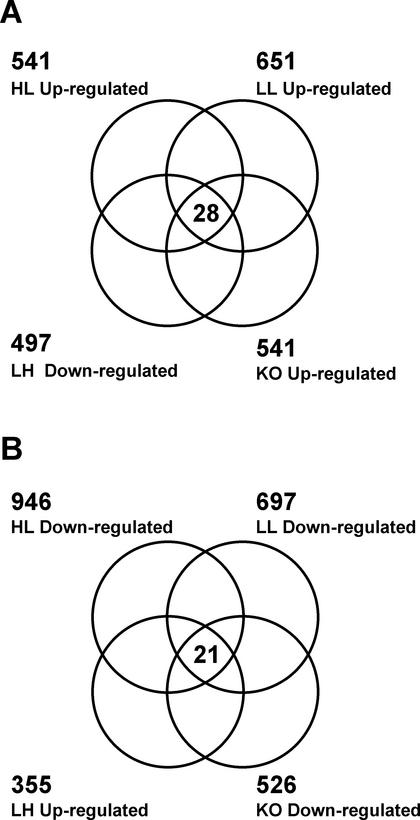
Four different comparisons were made to sort out the candidates of –S-responsive genes on the chip. Signal intensities of each gene were normalized with the median of all measurements on the chip. Fold increase or decrease of the normalized intensities were calculated in the comparisons of HL versus HH, LL versus HH, LH versus LL, and sel1-10 versus Ws. Genes that exhibit greater than 2-fold increase in the comparisons of HL versus HH, LL versus HH, and sel1-10 versus Ws, and genes that exhibit greater than 2-fold decrease in the comparisons LH versus LL were extracted as –S up-regulated genes. The Venn diagrams in Figure 3 indicate the numbers of genes selected from each comparison and the genes present in the intersections of four comparisons. A total of 28 –S up-regulated genes within the intersection show 2-fold increase in all four comparisons (Fig. 3A). Conversely, 21 genes appeared in the intersection of reverse comparisons as –S down-regulated genes (Fig. 3B). Hierarchal clustering of these –S-responsive genes clearly indicated strong correlations between the sel1-10 mutant (KO) and the –S-treated plants (HL and LL; Fig. 4). Table II indicates the fold changes of mRNA levels of selected genes present in the –S up-regulated cluster. The mRNA levels quantified by real-time RT-PCR were generally consistent with the results of the GeneChip experiments. The actual signals obtained from the chip experiments are provided as web supplemental data (see Supplemental Table II at www.plantphysiol.org). Conversion between the Affymetrix probe names and TIGR/MIPS locus names was carried out through the Microarray Element Search of The Arabidopsis Information Resource database (http://www.Arabidopsis.org/tools/bulk/microarray/index.html).
Figure 3.
Venn diagrams of comparisons between –S treatments and the sel1-10 mutant. Sets of experiment are described in Table I. Genes that show more than 2-fold changes were selected from each treatment. A, Intersection of –S up-regulated genes. B, Intersection of –S down-regulated genes.
Figure 4.
Hierarchal clustering of –S-regulated genes. Normalized expression levels of 28 –S up-regulated genes and 21 –S down-regulated genes are presented as relative up-regulation (red) and down-regulation (blue) in six experiments (HL, LL, HH, LH, KO, and Ws) shown in Table I.
Table II.
– S up-regulated genes in roots
Fold changes of mRNA levels were quantified by real-time RT-PCR. The fold change values were derived from the average of triplicate measurements of mRNA levels. Annotation was derived from The Institute for Genomic Research (TIGR; http://www.tigr.org/tdb/e2k1/ath1/) and the Munich Information Center for Protein Sequences (MIPS; http://mips.gsf.de/proj/thal/) databases.
