Abstract
The screening for mutants and their subsequent molecular analysis has permitted the identification of a number of genes of Arabidopsis involved in the development and functions of the gynoecium. However, these processes remain far from completely understood. It is clear that in many cases, genetic redundancy and other factors can limit the efficiency of classical mutant screening. We have taken the alternative approach of a reverse genetic analysis of gene function in the Arabidopsis gynoecium. A high-throughput fluorescent differential display screen performed between two Arabidopsis floral homeotic mutants has permitted the identification of a number of genes that are specifically or preferentially expressed in the gynoecium. Here, we present the results of this screen and a detailed characterization of the expression profiles of the genes identified. Our expression analysis makes novel use of several Arabidopsis floral homeotic mutants to provide floral organ-specific gene expression profiles. The results of these studies permit the efficient targeting of effort into a functional analysis of gynoecium-expressed genes.
The gynoecium is the fourth and innermost whorl of a typical bisexual flower. It is composed of the female reproductive organs, or carpels, and encloses the ovules, which develop into seeds after fertilization. The gynoecium may be composed of simple, unfused carpels, although in most species it is syncarpic, i.e. composed of several carpels fused together. The gynoecium functions to protect the ovules and to allow the operation of pollen-pistil incompatibility mechanisms. After fertilization, it develops into a fruit that participates in seed dissemination.
In Arabidopsis, the gynoecium is a complex syncarpic structure. This first develops as an open-ended tube from a primordial dome in the center of the floral meristem. A vertical septum then forms internally from either side of the gynoecial tube, and the two halves of this septum fuse to divide the structure into two loculi. Placental tissues develop in the zones where the vertical septum and gynoecial wall meet to generate two rows of ovule primordia within each loculus. Each ovule consists of a seven-celled embryo sac of the Polygonum type (Fahn, 1975), together with a small nucellus and two covering integuments. Cell division occurring at the distal end of the gynoecial cylinder forms the style and stigma tissues. The stigma consists of a pappillate epidermal cell layer with a modified external wall and cuticle. This tissue receives and permits the germination of compatible pollen grains. After the penetration of the stigma by pollen tubes, a transmitting tissue in the style and vertical septum functions to guide the pollen tubes toward the ovules where fertilization takes place. After fertilization, the Arabidopsis gynoecium develops into a two-chambered, capsular fruit, termed a silique. This structure opens at maturity to release its seeds by rupture along four zones of dehiscence in the silique wall situated on either side of the vertical septum. Detailed descriptions of gynoecium development in Arabidopsis are given by Bowman (1994) and Sessions (1997).
Relatively few genes have so far been identified that play important roles in the functional processes of the Arabidopsis gynoecium such as pollen reception, pollen tube guidance, and fertilization (for review, see Wilhelmi and Preuss, 1999; Faure and Dumas, 2001). By contrast, mutant screening and subsequent molecular analysis has been very successful in the identification of genes that control gynoecium development (for review, see Bowman et al., 1999, 2001; Ferrandiz et al., 1999) and ovule development (for review, see Schneitz et al., 1998). Despite the success of these mutagenesis-based approaches, some of the genes now known to influence gynoecium development had to be identified by the alternative reverse genetic approach. In these cases, genetic redundancy between similar genes led to a lack of mutant phenotypes in single-mutant plants. Examples of such genetic redundancy are to be found in two groups of MADS box genes controlling flower development: the genes SHATTERPROOF1 and -2, which are necessary for dehiscence zone development (Liljegren et al., 2000), and the genes SEPALLATA1, -2, and -3, which are necessary for the specification of floral organ identity (Pelaz et al., 2000; Honma and Goto, 2001).
To identify further genes that play important roles in the development and functions of the gynoecium, we have taken a reverse genetic approach by firstly performing a differential expression screen between two floral homeotic mutants to identify genes that are up-regulated in the Arabidopsis gynoecium. The availability of large collections of T-DNA and transposon insertion lines (Bouché and Bouchez, 2001), together with efficient techniques for the suppression of gene activity based on posttranscriptional gene silencing (PTGS; Wesley et al., 2001), then makes possible the reverse genetic analysis of the genes of interest and of closely related sequences with which these may show redundant interactions. Here, we present the first stage in a reverse genetic analysis: the results of a differential screen to identify novel genes that are up-regulated in the Arabidopsis gynoecium. We describe in detail the homologies and expression patterns of the genes identified, and for certain identified genes, we discuss possible functions.
RESULTS
The Identification of Genes Up-Regulated in the Arabidopsis Gynoecium by Fluorescent Differential Display (FDD) Analysis
To identify genes that were specifically upregulated in tissues of the gynoecium, we performed a differential screen of gene expression between the inflorescences of two floral homeotic mutants of Arabidopsis, pistillata-1 (pi-1) and agamous-1 (ag-1). Flowers of the pi-1 mutant are composed of carpel and sepal organs, whereas those of the ag-1 mutant are composed of sepals and petals. Genes specifically up-regulated in pi-1 inflorescences were, therefore, expected to be also up-regulated in the wild-type (wt) Arabidopsis gynoecium. Our studies focused mainly on early flower developmental stages to identify genes involved in early gynoecium development. A total of 360 PCR primer combinations were used to amplify an estimated 18,000 reverse transcriptase (RT)-PCR products from inflorescences of pi-1 and ag-1 mutants (data not shown), which included flower buds at up to stage 10 of flower development (Bowman, 1994). Key events in the gynoecium at stage 10, which precedes female meiosis, include the elongation of the developing ovules, the closure of the end of the gynoecial cylinder, and the fusion of the two halves of the vertical septum (Bowman, 1994). From the PCR products generated from early flower bud developmental stages, 20 pi-1-up-regulated bands were observed on FDD gels (approximately 0.06 RTPCR products per primer combination). Of these, 17 RT-PCR products were successfully purified and cloned in plasmid cloning vectors. In addition, a second FDD analysis on ag-1 and pi-1 material including later bud stages was performed. For this analysis, 60 PCR primer combinations were used together with RNA samples derived from inflorescences containing all flower bud developmental stages up to the mature flower stage 13 (Bowman, 1994). From these later samples, gene expression differences between pi-1 and ag-1 mutants were much more numerous than at early bud developmental stages. Forty-five clearly pi-1-upregulated bands (approximately 0.75 RT-PCR products per primer combination) were observed on FDD gels using these samples. Of these, 12 pi-1-upregulated bands were cloned in plasmid vectors for further analysis.
Sequencing of the total number of 29 RT-PCR products cloned after FDD analysis demonstrated the presence of 22 distinct sequences, the other seven having been amplified by more than one combination of PCR primer. One of these 22 unique PCR products proved, by searching of the complete Arabidopsis genome sequence database and by Southern blotting (data not shown), to show no homology to Arabidopsis DNA. Two others were derived from the Arabidopsis chloroplast genome and a further two represented nuclear rRNA genes. The remaining 18 PCR products represented the 3′ ends of known or predicted protein-encoding genes from the Arabidopsis nuclear genome. Further analysis of four of these by northern blotting (data not shown) failed to provide clear evidence of up-regulation in pi-1 over ag-1 mutant inflorescences. The remaining 14 PCR products represent partial cDNA sequences that were confirmed by subsequent analyses to be up-regulated in inflorescences of the pi-1 mutant. These cDNAs were termed Pup (for pistillata-up-regulated) 1 to 14 and are described in Table I, which includes details of the stages of development from which they were identified during FDD analysis. For each of the 14 Pup cDNAs, full-length gene-coding regions predicted from the complete Arabidopsis genome sequence were amplified using RT-PCR from wt inflorescence RNA. These putatively full-length cDNAs were cloned and partially sequenced to demonstrate their authenticity. In several cases, where full-length cDNAs had not previously been characterized, sequencing of the full-length amplified cDNAs was performed. These novel full-length cDNA sequence data have been deposited with the EMBL database (Table I). The homologies shown by the genes identified in this study are discussed in detail below, together with descriptions of their expression profiles.
Table I.