| Locus | Annotation | HL up | LL up | KO up | LH down |
|---|---|---|---|---|---|
| At2g44460 | Putative thioglucosidase | 8.8 × 103 | 5.8 × 104 | 1.2 × 104 | 6.3 × 10-4 |
| At1g23730 | Putative carbonic anhydrase | 67.0 | 313.5 | 51.0 | 0.1 |
| At4g08620 | Sulfate transporter SULTR1;1 | 38.9 | 80.3 | 5.2 | 0.1 |
| At5g10180 | Sulfate transporter SULTR2;1 | 23.1 | 12.6 | 2.0 | 0.1 |
| At4g31330 | Predicted protein | 20.9 | 34.9 | 13.4 | 0.1 |
| At3g60140 | Putative thioglucosidase | 12.6 | 181.1 | 4.6 | 0.1 |
| At1g75280 | Putative NADPH oxidoreductase | 8.6 | 13.5 | 5.0 | 0.2 |
| At2g43500 | Similar to Lotus japonicus nodule inception protein | 7.4 | 74.1 | 1.9 | 0.6 |
| At1g62180 | APS reductase APR2 | 5.1 | 4.1 | 2.7 | 0.3 |
| At4g28110 | myb Family transcription factor MYB41 | 4.3 | 6.9 | 1.3 | 1.1 |
| At4g02710 | Unknown protein | 4.3 | 4.2 | 1.1 | 0.7 |
| At2g34240 | Hypothetical protein (zinc finger motif) | 4.2 | 5.4 | 0.7 | 0.7 |
| At4g04830 | Similar to methionine sulfoxide reductase | 3.5 | 3.3 | 3.3 | 0.1 |
| At1g76680 | 12-Oxophytodienoate reductase (OPR1) | 3.4 | 3.6 | 1.8 | 0.3 |
| At2g22040 | WD-40 repeat protein family | 2.8 | 2.6 | 1.7 | 0.9 |
| At2g28500 | Putative protein | 2.4 | 2.6 | 1.8 | 0.8 |
–S Up-Regulated Genes in Roots
The profiles of –S up-regulated genes indicated recruitment of genes for sulfate uptake and assimilation (Fig. 4; Table II). It is suggested that induced expression of SULTR1;1 (At4g08620) and SULTR2;1 (At5g10180) may enhance the sulfate uptake capacities of root epidermis and pericycle cells, respectively (Takahashi et al., 2000). APS reductase is the key pathway enzyme of reductive sulfate assimilation that converts APS to sulfite (Leustek et al., 2000). Up-regulation of one of the APS reductase isoforms, APR2 (At1g62180), suggests that activation of this key metabolic pathway facilitates efficient synthesis of Cys under –S. Furthermore, two thioglucosidase-like genes (At2g44460 and At3g60140) were present in the cluster, suggesting a rapid turnover of glucosinolate under –S. The results suggest that glucosinolates accumulated in root tissues may serve as alternative source for sulfur assimilation. Sulfate spontaneously released from the aglycon of glucosinolate is recycled after degradation by thioglucosidase (Wittstock and Halkier, 2002).
We identified a set of genes related to the oxidative stress response (Fig. 4; Table II). It is suggested that plants may suffer from oxidative stress during shortage of GSH (Noctor and Foyer, 1998; Asada, 1999). Putative Met sulfoxide reductase (At4g04830) present in the cluster of –S up-regulated genes is suggested to repair oxidative damage of proteins (Gustavsson et al., 2002). Concomitantly, a heat shock protein, HSP21 (At4g27670), the potential target substrate of Met sulfoxide reductase, was abundantly expressed under –S. Putative NADPH oxidoreductase (At1g75280) is the closest homolog of the –S-inducible isoflavone reductase-like gene in maize (Petrucco et al., 1996). The exact function of this putative oxidoreductase is yet to be characterized, though it is suggested to play a role in preservation of reductants or synthesis of antioxidants. In addition, we found significant overexpression of carbonic anhydrase (At1g23730) under –S. Among the six isoforms of carbonic anhydrase in Arabidopsis, probes for five isoforms were present on the chip, and At1g23730 was the only one that showed significant response to –S. This carbonic anhydrase may have a specific function to alleviate oxidative stress by fueling the respiratory cycle. Synthesis of jasmonate was up-regulated corollary of oxidative stress triggered by shortage of GSH. The expression of 12-oxophytodienoate reductase (OPR1; At1g76680), the enzyme of the jasmonate synthetic pathway, was increased by –S transfer and in the sel1-10 mutant. Presence of a putative disease resistance protein (At3g46730) may implicate activation of downstream defense response through the jasmonate signaling pathway (Creelman and Mullet, 1997; Turner et al., 2002).
–S Down-Regulated Genes in Roots
As mentioned above, we have identified up-regulation of two thioglucosidase genes that potentially serve for degradation of glucosinolates. Within the list of –S down-regulated genes (Fig. 4), we found CYP79B3 P450 (At2g22330) that catalyzes N-hydoxylation of Trp at the first step of indole glucosinolate synthesis (Wittstock and Halkier, 2002). It is suggested that both the synthesis and degradation of glucosinolate are regulated under –S. Our results further suggested that modification of metabolic fluxes in other secondary sulfur metabolisms can also minimize consumption of essential sulfur source for primary assimilation. Appearance of steroid sulfotransferase (At4g26280) suggests repression of sulfation (Rouleau et al., 1999). The synthesis of thiamine was apparently affected by –S as indicated by down-regulation of the putative protein (At4g34020) similar to bacterial 4-methyl-5(β-hydroxymethyl)-thiazole kinase (Mizote et al., 1999). Down-regulation of metallothionein-like protein (At2g23240) may indicate recycling of Cys under –S.
–S Up-Regulated Genes in Leaves
Dysfunction of SULTR1;2 in the sel1-10 mutant displayed a clear view of oxidative stress response in leaf tissues. We carried out GeneChip hybridizations and selected up-regulated genes that exhibit greater than 3-fold abundant expression in the sel1-10 mutant than in the wild type. Only the “present” genes in the absolute call of the baseline data have been selected for this one-to-one comparison. A total of 13 genes showed greater than 3-fold abundant expression in the mutant than in the wild type on chip experiments. Quantification of mRNA levels by real-time RT-PCR revealed that 11 genes presented in Table III show more than 3-fold abundant accumulation of mRNA in the sel1-10 mutant.
Table III.
– S up-regulated genes in the leaves of sel1-10 mutant
Fold-increase of mRNA levels were quantified by real-time RT-PCR. The fold change values were derived from the average of triplicate measurements of mRNA levels. Annotation was derived from TIGR (http://www.tigr.org/tdb/e2k1/ath1/) and MIPS (http://mips.gsf.de/proj/thal/) databases.
| Locus | Annotation | KO up |
|---|---|---|
| At3g45140 | Lipoxygenase AtLOX2 | 29.9 |
| At4g12480 | pEARLI1 | 15.1 |
| At5g24770 | Vegetative storage protein (Vsp2) | 12.2 |
| At1g19670 | Coronatine-induced protein 1 | 7.6 |
| At4g23600 | Tyrosine transaminase-like protein | 6.3 |
| At4g08870 | Putative arginase | 6.1 |
| At1g72930 | Disease resistance RPP5 like protein | 5.5 |
| At5g42650 | Allene oxide synthase | 4.2 |
| At2g14610 | PR-1-like protein | 3.7 |
| At5g64120 | Peroxidase | 3.6 |
| At4g30170 | Peroxidase ATP8a | 3.2 |
In the sel1-10 mutant, we found abundant accumulation of lipoxygenase AtLOX2 (At3g45140) that initiates oxidative modification of membrane lipids for synthesis of jasmonate (Creelman and Mullet, 1997; Turner et al., 2002). The expression of allene oxide synthase (At5g42650), the key enzyme of jasmonate synthesis, was also significantly increased in the mutant. Furthermore, induction of coronatine-induced protein 1 (At1g19670) and vegetative storage protein Vsp2 (At5g24770) suggests activation of the jasmonate signaling cascade. Tyr transaminase (At4g23600) presented here has been identified as a coronatine-induced protein and is responsive to jasmonate (Lopukhina et al., 2001). It is suggested that 4-hydroxyphenyl-pyruvate, the product of Tyr transaminase, is utilized for the synthesis of antioxidants, α-tocopherol, and plastoquinone. Alternatively, the phenolic compounds synthesized from 4-hydroxyphenyl-pyruvate may serve for the cross-linking of cell walls upon pathogen attack. In addition, we identified up-regulation of the disease resistance protein (At1g72930), the PR-1 like protein (At2g14610), and peroxidase (At5g64120, At4g30170), suggesting activation of defense response through oxidative stress.