Details of the cDNA sequences Pup1-14, identified as up-regulated in the Arabidopsis gynoecium, and of their associated genes Details of the full-length cDNAs referred to can be found at http://signal.salk.edu/SSP/index.html.
| cDNA | Published Name | Unique Gene Identifier No. | Homology or Classification | Full-Length cDNA | Stages of Development Used in FDD Analysis | Main Tissues of Expression in Flowers and Siliques |
|---|---|---|---|---|---|---|
| Pup1 | ARGONAUTE9 (AGO9) | At5g21150 | ARGONAUTE family genes | EMBL accession no. AJ544236 | Before stage 11 | Ovules and anther sporogenous tissues |
| Pup2 | At5g53870 | Phytocyanins | EMBL accession no. AJ544237 | All stages | Embryo sac | |
| Pup3 | At2g34870 | Pro-rich proteins | Ceres 29605 | All stages | Epidermis of vertical septum | |
| Pup4 | At5g24420 | Phosphogluconolactonases | Ceres 13806 | Before stage 11 | Ovary and ovules | |
| Pup5 | At1g72290 | Kunitz protease inhibitors | Ceres 106020 | All stages | Transmitting tissue | |
| Pup6 | At5g14700 | Cinnamyl coA reductases | Ceres 17229 | Before stage 11 | Seed integuments | |
| Pup7 | Thi2.1 | At1g72260 | Thionins | Ceres 26029 | All stages | Integuments of ovules and seeds |
| Pup8 | At5g23960 | δ-Cadinene synthases | EMBL accession no. AJ544238 | All stages | Mesocarp of silique wall | |
| Pup9 | VSP1 | At5g24780 | Vegetative storage proteins | Ceres 32606 | Before stage 11 | Ovary wall |
| Pup10 | MBP2 | At1g52030 | Myrosinase-binding proteins and other lectins | SSP R12767 | Before stage 11 | Inner epidermis of ovary; ovule integuments; tapetum |
| Pup11 | At3g16470 | Myrosinase-binding proteins and other lectins | Ceres 30003 | Before stage 11 | Inner epidermis of ovary; ovule integuments; vasculature of gynoecium and stamens | |
| Pup12 | At2g39330 | Myrosinase-binding proteins and other lectins | Ceres 39069 | Before stage 11 | Ovary wall; vasculature of gyneocium, stamens, and petals | |
| Pup13 | At1g69870 | Peptide transporters | Ceres 22243 | All stages | Transmitting tissue; vasculature of floral organs; anther wall | |
| Pup14 | At1g52400 | β-Glucosidases | Ceres 17229 | Before stage 11 | Inner epidermis of ovary; ovule integuments; vasculature of gynoecium and stamens |
Expression Profiles of Genes Up-Regulated in the Arabidopsis Gynoecium
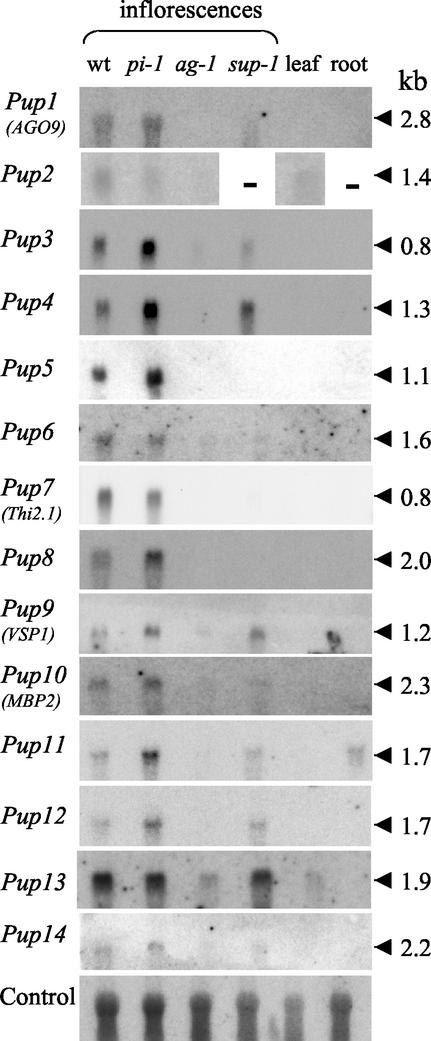
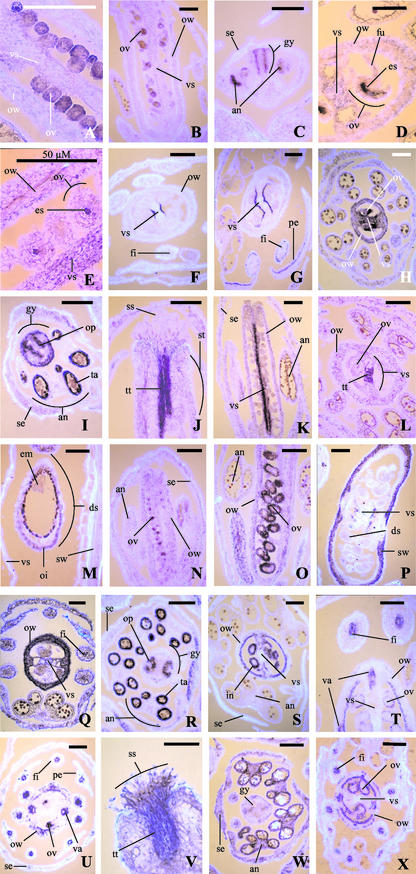
The expression profiles of the 14 Pup cDNAs identified from pi-1 flowers by FDD analysis were investigated by a combination of northern blotting and in situ hybridization. As floral organs of Arabidopsis cannot easily be dissected in adequate quantities for northern blotting, we used RNA samples from entire inflorescence tissues of wt plants and of three floral homeotic mutants whose flowers contain different combinations of floral organs. The floral homeotic mutants used in these analyses were pi-1 (flowers containing sepals and carpels), ag-1 (flowers containing sepals and petals), and superman-1 (sup-1; flowers containing sepals, petals, and stamens, with occasionally a vestigial gynoecium). The results of northern blotting for the 14 sequences analyzed, shown in Figure 1, demonstrate stronger hybridization signals for all of the cDNAs presented from pi-1 than from ag-1 flower RNA, thereby validating the results of the FDD screen. The detailed tissue-specific expression patterns of the 14 cDNAs were also investigated by non-radioisotopic in situ hybridizations to wt Arabidopsis flower bud, flower, and silique tissues, the results of which are presented in Figure 2. In situ hybridizations indicate that the majority of the Pup cDNA sequences show expression patterns largely confined to the gynoecium. In all cases, sense-strand cDNA control probes were used (data not shown) to verify the signals apparent with antisense cDNA probes. The detailed expression patterns shown by the 14 Pup cDNAs, as deduced from comparison of northern and in situ hybridization data, are as follows.
Figure 1.
Northern blots probed with full-length predicted coding regions of the cDNAs Pup1 to Pup14. The blots contain total RNA samples (or polyadenylated RNA samples in the case of Pup2) extracted from Arabidopsis plants of the Landsberg erecta (wt) genetic background and three mutant lines pi-1, ag-1, and sup-1. A control hybridization to demonstrate RNA loading in all tracks is shown using a cDNA probe corresponding to the gene At2g27040, encoding a close homolog of the Pup1/AGO9 cDNA, which is expressed in all Arabidopsis tissues so far tested. Molecular sizes of hybridizing transcripts, calculated from tracks of RNA size markers, are shown on the right.
Figure 2.
Non-radioisotopic in situ hybridizations using antisense-strand riboprobes corresponding to full-length predicted gene-coding regions of cDNAs Pup1 to Pup14. Hybridizations were performed on tissues of flower buds, mature flowers, and siliques of Arabidopsis plants of the Landsberg erecta genetic background. Probe hybridization signals corresponds to dark blue/violet staining, whereas counterstaining of cellulose-containing material shows as bright fluorescence. A, Longitudinal section (LS) of ovary showing Pup1/AGO9 signals in ovules at early stage 12. B, LS of flower bud showing Pup1/AGO9 signals in ovules at stage 13. C, LS of flower bud showing Pup1/AGO9 signals in placentae/ovule primordia and in anther loculi at stage 8. D, Transverse section (TS) of ovary showing a Pup2 signal in the embryo sac at stage 13. E, LS of ovary showing Pup2 signals in embryo sacs at stage 13. F, TS of flower bud showing Pup3 signals in epidermal cells of the vertical septum at late stage 12. G, TS of flower bud showing Pup3 signals in epidermal layers of the vertical septum, petals (abaxial surface), and filaments at stage 13. H, TS of flower bud showing Pup4 signals in the ovary and ovules at stages 11 to 12. I, TS (oblique) of flower bud showing Pup4 signals in the gynoecium wall, placentae/ovule primordia, and tapetum at stages 9 to 10. J, LS of upper gynoecium showing a Pup5 signal in the stylar transmitting tissue at stage 13. K, LS of flower bud showing a Pup5 signal in the vertical septum at stage 13. L, TS of flower bud showing a Pup5 signal in the transmitting tissue of the vertical septum at stage 13. M, LS of silique showing a Pup6 signal in the outer integument of the seed, approximately 3 d after fertilization. N, LS of flower bud showing Pup7/AtTH1 signals in ovules at early stage 12. O, LS of ovary showing Pup7/Thi2.1 signals in ovule integuments at stages 12–13. P, TS (oblique) of silique showing a Pup8 signal in mesocarp cell layers, 1 to 2 d after fertilization. Q, TS of flower bud showing Pup9/VSP1 signals in the ovary wall and stamen filaments at stage 13. R, TS of flower bud showing Pup10 signals in the placentae/ovule primordia of the gynoecium and in the tapetum at stages 9 to 10. S, TS of flower bud showing Pup10 signals in the inner epidermis of the ovary and in the integuments at stage 13. T, TS (oblique) flower bud showing Pup11 signals in the inner epidermis of the ovary, in the integuments, and in the vasculature of the gynoecium and filaments at stage 13. U, TS of flower bud showing Pup12 signals in the ovules, in the valves of the ovary and in the vasculature of the gynoecium, filaments, and petals at early stage 12. V, LS of upper gynoecium showing general Pup13 signals, although stronger in the stigmatic epidermis and stylar transmitting tissue at stage 13. W, TS of flower bud showing general Pup13 signals, although stronger in the anther walls and vascular tissue of floral organs at stage 10. X, TS of flower bud showing Pup14 signals in the epidermal cell layers of the ovary, in the ovule integuments, and in the vasculature of the gynoecium and filaments at stage 13. an, Anther; ds, developing seed; em, embryo; es, embryo sac; fi, filament of stamen; fu, funiculus; gy, gynoecium; in, integuments; oi, outer integument; op, ovule primordia; ov, ovule; ow, ovary wall; pe, petal; se, sepal; ss, stigmatic surface; st, style; sw, silique wall; ta, tapetum; tt, transmitting tissue; va, vasculature; vs, vertical septum. All scale bars = 100 μm, except where otherwise indicated.
The Pup1 cDNA corresponds to the gene ARGONAUTE9 (AGO9) of the ARGONAUTE gene family. This family consists of 10 genes in Arabidopsis, three of which, ARGONAUTE (AGO1), ZWILLE (ZLL, also known as PINHEAD), and ARGONAUTE4 (AGO4), have been functionally characterized. Mutations in AGO1 (Bohmert et al., 1998) and ZLL (Moussian et al., 1998; Lynn et al., 1999) lead to developmental perturbations in both vegetative and reproductive tissues. Additionally, AGO1 has been found to be necessary for PTGS (Fagard et al., 2000). AGO4 is required for the methylation-dependent silencing of the SUPERMAN gene in a clark kent epigenetic mutant and for the accumulation of certain classes of small interfering RNAs (Zilberman et al., 2003). Northern hybridization (Fig. 1) shows Pup1/AGO9 to be expressed in wt and pi-1 mutant inflorescences and unexpressed in ag-1 inflorescences and in wt vegetative tissues, suggesting the up-regulation of Pup1/AGO9 in the gynoecium. A Pup1/AGO9 hybridization signal on northern blots is also revealed in sup-1 inflorescence RNA, suggesting this gene to be also expressed in stamens (Fig. 1). In situ hybridization shows that Pup1/AGO9 is expressed principally in the ovule (Fig. 2, A and B). The Pup1/AGO9 gene appears to have a uniform level of expression throughout each ovule up to stage 12 (Fig. 2A) and lower and less uniform expression in ovule tissues at stage 13 (Fig. 2B) and later. A Pup1/AGO9 in situ hybridization signal is also apparent in the sporogenous tissue contained in the loculi of the anthers at around stage 8 of flower development (Fig. 2C), which is before pollen meiosis. This result is in accordance with the Pup1/AGO9 signal detected on northern blots of sup-1 inflorescence RNA (Fig. 1). At this earlier stage of development, Pup1/AGO9 expression is also apparent in two zones of the gynoecial tube corresponding to the placentae and ovule primordia (Fig. 2C), demonstrating that expression of this gene in female tissues also commences early in development. AGO9 is the first member of the ARGONAUTE family for which expression specifically in reproductive tissues has been demonstrated.
The Pup2 cDNA encodes a phytocyanin protein and is homologous to a number of early nodulin (ENOD) genes expressed in the developing root nodules of species of the Leguminosae (Greene et al., 1998). Northern blotting of total RNA samples failed to detect expression of the Pup2 gene (data not shown). However, northern blotting of polyadenylated RNA samples demonstrated weak Pup2 expression specifically in wt and pi-1 flowers and in leaves (Fig. 1). In situ hybridization (Fig. 2, D and E) shows the precise location of Pup2 expression in flowers to be the embryo sac, with the signal being more apparent toward the chalazal pole, opposite to the micropyle where pollen tubes enter (Fig. 2D). In situ hybridization to leaf tissue (data not shown) failed to reveal any Pup2 expression. Pup2 expression in leaves, which was apparent on northern blots, may therefore be below the limit of detection of in situ analysis, or be at a uniform level in all leaf cells and therefore difficult to locate. Database searching indicates the Pup2 cDNA to be a member of a gene family containing approximately 35 genes in Arabidopsis. However, no other Arabidopsis-predicted protein shows particularly close similarity to that predicted from the Pup2 cDNA, the closest being the protein predicted from the gene At4g27520 (40.4% amino acid identity).
The Pup3 cDNA encodes a short, Pro- or Hyp-rich peptide. Hydropathy predictions (Kyte and Doolittle, 1982) for this predicted peptide (data not shown) indicate a high probability of an N-terminal signal sequence with a consensus peptide cleavage site, allowing the possibility that the mature peptide may be secreted from the cell. Northern blotting demonstrates expression of the Pup3 gene to be entirely flower specific and mostly confined to the gynoecium, because hybridization signals are very weak from ag-1 and sup-1 mutants by comparison with those from wt plants and pi-1 mutants (Fig. 1). In situ hybridizations (Fig. 2, F and G) demonstrate expression of Pup3 in the gynoecium to be confined to the epidermal layers of the vertical septum. Expression of this gene commences in the latter half of stage 12 of flower development in the central region of the epidermis of the vertical septum (Fig. 2F). Pup3 expression then spreads to all epidermal cells of the vertical septum in fully mature flowers at stage 13 (Fig. 2G). This gene is additionally, but more weakly, expressed in the adaxial epidermis of the petal and the epidermis of the stamen filaments at the fully mature stage 13 of flower development (Fig. 2G). Although several distinct gene families encoding Pro and Hyp-rich proteins are known to be expressed in plant reproductive tissues (Sommer-Knudsen et al., 1997), the protein encoded by Pup3 belongs to a novel family containing only three members in Arabidopsis, the other two being the genes At1g30875 and At3g02670.
The Pup4 gene putatively encodes a phosphogluconolactonase, the second enzyme in the oxidative pentose phosphate pathway. This enzyme is responsible for the conversion of d-glucono-δ-lactone-6-phosphate into 6-phosphogluconic acid. Northern blotting (Fig. 1) shows the Pup4 mRNA to be present uniquely in inflorescence tissues, with expression specifically in wt, pi-1, and sup-1 inflorescences, suggesting expression only in the gynoecium and stamens. In situ hybridization (Fig. 2H) confirms the northern-blot results and demonstrates expression of the Pup4 gene in all parts of the gynoecium, with strongest expression in the developing ovules. A strong Pup4 signal is also apparent in the tapetum of the anthers at around stages 9 to 10 of flower development (Fig. 2I). At this earlier stage of development, gynoecial expression of Pup4 seems to be particularly strong in the ovary wall and in two internal zones corresponding to the placentae and developing ovule primordia (Fig. 2I). The specific expression of a phosphogluconolactonase in rapidly developing reproductive tissues is indicative of high activity in the pentose phosphate pathway. This pathway provides NADPH in non-photosynthetic tissues, generates Rib-5-phosphase for nucleic acid synthesis and contributes to the production of shikimic acid, which is a precursor of aromatic ring compounds (Goodwin and Mercer, 1983; Miclet et al., 2001). Database searching indicates the Pup4 gene to belong to a family of five genes in Arabidopsis. Pup4, therefore, seems to be a differentially regulated member of the phosphogluconolactonase gene family that functions uniquely in tissues within the third and fourth floral whorls.