In addition to the jasmonate response, we identified significant up-regulation of pEARLI1 (At4g12480) in the leaves of the mutant (Table III). The pEARLI1 protein contains a protease inhibitor/seed storage/lipid transfer protein family signature and is induced in response to aluminum stress (Richards and Gardner, 1995). The observed induction of pEARLI1 suggests oxidative stress as a common factor between –S and aluminum toxicity that triggers gene expression. A putative arginase (At4g08870) was abundantly expressed in the leaf of the sel1-10 mutant. Arabidopsis lacks Orn decarboxylase; hence, polyamines are exclusively synthesized from Arg (Hanfrey et al., 2001). Degradation of Arg may reduce accumulation of polyamines. This in turn may prevent generation of peroxides from the catabolic diamine oxidation of polyamines (Tiburcio et al., 1997).
DISCUSSION
The analysis of the sel1-10 mutant in the present study revealed that dysfunction of SULTR1;2 significantly restricts the uptake of sulfate and plant growth (Fig. 1). The sel1-10 mutant accumulated SULTR1;1 mRNA (Fig. 2A), suggesting a compensatory regulation of this –S-inducible isoform. SULTR1;1 accumulated in the mutant irrespective of the external sulfur conditions, indicating that SULTR1;1 mRNA is induced in response to the internal sulfur status. Disruption of SULTR1;2 allowed full induction of SULTR1;1 mRNA; however, the growth was not completely recovered, and the sulfate content in the sel1-10 mutant reached only 10% to 20% of the wild type. Shortage of sulfur was generally reflected on the abundances of Cys, Met, and GSH. It is suggested that growth defect of the mutant is caused by insufficient synthesis of sulfur metabolites.
Sulfur-containing metabolites have various functions in plants (Leustek and Saito, 1999). Cys and Met synthesized from the reductive sulfur assimilation pathway are the essential constituents of proteins. Cys is incorporated into GSH that is one of the major redox controllers and plays significant roles in scavenging reactive oxygen species through the GSH-ascorbate cycle (Noctor and Foyer, 1998; Asada,1999). Flavonoids, steroids, polysaccharides, and lipids are modified by sulfation and sulfonation (Varin et al., 1997; Benning, 1998). These reactions provide an array of secondary sulfur metabolites in plants. During –S, plants activate the uptake of sulfate to acquire sulfur source for Cys synthesis. Reductive assimilation pathway can be up-regulated for efficient utilization of sulfur. Under such conditions, the synthesis of secondary sulfur metabolites may be halted and remobilized to obtain sustainable amounts of sulfur for primary assimilation. In the present study, we successfully demonstrated the metabolic response of Arabidopsis to –S stress from the transcriptome profiling of the sel1-10 mutant and –S-treated plants.
Up-regulation of sulfate transporters and APS reductase is assumed to be the most important step that may enhance the capacities of primary sulfate uptake and reductive sulfate assimilation under –S (Takahashi et al., 1997, 2000; Leustek and Saito, 1999; Leustek et al., 2000). The –S up-regulated genes included sulfate transporters: SULTR1;1 (At4g08620) and SULTR2;1 (At5g10180; Fig. 4; Table II). These transporters facilitate sulfate uptake in root epidermis and pericycle cells, respectively (Takahashi et al., 2000). The APR2 isoform of APS reductase (At1g62180) was abundantly expressed in –S-treated plants and in the sel1-10 mutant. The activity of APS reductase, particularly the APR1 isoform, is regulated by the cellular redox status (Bick et al., 2001). It is suggested that both the induction of APR mRNA accumulation and APR1 enzyme activities contribute to the sulfate assimilation under –S. We identified up-regulation of two thioglucosidase genes (At2g44460 and At3g60140) and down-regulation of CYP79B3 N-hydroxylase (At2g22330) that catalyzes the first step of indole glucosinolate biosynthesis. This clearly suggests remobilization of glucosinolates under –S. Overexpression of thioglucosidase may force degradation of glucosinolates and generates unstable aglycons spontaneously releasing sulfate (Wittstock and Halkier, 2002). Plants may pull out sustainable amount of sulfate from this catabolic cycle of glucosinolates to undergo shortage of sulfur. Degradation of indole glucosinolates in this reaction generates indole-3-acetonitrile, the precursor of indole-3-acetic acid. Induction of NIT3 nitrilase under –S (Kutz et al., 2002) considerably participates in converting indole-3-acetonitrile to auxin. Our data further demonstrated a metabolic shift of sulfur source from other secondary sulfur metabolisms. Down-regulation of sulfation and thiamine synthesis was apparent under –S. Steroid sulfotransferase (At4g26280) and 4-methyl-5(β-hydroxymethyl)-thiazole kinase-like protein (At4g34020) were less abundant in the sel1-10 mutant and –S-treated plants. Down-regulation of the metallothionein-like protein (At2g23240) presumably prevents the loss of Cys.