Pup5 encodes a protein that shows 57.2% amino acid sequence identity to a heat shock-induced kunitz proteinase inhibitor from cauliflower (Brassica oleracea; Annamalai and Yanagihara, 1999). The Pup5 mRNA appears to be entirely specific to gynoecial tissues, giving signals only in pi-1 and wt inflorescence tissues on northern blotting (Fig. 1). In situ hybridization reveals this gene to be expressed in the transmitting tissue of the style (Fig. 2J) and vertical septum (Fig. 2, K and L), the route taken by pollen tubes toward the ovules.
The Pup6 cDNA encodes a putative cinnamoyl CoA reductase, the enzyme which catalyzes the first committed step in the production of lignin by the conversion of cinnamoyl CoAs to their respective cinnamaldehydes (Lauvergeat et al., 2001). Northern analysis (Fig. 1) indicates the Pup6 cDNA to be entirely flower specific and, from its much higher expression in wt and pi-1 inflorescences as compared with ag-1 and sup-1 inflorescences, to be largely gynoecium-specific. Although the Pup6 mRNA was identified from relatively early stages of flower development, no strong expression signal could be found in young flower buds by in situ hybridization (data not shown). By contrast, the Pup6 cDNA was found to be expressed in the outer integument of the developing seeds (Fig. 2M). Its expression seems to be strongest in the subepidermal cell layer. A natural coloration of the tissue, possibly due to an accumulation of phenolic compounds (Western et al., 2000), obscures any possible Pup6 expression in the inner cell layer of the inner epidermis (Fig. 2M).
Pup7 corresponds to the published AthTH1 cDNA encoding a thionin (Epple et al., 1995). The corresponding gene, Thi2.1 (Bohlmann et al., 1998), has been previously shown by northern blotting (Epple et al., 1995) and by GUS reporter gene studies (Vignutelli et al., 1998) to be expressed constitutively in flowers and siliques. In addition to its constitutive expression in reproductive tissues, Thi2.1 expression can be induced in vegetative tissues by wounding and infection (Bohlmann, et al., 1998; Vignutelli et al., 1998). Northern blotting (Fig. 1) performed in the current study confirms the findings of Epple et al. (1995), showing signals only in wt and pi-1 inflorescence tissue, suggesting up-regulation in the gynoecium. In situ hybridization demonstrates the exact location of Pup7/Thi2.1 expression to be the integuments of the ovule, a result that was not apparent from the study of the promoter activity of this gene (Vignutelli et al., 1998), which appeared to show a rather more generalized expression in the ovary. Expression of Pup7/Thi2.1 commences at stage 11 (Fig. 2N), during the formation of inner and outer integument primordia, and then increases throughout stages 12 and 13 to a maximum level in the integuments of the mature ovule at late stage 13 (Fig. 2O).
The Pup8 cDNA encodes a putative δ-cadinene synthase, the enzyme that catalyzes the conversion of farnesyl diphosphate into δ-cadinene, a committed step in the production of sesquiterpene phytoalexins (Benedict et al., 2001). Northern blotting (Fig. 1) indicates Pup8 expression to be entirely flower-specific. Furthermore, the expression of this gene is limited to wt and pi-1 mutant inflorescences, with no detectable expression in ag-1 or sup-1 inflorescences, suggesting an entirely gynoecium-specific expression profile. In situ hybridization (Fig. 2P) demonstrates expression of the Pup8 gene only in post-fertilization stages of development. Its expression is limited to the wall of the silique and seems to be particularly abundant in the mesocarp cell layers. No Pup8 hybridization signals are apparent in the ovary wall before stage 14 of flower development (data not shown).
The Pup9 cDNA is identical to the previously identified vegetative storage protein1 (VSP1)cDNA, encoding a vegetative storage protein (Utsugi et al., 1998). VSP1 is located at a distance of 6 kb from its close homolog VSP2 on Arabidopsis chromosome 5. Vegetative storage proteins were first identified by their accumulation in the vacuoles of leaf mesophyll cells of soybean (Glycine max) plants that had been depodded and thereby deprived of a nutrient sink (Wittenbach, 1983). It is thought that VSPs represent a protein reserve in the mature or developing plant and are termed vegetative to distinguish them from the storage proteins that accumulate in seeds. The promoter activity of the Pup9/VSP1 gene in Arabidopsis has been previously investigated using a β-glucuronidase reporter gene strategy (Utsugi et al., 1998). The Pup9/VSP1 promoter was found to be active in gynoecium tissues, although no promoter activity in the stamens or elsewhere in the flower was noted. Northern blotting in the present work indicates Pup9/VSP1 expression only in inflorescence tissues (Fig. 1). Hybridization signals are strongest from pi-1 mutant inflorescences, with considerable expression also from wt and sup-1 mutant inflorescences. A slight hybridization signal is also apparent in ag-1 mutant inflorescence tissue. Taken together, these data suggest a strong Pup9/VSP1 expression in the gynoecium and in stamens and a weak Pup9/VSP1 expression elsewhere in the flower. In situ hybridization of Pup9/VSP1 confirms the findings of northern analysis, showing a high level of expression in the ovary wall and the inner and outer epidermal cell layers of the ovary (Fig. 2Q). The high levels of Pup9/VSP1 expression apparent in sup-1 inflorescences by northern blotting (Fig. 1) are in agreement with in situ hybridization signals detected in the vasculature of the stamen filaments (Fig. 2Q). Although Pup9/VSP1 seems less highly expressed in filaments that in the gynoecium, a considerable level of expression in sup-1 mutants would be apparent on northern blots, because sup-1 flowers contain large numbers of stamens. In addition to high levels of Pup9/VSP1 in the gynoecium and stamen filaments, a general low level of hybridization of Pup9/VSP1 to all floral tissues is apparent (Fig. 2Q). This observation is in agreement with the low level of Pup9/VSP1 expression in ag-1 inflorescences observed on northern blots (Fig. 1).
The three cDNAs Pup10, Pup11, and Pup12 are homologous to genes encoding various classes of lectins, including myrosinase-binding proteins (MBPs), β-glucosidase-binding proteins (Rask et al., 2000), and jacalin lectin from the seeds of Artocarpus integrifolia (Young et al., 1991). The Pup10 cDNA has been previously characterized (Capella et al., 2001) and termed MPB2 on the grounds of similarity between its predicted protein and a characterized MBP from canola (Brassica napus; Taipalensuu et al., 1997). The Pup2/MBP2-predicted protein shows 47.2% amino acid similarity to this MBP from canola and is the most similar predicted protein to the canola MBP that exists in the Arabidopsis predicted proteome. However, no evidence as yet exists to indicate that the Pup10-predicted protein binds to myrosinases either in vitro or in vivo. Because many other classes of lectins share close similarity to MBPs and because database searching indicates Pup10, Pup11, and Pup12 to belong to a family of approximately 50 genes in Arabidopsis, the designation of Pup10 as encoding an MBP must await further evidence.
MBPs bind to myrosinases, which are glycosylated enzymes that show a thioglucosidase activity and are implicated in defense against insects, especially in the Brassicaceae and closely related families. Myrosinases catalyze the release of thiocyanates and other toxic compounds from glucosylated precursors, termed glucosinolates. Myrosinase enzymes are known to be constitutively present in specialized myrosinase cells, particularly in the seed and seedling, and are also inducible in the mature plant by wounding (Rask et al., 2000). MPBs are capable of binding to myrosinases and colocalize with these in some plant tissues (Geshi et al., 1998). The role of MPBs is not known, although it has been suggested that they may be capable of modulating myrosinase activity (Andreasson et al., 1999) or may show direct toxicity toward insects (Rask et al., 2000). Although the Pup10 and Pup11-predicted peptides show reasonable levels of similarity to a characterized MBP from canola (Taipalensuu et al., 1997), the putative lectin predicted from the Pup11 cDNA is considerably less similar to this protein of known myrosinasebinding activity (40.4% amino acid identity).
Northern blotting of the Pup10, Pup11, and Pup12 cDNAs (Fig. 1) demonstrates these to be expressed in wt and pi-1 inflorescences and to be expressed very lowly or to be unexpressed in ag-1 inflorescences, suggesting up-regulation in the gynoecium. All three of these genes additionally show hybridization to sup-1 inflorescence RNA, suggesting expression in the stamens. None of the genes Pup10, Pup11, or Pup12 is constitutively expressed in leaf tissue, and of the three of them, only Pup11 was found to be expressed in roots (Fig. 1).
In situ hybridization demonstrates that Pup10 is expressed at stages 9 to 10 of flower development in two regions internal to the gynoecial cylinder corresponding to the developing placentae and ovule primordia (Fig. 2R). By the mature stage 13, Pup10 is expressed specifically in the ovule integuments, in the funiculus, and in the internal epidermis of the ovary (Fig. 2S). In addition to its expression in female tissues, Pup10 is expressed in the tapetum of the anthers at stages 9 to 10 of flower development, during and immediately following pollen meiosis (Fig. 2R). Tapetal expression of Pup10 (MBP2) is also apparent in the in situ analysis performed by Capella et al. (2001), although the precise location of the expression of this gene in the inner epidermis and ovule integuments, clearly demonstrated in the present work, is not clear from the in situ hybridization data presented by Capella et al. (2001).
Pup11, like Pup10, shows expression in the integuments and in the inner epidermis of the ovary (Fig. 2T). However, this gene is additionally expressed in the four vascular strands of the gynoecium and in the vasculature of the stamen filaments in flowers buds at stage 13 of development (Fig. 2T). Pup11, again unlike Pup10, is not expressed in the tapetum (data not shown).
Pup12 is principally expressed in the vasculature of the immature floral organs (Fig. 2U). Expression of this gene is apparent in the four vascular bundles of the gynoecium, in the vasculature of the stamen filaments, and in the mid-vein of the petals (Fig. 2U). Pup12 is also expressed in the developing ovules at stages 11 and 12 (Fig. 2U). However, the ovule expression of Pup12 ceases at more mature developmental stages such that it is not expressed in the integuments of the mature ovule, as are Pup10 and Pup11 (data not shown). Pup12 is more generally expressed in the tissues of the immature ovary wall (Fig. 2U) than are Pup10 and Pup11. Again unlike Pup10 and Pup11, this gene does not show specific expression in the inner epidermis of the mature ovary. Like Pup11 but unlike Pup10, Pup12 is not expressed in the tapetum (data not shown). The Pup10, Pup11, and Pup12 cDNAs therefore show precise and distinct cellular patterns of expression that do not correspond to the presence of specialized myrosinase cells. Although from their sequence homologies, the proteins encoded by these genes may be predicted to show probable lectin activities, any carbohydrate-containing molecules to which they may bind in vivo have yet to be identified.
The Pup13 cDNA encodes a putative peptide transporter protein. The peptide transporter of known activity that exhibits closest similarity to the Pup13-predicted protein (36.4% amino acid sequence identity) is encoded by the HvPTR1 gene of barley (Hordeum vulgare; West et al., 1998). This gene is expressed specifically in the barley embryo during seed germination and is thought to be involved in the mobilization of protein reserves. Database searching indicates that the Pup13 gene is a member of a very extensive Arabidopsis gene family containing over 50 members. Northern blotting indicates the Pup13 gene to be expressed in all tissues tested other than roots (Fig. 1). Its expression is highest in the inflorescences of wt plants and of pi-1 and sup-1 mutants, with lower expression in ag-1 mutant inflorescences and in wt leaves. These results suggest the up-regulation of Pup13 expression in the gyneocium and stamens, with lower levels of expression in the perianth organs and the aboveground vegetative organs. In situ hybridization to mature flowers indicates some Pup13 expression in all flower tissues, with higher signals in the stigmatic epidermis (Fig. 2V) and the transmitting tissue of the style (Fig. 2V) and vertical septum (data not shown). At an earlier stage of flower development, higher levels of Pup13 expression are also apparent in the anther wall layers (Fig. 2W), explaining the high expression levels of this gene apparent on northern blotting of RNA from sup-1 mutant inflorescences (Fig. 1). In flower buds at stage 10, Pup13 is not expressed at a detectable level in the tapetum or sporogenous tissues (Fig. 2W). Pup13 is, however, highly expressed in the vasculature of the gynoecium, stamen filaments, and sepals at this same developmental stage (Fig. 2W).
The Pup14 cDNA encodes a putative β-glucosidase. β-Glucosidases are known to play a large variety of roles in plants by liberating many different classes of molecules from inactive glucosylated precursor forms. Databases searching indicates the protein of known function most closely resembling the Pup14-predicted protein (71.2% amino acid identity) to be a β-glucosidase that specifically degrades zeatin-o-glucosides in canola seeds (Falk and Rask, 1995). Another enzyme showing a similar activity related to hormone metabolism is involved in the release of active cytokinin during the development of maize (Zea mays) embryos (Brzobohaty et al., 1993). Several other β-glucosidases of unknown functions and substrate specificities have previously been found to be differentially regulated in response to environmental conditions such as phosphate starvation (Malboobi and Lefebvre, 1997) or to be developmentally upregulated in reproductive tissues such as the Arabidopsis tapetum (Rubinelli et al., 1998). β-Glucosidases show high levels of sequence similarity to thioglucosidases (myrosinases). One amino acid position within the active sites of these two classes of enzymes is occupied invariably by a Glu residue in β-glucosidases and by a Gln residue in myrosinases (Rask et al., 2000). This amino acid residue corresponds to position 207 in the Pup14-predicted peptide (data not shown), where the presence of Glu indicates a probable β-glucosidase activity.
Northern blotting (Fig. 1) indicates Pup14 to be entirely flower-specifically expressed. It is expressed in wt, pi-1, and sup-1 mutant inflorescences and also shows a very slight hybridization to ag-1 inflorescence RNA, suggesting up-regulation of this gene in the gynoecium and in stamens, with some expression elsewhere in the flower. In situ hybridization confirms the findings of northern blotting and demonstrates expression of Pup14 in various tissues of the mature flower bud. The Pup14 gene is strongly expressed in the integuments of the ovules and in the inner epidermis of the ovary and less strongly expressed in the outer epidermis of the ovary (Fig. 2X). In addition, it is strongly expressed in the four vascular strands of the gynoecium and in the vasculature of the stamen filaments and petals (Fig. 2X). The Pup14 expression pattern is therefore complex, involving several distinct tissues. This expression pattern resembles very closely that of the gene Pup11, also characterized in the present study, which shows similarity to myrosinase-binding and β-glucosidasebinding proteins and other lectins.
DISCUSSION
The FDD Analysis of Floral Homeotic Mutants Proves an Efficient Method for the Identification of Novel Gynoecium-Specific Genes
We have identified 14 genes that are up-regulated in tissues of the Arabidopsis gynoecium by the comparison of the genes expressed in the inflorescences of two Arabidopsis floral homeotic mutants, pi-1 and ag-1, using the technique of FDD. Eight of the 14 sequences described here were identified from early stages of flower development. However, in situ hybridization demonstrated all of these cDNAs to be also expressed at later developmental stages, suggesting that our intention to clone genes specifically expressed early in gynoecium development did not work as well as intended.
Most of the sequences identified in the present study were at least moderately highly expressed. All except one of them, Pup2, could be detected by northern blotting of total rather than polyadenylated RNA. In addition, none of the low-expressed genes already known to control gynoecium development (Bowman et al., 1999; Ferrandiz et al., 1999) was re-identified in the present study. This suggests that FDD may be more efficient for the detection of moderately or highly expressed rather than low-expressed mRNAs.
Comparison of pi-1 and ag-1 inflorescences containing flower buds up to developmental stage 10 in the present study yielded approximately 0.06 pi-1 mutant-up-regulated RT-PCR products per PCR primer combination. Inflorescences additionally containing buds up to developmental stage 13, however, yielded the much higher number of approximately 0.75 pi-1 mutant-up-regulated RT-PCR products per PCR primer combination. This suggests a large (approximately 12.5-fold) increase in the expression of moderately and highly abundant mRNAs during the latter phases of gynoecium and ovule development. Only a small proportion of the differential genes detected by FDD at later stages of flower development were cloned in the present study. Having already been identified in FDD analysis, these other differential mRNAs should, however, be amenable to cloning at a later time.
Despite the potential drawback of limited sensitivity, our results indicate the FDD technique to represent a substantial improvement over conventional differential display using radioactive detection (Liang and Pardee, 1992). An earlier study of gynoecium-specific gene expression in Arabidopsis using a similar mutant-based strategy in combination with conventional differential display succeeded in identifying only two gynoecium-expressed genes (Yung et al., 1999), one of which had been previously characterized. Although FDD may continue to provide the most efficient available means of differential screening that is of applicability to any eukaryotic organism, it is probable that microarray-based methods, due to their potentially greater convenience and sensitivity, will supersede the use of FDD in organisms for which complete genome sequences and/or large expressed sequence tag collections are available.