Under –S conditions, plants may undergo oxidative stress caused by the shortage of GSH (Noctor and Foyer, 1998; Asada, 1999). Induction of oxidative stress in –S plants was clearly demonstrated (Fig. 4; Tables II and III). The data presented here suggest activation of jasmonate-mediated metabolic processes and related responses (Creelman and Mullet, 1997; Turner et al., 2002). The overaccumulation of AtLOX2 lipoxygenase (At3g45140), 12-oxophytodienoate reductase (At1g76680), and allene oxide synthase (At5g42650) indicates that jasmonate synthesis is significantly induced by –S. Induction of vegetative storage protein Vsp2 (At5g24770), coronatine-induced protein 1 (At1g19670), and Tyr transaminase (At4g23600; Table III) suggests jasmonate response. 4-Hydroxyphenyl-pyruvate generated by Tyr transaminase is converted to antioxidants, α-tocopherol, and plastoquinone, serving for mitigation of oxidative stress (Lopukhina et al., 2001). An alternative contribution of this enzyme is strengthening of the cell walls upon pathogen attack. It is suggested that phenolic compounds synthesized from 4-hydroxyphenyl-pyruvate maintain rigidity of cell walls by cross-linking. Furthermore, induction of putative disease resistance proteins (At3g46730 and At1g72930), PR-1-like protein (At2g14610), and peroxidase (At5g64120 and At4g30170) suggests activation of downstream defense response through the jasmonate signaling pathway.
We identified genes that potentially detoxify or alleviate the function of reactive oxygen species (Fig. 4; Tables II and III). A putative NADPH oxidoreductase (At1g75280) is the known –S-inducible gene orthologous to the isoflavone reductase-like gene in maize. The exact role of this putative oxidoreductase is unclear but is expected to function for the synthesis of antioxidants based on the sequence similarities to isoflavone reductase (Petrucco et al., 1996). Alternatively, this protein may play a role in restoration of NADPH under oxidative conditions. Induction of putative Met sulfoxide reductase (At4g04830) may protect and repair oxidative deformation of HSP21 (At4g27670), thereby maintaining the chaperone-like function of HSP21 (Gustavsson et al., 2002). Up-regulation of a putative arginase (At4g08870) presumably decreases production of peroxides from polyamines (Tiburcio et al., 1997). Arg is the initial substrate of polyamine synthesis in Arabidopsis (Hanfrey et al., 2001). Expression of arginase may reduce accumulation and subsequent catabolic degradation of polyamines, preventing generation of an unnecessary amount of peroxides. In addition, we found significant overexpression of carbonic anhydrase (At1g23730) in –S-treated roots. Increased expression of carbonic anhydrase provides bicarbonate to phosphoenolpyruvate carboxylase, which may allow up-regulation of synthesis of organic acids and the respiratory cycle. This also may prevent production of reactive oxygen species. pEARLI1, identified in the leaf of sel1-10 mutant, is an aluminum stress-inducible gene (Richards and Gardner, 1995). Coincidental up-regulation of pEARLI1 in –S plants suggests oxidative stress response of this unknown gene.