A Member of the ARGONAUTE Gene Family Functions in Developing Ovules and Sporogenous Tissue
The principal aim of this work has been to identify target genes for a reverse genetic functional analysis of processes occurring in the gynoecium. Our studies have identified the Pup1 cDNA as a gene with potential functions in ovules and the sporogenous tissues of the anther. Pup1 is a previously unstudied member of the ARGONAUTE gene family, designated after a homology search of the Arabidopsis genome by Morel et al. (2002) as AGO9. ARGONAUTE genes encode proteins containing PAZ and PIWI domains (Cerutti et al., 2000) and are known from plants, fungi, and animals (Fagard et al., 2000). These genes are variously involved in development and in a group of related processes termed PTGS in plants, RNA interference in animals, and quelling in fungi. The effect of PTGS can be seen in the silencing of reporter genes in transgenic plants by the specific destruction of their transcripts (for review, see Vaucheret and Fagard, 2001). Plants that are defective in PTGS due to mutations in the AGO1 gene have been shown to be less resistant to viruses (Morel et al., 2002), and it is hypothesized that the natural function of PTGS and related processes is in virus resistance.
In the present work, we show Pup1/AGO9 to be a transcribed gene that is expressed specifically in the ovule and in the sporogenous tissues of the anther loculus. In both of these tissues Pup1/AGO9 expression commences before meiosis. Both ovules and sporogenous tissues are in phases of rapid development and, in addition, both of these tissues might represent control points for the prevention of the vertical transmission of viruses. Several mutant alleles with differential effects on development and PTGS are known for the previously investigated AGO1 gene. In the ago1-27 allele, a mutation converting the predicted Ala-992 residue to Val causes a complete loss of PTGS, but has only a slight effect on plant development (Morel et al., 2002). This Ala residue is not conserved with the wt Pup1/AGO9-predicted peptide, which may suggest that Pup1/AGO9 is not involved in PTGS. Further studies, however, will be required to determine whether Pup1/AGO9 functions in ovule and pollen development, in PTGS for virus resistance, or in both of these processes.
FDD Identifies Several Classes of Genes with Potential Roles in the Development or Essential Functions of the Gynoecium
In addition to Pup1/AGO9, our studies have identified a number of other genes that present homologies and expression patterns suggesting potential roles either in the development of the gynoecium or in its essential processes. These genes also represent interesting candidates for reverse genetic functional analysis. The Pup2 gene is expressed specifically in the Arabidopsis embryo sac and is homologous to various early nodulin (ENOD) genes including ENOD5 from pea (Pisum sativum; Scheres et al., 1990), ENOD55 from soybean (de Blank et al., 1993), and ENOD16 and ENOD20 from Medicago truncata (Greene et al., 1998). These ENOD genes encode proteins of the phytocyanin class and, like other classes of ENOD genes, are expressed early in the development of nitrogen-fixing root nodules in species of the Leguminosae. The phytocyanins include a group of chloroplast copper-binding proteins termed plastocyanins that function as photosynthetic electron transporters. However, the known ENOD phytocyanins lack the ligands necessary for binding copper that are present in plastocyanins (Greene et al., 1998). The cellular or intracellular locations of the plastocyanin-like ENOD proteins and their functions during the process of nodulation are unknown. Multiple sequence alignment with plastocyanin proteins (data not shown) indicates that the Pup2-predicted phytocyanin also lacks three out of four of the residues that coordinate copper in plastocyanins (for review, see Greene et al., 1998), strongly suggesting that this predicted protein would be unable to bind copper.
The Pup3 cDNA encodes a short Pro- or Hyp-rich peptide that presents a very specific expression pattern in the epidermis of the vertical septum during the later stages of flower development. Pro- and Hyp-rich proteins are frequently secreted from the cell and often accumulate in the cell wall. This may be the case for the Pup3-predicted peptide, which shows a putative N-terminal signal peptide sequence and consensus cleavage site. Hyp-rich peptides are involved in many processes in plant reproductive tissues, including some cell-signaling processes, reviewed by Sommer-Knudsen et al. (1997). Because Pup3 is expressed late in flower development, it may not be involved in developmental processes that would alter the pattern of cell division and differentiation in the vertical septum. Pup3 is, however, expressed in a cell layer that must be traversed by growing pollen tubes immediately before fertilization.
Pup14 encodes a β-glucosidase with greatest similarity to an enzyme involved in the release of active cytokinin in canola seeds. Our studies show Pup14, by contrast, to be expressed in several different tissues of the developing Arabidopsis gynoecium. Our studies have serendipitously identified, along with Pup14, three genes encoding proteins that are similar to lectins known to bind to myrosinases, β-glucosidases, and other glycoproteins. The β-glucosidase from canola to which Pup14 shows close similarity was originally copurified together with two low Mr proteins (Falk and Rask, 1995) that may be examples of β-glucosidase-binding proteins from B. napus. One of the putative lectin-encoding cDNAs identified in the present study, Pup11, shows a pattern of expression very similar to that of the Pup14-predicted β-glucosidase. Further studies will be necessary to determine the substrate specificity of the predicted Pup14 β-glucosidase, its possible in vivo functions, and any potential interaction with the protein encoded by the Pup11 cDNA.
Many Classes of Defense-Related Genes Are Up-Regulated in the Gynoecium
One of the primary functions of the carpel appears to be the protection of the ovules and seeds that are contained within it. In addition to providing a physical barrier to insects and microbial pathogens, the carpel tissues and the integuments of the ovules and seeds of Arabidopsis have been shown in the present study to be the location of expression of many genes that are specifically related to plant defense. Pup5 encodes a putative proteinase inhibitor expressed in the transmitting tissue of the style and vertical septum. Proteinase inhibitors from the Brassicaceae have been shown to be active against enzymes of both fungal and animal origin (Lorito et al., 1994), whereas another class of Ser proteinase inhibitor expressed in the stigmas of Nicotiana alata (Atkinson et al., 1993) has activity against insect proteinases (Heath et al., 1997). Because the Pup5-encoded putative proteinase inhibitor is very precisely expressed along the transmitting tissue, a potential route of entry route for fungal hyphae, the natural function of Pup5 may be principally in antifungal defense.
Pup6 encodes a putative cinnamyl CoA reductase, expressed principally in the subepidermal cell layer of the outer seed integument, potentially involved in the production of lignin. Lignins of various structures and chemical compositions are known to play a variety of structural and defensive roles in plants tissues (Nicholson and Hammerschmidt, 1992). The epidermal cell layer of the Arabidopsis outer seed integument is specialized for the production of mucilage and sheds its outer cell wall on wetting (Western et al., 2000; Windsor et al., 2000). The expression of Pup6 in the subepidermal layer, therefore, suggests the presence of a lignified barrier below the outer mucilage-secreting cell layer. Pup7/Thi2.1 encodes a thionin that, like Pup6, is also specific to the integuments. Because thionins are implicated in plant defense, it is likely that Pup7/Thi2.1 also plays a role in the protection of the seeds.
Pup8 encodes a putative δ-cadinene synthase that is specific to the mesocarp cell layers of the silique wall. Pup8 can therefore be predicted to contribute to the production of sesquiterpene phytoalexins in the silique wall. These compounds are secondary plant products that play a defensive role against fungal pathogens (Harborne, 1993).
In addition to the above-mentioned genes that are likely to be primarily involved in plant defense, defensive roles cannot be excluded for some or all of the genes Pup10, Pup11, and Pup12, encoding putative lectins that may bind to myrosinases, β-glucosidases, or other carbohydrate-containing molecules. A defensive role is also possible for Pup1/AGO9, which may be involved in PTGS for viral resistance, in addition to a possible function in ovule and pollen development.
The Continued Reverse Genetic Analysis of Gene Function in the Gynoecium
The data presented here mainly relate to the position and timing of specific gene expression in the gynoecium and other Arabidopsis floral tissues. This information, together with the complete Arabidopsis genome sequence and efficient methods for insertional mutagenesis and gene knock-outs through RNA interference, facilitates the next stage in our analyses. The present work will enable the efficient targeting of effort into a reverse genetic analysis of gene function in the Arabidopsis gynoecium.
MATERIALS AND METHODS
Plant Material
Seed stocks of wt and mutant lines of Arabidopsis were originally obtained from the Nottingham Arabidopsis Seed Centre Seed Bank (Nottingham, UK). Both the wt plants and the mutants analyzed, pi-1, ag-1 (Bowman et al., 1989), and sup-1 (Bowman et al., 1992), were obtained in the Landsberg erecta genetic background. Arabidopsis plants were grown in peat-based potting compost in a growth chamber at 22°C with an 8-h photoperiod per 24 h. Flowering was induced by transfer to a greenhouse equipped with supplementary lighting to increase the photoperiod to ≥14 h.
FDD Analysis
Differential screening to identify mRNAs that were up-regulated in inflorescences of pi-1, compared with ag-1, mutants of Arabidopsis was performed by the semi-automated method of FDD, as described by Kuno et al. (2000). This method is based on the RT-PCR amplification of the 3′ extremities of mRNAs derived from different tissue samples. Reverse transcriptase PCR amplifications for FDD analysis employed three different fluorescent oligo(dT) primers, respectively containing dA, dC, and dG residues immediately 3′ to an oligo(dT) sequence. These 3′ residues were included to anneal with the last base of mRNAs, adjacent to their poly(A) tails, thereby selecting three subgroups of mRNAs. The three fluorescent 3′ primers were separately used in conjunction with 120 randomly chosen 10-mer primers (Operon Technologies, Alameda, CA). The mixtures of products resulting from PCR amplifications were analyzed on an automated DNA sequencer (SQ5500T, Hitachi, Tokyo). The molecular cloning of differentially expressed RT-PCR products identified by FDD analysis was performed using a Southern blotting-based system as described by Scutt et al. (2002).
Sequence Analysis and PCR Amplification of Full-Length cDNAs
The cloned RT-PCR products identified by FDD analysis, representing the 3′ extremities of transcribed sequences, were sequenced and their respective genes were located in the complete Arabidopsis genome sequence by BLAST searching (Altschul et al., 1990). Full-length cDNAs corresponding to the identified genes were then amplified by RT-PCR (McPherson et al., 1995) from inflorescence RNA of either the Landsberg or Columbia ecotypes of Arabidopsis using primers designed from the 5′ and 3′ extremities of their respective predicted protein-coding regions. In cases where full-length cDNAs had not previously been identified, these were fully sequenced to confirm or correct the coding sequence predictions accompanying the complete Arabidopsis genome sequence.
Northern-Blot Analysis
Plants used for northern-blot analysis were of the Landsberg erecta genetic background, with or without additional mutations in floral homeotic genes, as indicated. RNA was extracted from plant tissues either by a method based on hot SDS and phenol (Scutt, 1997) or using Trisol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Where required, polyadenylated RNA samples were purified using a PolyATract RNA extraction kit (Promega, Madison, WI). Inflorescence tissues included all stages of flower bud and flower development up to stage 14, the fertilization stage of wt flowers (Bowman, 1994). Inflorescence tissues from three homeotic mutant lines were also included in northern analyses. Of these, the pi-1 and ag-1 mutations showed high penetrance, giving uniform mutant phenotypes. The sup-1 mutation, by contrast, showed low penetrance, giving highly plastic phenotypic effects. For this reason, RNA used in northern blotting from sup-1 mutants was extracted only from plants that showed an extreme sup-1 phenotype. This phenotype corresponded to a greatly increased number of stamens and a gynoecium that was reduced to a thin filament-like structure or was absent (Bowman et al., 1992). Total RNA samples of 10 μg or polyadenylated RNA samples of 4 μg were analyzed on formaldehyde-containing agarose gels (Sambrook et al., 1989) and capillary blotted onto Hybond-N membranes (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK) according to the manufacturer's instructions. Northern hybridizations with radiolabeled full-length cDNA probes were carried out as described by Scutt et al. (1997) and washed to high stringency in solutions containing 0.1× SSPE (Sambrook et al., 1989) and 0.1% (w/v) SDS at 65°C. This washing stringency has been demonstrated (data not shown) to discriminate between homologous sequences of 4 kb sharing an average of >95% nucleic acid sequence similarity.
In Situ Hybridization
In situ hybridizations were performed on flower bud, mature flower, and silique material of plants of the Landsberg erecta genetic background. Riboprobes of full-length gynoecium-expressed cDNAs were labeled by incorporation of digoxygenin-conjugated ribonucleotides during in vitro transcription reactions. These reactions employed templates of linearized plasmids containing T3, T7, or SP6 RNA polymerase promoter sites flanking the full-length, gynoecium-expressed cDNAs. Riboprobes corresponding to antisense cDNA strands were used to detect the presence of homologous mRNAs, whereas the corresponding sense strands were generated using alternative RNA polymerases for use as negative controls. Tissue fixation, embedding in Paraplast Extra (Sherwood Medical, St. Louis), sectioning, and in situ hybridization was carried out as described by Bradley et al. (1993). Sections were counterstained with 0.1% (w/v) Calcofluor White MR2 (Sigma-Aldrich, San Luis Obispo, CA) to visualize cellulose-containing material under UV fluorescence microscopy. The sections were then dried and rendered permanent in Entellan reagent (Merck, Darmstadt, Germany). Photomicrographs were taken on daylight color reversal film using mixed UV and visible illumination on a fluorescence microscope (Optiphot-2, Nikon, Tokyo).
Acknowledgments
We thank the technical staff of the Reproduction et Développement des Plantes laboratory for help with plant culture and for general technical assistance. We also acknowledge the technical assistance of the DNA sequencing service of the Département de Biologie, Ecole Normale Supérieure de Lyon.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.017798.
The laboratory of Reproduction et Développement des Plantes is funded jointly by the Centre National de la Recherche Scientifique, the Institut National de la Recherche Agronomique, the Ecole Normale Supérieure de Lyon and the Université Claude Bernard-Lyon. C.P.S. was funded during this work formerly by a European Community Marie-Curie Fellowship and latterly as a Centre National de la Recherche Scientifique researcher. This work was supported in part by Hitachi (Advanced Research Laboratory grant no. B2023 to M.F.) and by the Program for the Promotion of Basic Research Activities for Innovative Bioscience (grant to M.F.).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Andreasson E, Taipalensuu J, Rask L, Meijer J (1999) Age-dependent wound induction of a myrosinase-associated protein from oilseed rape (Brassica napus). Plant Mol Biol 41: 171–180 [DOI] [PubMed] [Google Scholar]
- Annamalai P, Yanagihara S (1999) Identification and characterization of a heat-stress induced gene in cabbage encodes a kunitz type protease inhibitor. J Plant Physiol 155: 226–233 [Google Scholar]
- Atkinson AH, Heath RL, Simpson RJ, Clarke AE, Anderson MA (1993) Proteinase inhibitors in Nicotiana alata stigmas are derived from a precursor protein which is processed into five homologous inhibitors. Plant Cell 5: 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict CR, Lu JL, Pettigrew DW, Liu JG, Stipanovic RD, Williams HJ (2001) The cyclization of farnesyl diphosphate and nerolidyl diphosphate by a purified recombinant δ-cadinene synthase. Plant Physiol 125: 1754–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlmann H, Vignutelli A, Hilpert B, Miersch O, Wasternack C, Apel K (1998) Wounding and chemicals induce expression of the Arabidopsis thaliana gene Thi2.1, encoding a fungal defense thionin, via the octadecanoid pathway. FEBS Lett 437: 281–286 [DOI] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C (1998) AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J 17: 170–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Bouchez D (2001) Arabidopsis gene knockout: phenotypes wanted. Curr Opin Plant Biol 4: 111–117 [DOI] [PubMed] [Google Scholar]
- Bowman J (1994) Arabidopsis: An Atlas of Morphology and Development. Springer-Verlag, New York
- Bowman JL, Baum SF, Eshed Y, Putterill J, Alvarez J (1999) Molecular genetics of gynoecium development in Arabidopsis. Curr Topics Dev Biol 45: 155–205 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Eshed Y, Baum S, Emery JF, Floyd SK, Alvarez J, Hawker NP, Lee J-Y, Siegfried KR, Khodosh R et al. (2001) The story of crabs claw (or how we learned to love the mutagen). Flowering Newslett 31: 3–11 [Google Scholar]
- Bowman JL, Sakai H, Jack T, Weigel D, Mayer U, Meyerowitz EM (1992) SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114: 599–615 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D, Carpenter R, Sommer H, Hartley N, Coen E (1993) Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the PLENA locus of Antirrhinum. Cell 72: 85–95 [DOI] [PubMed] [Google Scholar]
- Brzobohaty B, Moore I, Kristoffersen P, Bako L, Campos N, Schell J, Palme K (1993) Release of active cytokinin by a β-glucosidase localized to the maize root meristem. Science 262: 1051–1054 [DOI] [PubMed] [Google Scholar]