We identified numbers of putative transcription factors and proteins that participate in signaling cascades of stress response (Fig. 4; Table II). Interestingly, a nodule inception (NIN) protein homolog (At2g43500) was present in the cluster of –S up-regulated genes. The NIN protein is suggested to function as a transcription regulator during nodule development in L. japonicus (Schauser et al., 1999). Arabidopsis has several NIN homologs, suggesting conservation of this protein family in non-nodulating plant species; however, their functions are yet characterized. The –S inducibility of the Arabidopsis NIN homolog may suggest relevance of this putative regulator to sulfur nutrient acquisition. The functions of putative regulatory proteins identified in this study may include both the sulfur-specific response and the secondary effects caused by the shortage of sulfur. Our present results provided a set of candidates of regulatory genes that should be further elaborated to define their roles in sulfur assimilation.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis plants were cultured in a growth chamber controlled at 22°C under 16-h-light/8-h-dark cycles. The light intensity was 40 μmol m–2 s–1. The sel1-10 mutant and the background Ws wild-type plants were grown on GM agar medium (Valvekens et al., 1988) containing 1.7 mm sulfate as Murashige and Skoog salts (Murashige and Skoog, 1962). The Columbia ecotype was used for –S and +S transfer experiments. For the transfer experiments, MgSO4 was replaced with MgCl2 to control the sulfate concentration in the agar medium (Inaba et al., 1994).
The T-DNA insertion mutant (sel1-10) was isolated from 60,480-mutant population of the University of Wisconsin (http://www.biotech.wisc.edu/Arabidopsis/default.htm). PCR screening (Krysan et al., 1999) was carried out using oligonucleotide primers: 1;2-F (5′-ACGGTGGACATGTTCCGATGAAACCTTCA-3′) and 1;2-R (5′-TGCGACAAGTGTAGCTTGCCTATCACCAA-3′). Primers were designed according to the sequence of the Arabidopsis genome (Arabidopsis Genome Initiative, 2000). The integration site of the T-DNA was determined by sequencing PCR fragments amplified with the SULTR1;2-specific primers and T-DNA border primers. The mutant was backcrossed to Ws, and segregation of T-DNA insertion in F2 generation was analyzed by PCR using specific primers described above. A total of 60 F2 plants segregated into 3:1 ratio (45:15) for kanamycin resistance and 16 plants showed homozygous insertion of T-DNA in SULTR1;2, indicating a single insertion of T-DNA in SULTR1;2 in the parental mutant. Single insertion of T-DNA was further confirmed by Southern hybridization.
RT-PCR
Molecular biological experiments were carried out according to the standard protocols (Sambrook et al., 1989). Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). RT was carried out using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) as described previously (Yoshimoto et al., 2002). RT-PCR of SULTR1;1 and SULTR1;2 mRNAs in the sel1-10 mutant was carried out as described previously (Yoshimoto et al., 2002). α-Tubulin (Ludwig et al., 1987) was used as a constitutive control. PCR products were separated in agarose gels and stained with SYBR green (Takara, Tokyo). Signals were detected and quantified using FluorImager 595 (Molecular Dynamics, Sunnyvale, CA) with a 515- to 545-nm band-pass filter. PCR was carried out for 24 cycles where cDNAs were exponentially amplified using ExTaq DNA polymerase (Takara).
For real-time RT-PCR, total RNA was treated with DNase I (Invitrogen) and reverse transcribed with SuperScript II reverse transcriptase (Invitrogen). Specific primers for the –S up-regulated genes were designed using Primer Express (Applied Biosystems, Foster City, CA). Amplification was carried out using SYBR green PCR master mix and detected with the GeneAmp 5700 system (Applied Biosystems). Triplicate measurements were carried out to determine the mRNA abundance of each gene in each sample. The relative increase or decrease of mRNA abundance between the two samples was determined by comparing the threshold cycle values. Equality of RNA preparation was confirmed by constitutive expression of ubiquitin (UBQ2, accession no. J05508).
GeneChip Hybridization and Data Analysis
Total RNA was extracted using RNeasy Plant Mini Kit (Qiagen). Preparation of labeled target cRNA was carried out following the technical manual of Arabidopsis Genome GeneChip array (Affymetrix). Double-stranded cDNA was prepared from 20 μg of total RNA using SuperScript Choice System (Invitrogen). The resultant cDNA was transcribed in vitro using BioArray High Yield RNA Transcript Kit (Enzo, New York). After purification and fragmentation, the labeled cRNA was hybridized to Arabidopsis Genome GeneChip array (Affymetrix) in a Hybridization Oven model 640 (Affymetrix). Washing and staining of chips were carried out using GeneChip Fluidics Station model 400. Scanning was carried out with the Gene Array Scanner (Agilent Technologies, Palo Alto, CA). The Microarray Suite 5.0 (Affymetrix) and Genespring 4.2 (Silicon Genetics, Redwood City, CA) were used for data analysis. Raw signals of each gene were normalized with the median of all measurements on the chip. Fold changes of signal intensities were calculated from the normalized data. In the case of root RNA samples, genes with greater than 2-fold changes in four different sets of comparisons (Fig. 3) were pulled out for further analysis. A total of 28 for the –S up-regulated genes and 21 for the –S down-regulated genes, respectively, passed the selection as described in the results (Figs. 3 and 4). For one-to-one comparison of leaf RNA samples of sel1-10 mutant and wild type, we first carried out selection of genes with “present” values in the absolute call of the baseline data. Among them, a total of 13 genes showed greater than 3-fold abundant expression in the mutant. The mRNA levels were quantified by real-time RT-PCR, and 11 genes showed greater than 3-fold abundant expression in the mutant as described in the results (Table III).