- Capella AN, Menossi M, Arruda P, Benedetti CE (2001) COI1 affects myrosinase activity and controls the expression of two flower-specific myrosinase-binding protein homologues in Arabidopsis. Planta 213: 691–699 [DOI] [PubMed] [Google Scholar]
- Cerutti L, Mian N, Bateman A (2000) Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the PIWI domain. Trends Biochem Sci 25: 481–482 [DOI] [PubMed] [Google Scholar]
- de Blank C, Mylona P, Yang WC, Katinakis P, Bisseling T, Franssen H (1993) Characterization of the soybean early nodulin cDNA clone Gm-ENOD55. Plant Mol Biol 22: 1167–1171 [DOI] [PubMed] [Google Scholar]
- Epple P, Apel K, Bohlmann H (1995) An Arabidopsis thaliana thionin gene is inducible via a signal-transduction pathway different from that for pathogenesis-related proteins. Plant Physiol 109: 813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Boutet S, Morel JB, Bellini C, Vaucheret H (2000) AGO1, QDE-2, and RDZ-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci USA 97: 11650–11654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahn A (1975) Plant Anatomy, Ed 2. Pergamon Press, Oxford
- Falk A, Rask L (1995) Expression of a zeatin-o-glucoside-degrading β-glucosidase in Brassica napus. Plant Physiol 108: 1369–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure J-E, Dumas C (2001) Fertilization in flowering plants: new approaches for an old story. Plant Physiol 125: 102–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandiz C, Pelaz S, Yanofsky MF (1999) Control of carpel and fruit development in Arabidopsis. Annu Rev Biochem 68: 321–354 [DOI] [PubMed] [Google Scholar]
- Geshi N, Andreasson E, Meijer J, Rask L, Brandt A (1998) Co-localization of myrosinase- and myrosinase-binding proteins in grains of myrosin cells in cotyledon of Brassica napus seedlings. Plant Physiol Biochem 36: 583–590 [Google Scholar]
- Goodwin TW, Mercer EI (1983) Introduction to Plant Biochemistry, Ed 2. Pergamon, Oxford
- Greene EA, Erard M, Dedieu A, Barker DG (1998) MtENOD16 and 20 are members of a family of phytocyanin-related early nodulins. Plant Mol Biol 36: 775–783 [DOI] [PubMed] [Google Scholar]
- Harborne JB (1993) Introduction to Ecological Biochemistry, Ed 4. Academic Press, London
- Heath RL, McDonald G, Christeller JT, Lee M, Bateman K, West J, van Heeswijck R, Anderson MA (1997) Proteinase inhibitors from Nicotiana alata enhance plant resistance to insect pests. J Insect Physiol 43: 833–842 [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529 [DOI] [PubMed] [Google Scholar]
- Kuno N, Muramatsu T, Hamazato F, Furuya M (2000) Identification by large-scale screening of phytochrome-regulated genes in etiolated seedlings of Arabidopsis using a fluorescent differential display technique. Plant Physiol 122: 15–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Lauvergeat V, Lacomme C, Lacombe E, Lasserre E, Roby D, Grima-Pettenati J (2001) Two cinnamoyl-coA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry 57: 1187–1195 [DOI] [PubMed] [Google Scholar]
- Liang P, Pardee AB (1992) Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257: 967–971 [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed HY, Savidge B, Bowman JL, Yanofsky MF (2000) SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770 [DOI] [PubMed] [Google Scholar]
- Lorito M, Broadway RM, Hayes CK, Woo SL, Williams DL, Harman GE (1994) Proteinase inhibitors from plants as a novel class of fungicides. Mol Plant-Microbe Interact 4: 525–527 [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK (1999) The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126: 469–481 [DOI] [PubMed] [Google Scholar]
- Malboobi MA, Lefebvre DD (1997) A phosphate-starvation inducible β-glucosidase gene (PSR3.2) isolated from Arabidopsis thaliana is a member of a distinct subfamily of the BGA family. Plant Mol Biol 34: 57–68 [DOI] [PubMed] [Google Scholar]
- McPherson MJ, Hames BD, Taylor GR (1995) PCR 2: A Practical Approach. Oxford University Press, Oxford
- Miclet E, Stoven V, Michels PAM, Opperdoes FR, Lallemand JY, Duffieux F (2001) NMR spectroscopic analysis of the first two steps of the pentosephosphate pathway elucidates the role of 6-phosphogluconolactonase. J Biol Chem 276: 34840–34846 [DOI] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H (2002) Fertile hypomorphic argonaute1 (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian B, Schoof H, Haecker A, Jurgens G, Laux T (1998) Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J 17: 1799–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson RL, Hammerschmidt R (1992) Phenolic-compounds and their role in disease resistance. Annu Rev Phytopathol 30: 369–389 [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Rask L, Andreasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J (2000) Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol Biol 42: 93–113 [PubMed] [Google Scholar]
- Rubinelli P, Hu Y, Ma H (1998) Identification, sequence analysis and expression studies of novel anther-specific genes of Arabidopsis thaliana. Plant Mol Biol 37: 607–619 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsh EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Scheres B, Vanengelen F, Vanderknaap E, Vandewiel C, Vankammen A, Bisseling T (1990) Sequential induction of nodulin gene-expression in the developing pea nodule. Plant Cell 2: 687–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz K, Balasubramanian S, Schiefthaler U (1998) Organogenesis in plants: the molecular and genetic control of ovule development. Trends Plant Sci 3: 468–472 [Google Scholar]
- Scutt CP (1997) Differential screening. In E Hansen, G Harper, eds, Differentially-Expressed Genes in Plants: A Bench Manual. Taylor and Francis, London, pp 1–22
- Scutt CP, Jenkins T, Furuya M, Gilmartin PM (2002) Male specific genes from dioecious white campion identified by fluorescent differential display. Plant Cell Physiol 43: 563–572 [DOI] [PubMed] [Google Scholar]
- Scutt CP, Li Y, Robertson SE, Willis ME, Gilmartin PM (1997) Sex determination in dioecious Silene latifolia: effects of the y chromosome and the parasitic smut fungus (Ustilago violacea) on gene expression during flower development. Plant Physiol 114: 969–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions RA (1997) Arabidopsis (Brassicaceae) flower development and gynoecium patterning in wild type and ETTIN mutants. Am J Bot 84: 1179–1191 [PubMed] [Google Scholar]
- Sommer-Knudsen J, Clarke AE, Bacic A (1997) Proline- and hydroxyproline-rich gene products in the sexual tissues of flowers. Sex Plant Reprod 10: 253–260 [Google Scholar]
- Taipalensuu J, Eriksson S, Rask L (1997) The myrosinase-binding protein from Brassica napus seeds possesses lectin activity and has a highly similar vegetatively-expressed, wound-inducible counterpart. Eur J Biochem 250: 680–688 [DOI] [PubMed] [Google Scholar]
- Utsugi S, Sakamoto W, Murata M, Motoyoshi F (1998) Arabidopsis thaliana vegetative storage protein (VSP) genes: gene organization and tissue-specific expression. Plant Mol Biol 38: 565–576 [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Fagard M (2001) Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet 17: 29–35 [DOI] [PubMed] [Google Scholar]
- Vignutelli A, Wasternack C, Apel K, Bohlmann H (1998) Systemic and local induction of an Arabidopsis thionin gene by wounding and pathogens. Plant J 14: 285–295 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- West CE, Waterworth WM, Stephens SM, Smith CP, Bray CM (1998) Cloning and functional characterisation of a peptide transporter expressed in the scutellum of barley grain during the early stages of germination. Plant J 15: 221–229 [DOI] [PubMed] [Google Scholar]
- Western TL, Skinner DJ, Haughn GW (2000) Differentiation of mucilage secretory cells of the Arabidopsis outer seed coat. Plant Physiol 122: 345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmi LK, Preuss D (1999) The mating game: pollination and fertilization in flowering plants. Curr Opin Plant Biol 2: 18–22 [DOI] [PubMed] [Google Scholar]
- Windsor JB, Symonds VV, Mendenhall J, Llotd AM (2000) Arabidopsis seed coat development: morphological differentiation of the outer integument. Plant J 22: 483–493 [DOI] [PubMed] [Google Scholar]
- Wittenbach VA (1983) Effect of pod removal on leaf photosynthesis and soluble protein composition of field-grown soybeans. Plant Physiol 73: 121–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NM, Johnston RAZ, Watson DC (1991) The amino acid sequences of jacalin and the Maclura pomifera aglutinin. FEBS Lett 282: 382–386 [DOI] [PubMed] [Google Scholar]
- Yung MH, Schaffer R, Putterill J (1999) Identification of genes expressed during early Arabidopsis carpel development by mRNA differential display: characterisation of ATCEL2, a novel endo-1,4-β-d-glucanase gene. Plant J 17: 203–208 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]