Quantification of Sulfate, Thiols, and Met Contents
Plants tissues were ground in liquid nitrogen and extracted with 10 mm HCl. Crude extracts were centrifuged at 10,000g, and the supernatants were filtered through Ultrafree-MC 5000 NMWL Filter Unit (Millipore, Bedford, MA). Quantification of sulfate was carried out with the HP30 Capillary Electrophoresis system using inorganic anion buffer (Agilent Technologies). Thiols in the extract were reduced by dithiothreitol and labeled with mono-bromobimane (Molecular Probes, Eugene, OR). Bimane adducts of Cys and GSH were separated with a 4.6× 150-mm Symmetry C18 column (Waters, Milford, MA) on a Waters 2695 HPLC system and detected by Scanning Fluorescence Detector 474 (Waters). Met was derivatized by AccQ Fluor Reagent (Waters) and analyzed with a 4.6-× 150-mm AccQ Tag column (Waters) following the manufacturer's instructions.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center and the Arabidopsis Knockout Facility of University of Wisconsin Biotech Center for providing the pools of T-DNA insertion mutants. We are grateful to all colleagues in the laboratory for valuable suggestions and discussions.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.019802.
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan.
The online version of this article contains Web-only data. The supplemental material is available at http://www.plantphysiol.org.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Asada K (1999) The water-water cycle in chloroplasts: scavenging of active oxygen and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50: 601–639 [DOI] [PubMed] [Google Scholar]
- Benning C (1998) Biosynthesis and function of the sulfolipid sulfoquinovosyl diacylglycerol. Annu Rev Plant Physiol Plant Mol Biol 49: 53–75 [DOI] [PubMed] [Google Scholar]
- Bick JA, Setterdahl AT, Knaff DB, Chen Y, Pitcher LH, Zilinskas BA, Leustek T (2001) Regulation of the plant-type 5′-adenylyl sulfate reductase by oxidative stress. Biochemistry 40: 9040–9048 [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE (1997) Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 48: 355–381 [DOI] [PubMed] [Google Scholar]
- Grossman A, Takahashi H (2001) Macronutrient utilization by photosynthetic eukaryotes and the fabric of interactions. Annu Rev Plant Physiol Plant Mol Biol 52: 163–210 [DOI] [PubMed] [Google Scholar]
- Gustavsson N, Kokke BP, Harndahl U, Silow M, Bechtold U, Poghosyan Z, Murphy D, Boelens WC, Sundby C (2002) A peptide methionine sulfoxide reductase highly expressed in photosynthetic tissue in Arabidopsis thaliana can protect the chaperone-like activity of a chloroplast-localized small heat shock protein. Plant J 29: 545–553 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Roberts MA, Campbell EI, Wray JL (1996) Three members of a novel small gene-family from Arabidopsis thaliana able to complement functionally an Escherichia coli mutant defective in PAPS reductase activity encode proteins with a thioredoxin-like domain and “APS reductase” activity. Proc Natl Acad Sci USA 93: 13377–13382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanfrey C, Sommer S, Mayer MJ, Burtin D, Michael AJ (2001) Arabidopsis polyamine biosynthesis: absence of ornithine decarboxylase and the mechanism of arginine decarboxylase activity. Plant J 27: 551–560 [DOI] [PubMed] [Google Scholar]
- Inaba K, Fujiwara T, Hayashi H, Chino M, Komeda Y, Naito S (1994) Isolation of an Arabidopsis thaliana mutant, mto1, that overaccumulates soluble methionine: temporal and spatial patterns of soluble methionine accumulation. Plant Physiol 104: 881–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutz A, Muller A, Hennig P, Kaiser WM, Piotrowski M, Weiler EW (2002) A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant J 30: 95–106 [DOI] [PubMed] [Google Scholar]
- Leustek T, Martin MN, Bick J, Davies JP (2000) Pathways and regulation of sulfur metabolism revealed through molecular genetic studies. Annu Rev Plant Physiol Plant Mol Biol 51: 141–166 [DOI] [PubMed] [Google Scholar]
- Leustek T, Saito K (1999) Sulfate transport and assimilation in plants. Plant Physiol 120: 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopukhina A, Dettenberg M, Weiler EW, Hollander-Czytko H (2001) Cloning and characterization of a coronatine-regulated tyrosine amino-transferase from Arabidopsis. Plant Physiol 126: 1678–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig SR, Oppenheimer DG, Silflow CD, Snustad DP (1987) Characterization of the α-tubulin gene family of Arabidopsis thaliana. Proc Natl Acad Sci USA 84: 5833–5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizote T, Tsuda M, Smith DD, Nakayama H, Nakazawa T (1999) Cloning and characterization of the thiD/J gene of Escherichia coli encoding a thiamin-synthesizing bifunctional enzyme, hydroxymethylpyrimidine kinase/phosphomethylpyrimidine kinase. Microbiology 145: 495–501 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Noctor G, Foyer C (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49: 249–279 [DOI] [PubMed] [Google Scholar]
- Petrucco S, Bolchi A, Foroni C, Percudani R, Rossi GL, Ottonello S (1996) A maize gene encoding an NADPH binding enzyme highly homologous to isoflavone reductases is activated in response to sulfur starvation. Plant Cell 8: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KD, Gardner RC (1995) pEARLI 1 (accession no. L43080): an Arabidopsis member of a conserved gene family (PGR95-099). Plant Physiol 109: 14978539300 [Google Scholar]
- Rouleau M, Marsolais F, Richard M, Nicolle L, Voigt B, Adam G, Varin L (1999) Inactivation of brassinosteroid biological activity by a salicylate-inducible steroid sulfotransferase from Brassica napus. J Biol Chem 274: 20925–20930 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning, A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Shibagaki N, Rose A, McDermott JP, Fujiwara T, Hayashi H, Yoneyama T, Davies JP (2002) Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J 29: 475–486 [DOI] [PubMed] [Google Scholar]
- Smith FW, Ealing PM, Hawkesford MJ, Clarkson DT (1995) Plant members of a family of sulfate transporters reveal functional subtypes. Proc Natl Acad Sci USA 92: 9373–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FW, Hawkesford MJ, Ealing PM, Clarkson DT, Vanden Berg PJ, Belcher AR, Warrilow AGS (1997) Regulation of expression of a cDNA from barley roots encoding a high affinity sulfate transporter. Plant J 12: 875–884 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K (2000) The role of three functional sulfate transporters involved in uptake and translocation of sulfate in Arabidopsis thaliana. Plant J 23: 171–182 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Yamazaki M, Sasakura N, Watanabe A, Leustek T, Engler JA, Engler G, Van Montagu M, Saito K (1997) Regulation of sulfur assimilation in higher plants: a sulfate transporter induced in sulfate-starved roots plays a central role in Arabidopsis thaliana. Proc Natl Acad Sci USA 94: 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiburcio AF, Altabella T, Borrell A, Masgrau C (1997) Polyamine metabolism and its regulation. Physiol Plant 100: 664–674 [Google Scholar]
- Turner JG, Ellis C, Devoto A (2002) The jasmonate signal pathway. Plant Cell 14: S153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M (1988) Agrobacterium-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85: 5536–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varin L, Marsolais F, Richard M, Rouleau M (1997) Sulfation and sulfotransferases: VI. Biochemistry and molecular biology of plant sulfotransferases. FASEB J 11: 517–525 [DOI] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7: 263–270 [DOI] [PubMed] [Google Scholar]
- Yoshimoto N, Takahashi H, Smith FW, Yamaya T, Saito K (2002) Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J 29: 465–473 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.