Abstract
The CDPK-SnRK superfamily consists of seven types of serine-threonine protein kinases: calcium-dependent protein kinase (CDPKs), CDPK-related kinases (CRKs), phosphoenolpyruvate carboxylase kinases (PPCKs), PEP carboxylase kinase-related kinases (PEPRKs), calmodulin-dependent protein kinases (CaMKs), calcium and calmodulin-dependent protein kinases (CCaMKs), and SnRKs. Within this superfamily, individual isoforms and subfamilies contain distinct regulatory domains, subcellular targeting information, and substrate specificities. Our analysis of the Arabidopsis genome identified 34 CDPKs, eight CRKs, two PPCKs, two PEPRKs, and 38 SnRKs. No definitive examples were found for a CCaMK similar to those previously identified in lily (Lilium longiflorum) and tobacco (Nicotiana tabacum) or for a CaMK similar to those in animals or yeast. CDPKs are present in plants and a specific subgroup of protists, but CRKs, PPCKs, PEPRKs, and two of the SnRK subgroups have been found only in plants. CDPKs and at least one SnRK have been implicated in decoding calcium signals in Arabidopsis. Analysis of intron placements supports the hypothesis that CDPKs, CRKs, PPCKs and PEPRKs have a common evolutionary origin; however there are no conserved intron positions between these kinases and the SnRK subgroup. CDPKs and SnRKs are found on all five Arabidopsis chromosomes. The presence of closely related kinases in regions of the genome known to have arisen by genome duplication indicates that these kinases probably arose by divergence from common ancestors. The PlantsP database provides a resource of continuously updated information on protein kinases from Arabidopsis and other plants.
In eukaryotes, protein kinases are involved in regulating key aspects of cellular function, including cell division, metabolism, and responses to external signals. The completed sequence of the Arabidopsis genome provides the first opportunity to identify all of the protein kinases present in a model plant. The Arabidopsis genome encodes 1,085 typical protein kinases (M. Gribskov, unpublished data), which is about 4% of the predicted 25,500 genes (Arabidopsis Genome Initiative, 2000). Not only is the proportion of Arabidopsis kinase genes about twice that found in Brewer's yeast (Saccharomyces cerevisiae; Hunter and Plowman, 1997) or Caenorhabditis elegans (Plowman et al., 1999), but there are also major differences in the types of kinases found in plants. For example, receptor kinases in plants phosphorylate Ser and Thr residues, whereas in animals, the predominant type of receptor kinase phosphorylates Tyr residues. In addition, the two major types of kinases that decode calcium signals in animals (calmodulin-dependent protein kinases [CaMKs] and protein kinase C) appear to be missing or under-represented in plants.
Conversely, plants contain a number of kinase families that either are not found in animals or yeast or are highly divergent. For example, the calcium-dependent protein kinases (CDPKs) are found in vascular and nonvascular plants, in green algae, and also in certain protozoa (ciliates and apicomplexans; Hrabak, 2000). These kinases are calcium-regulated and are distinguished by a structural arrangement in which a calmodulin-like regulatory domain is located at the C-terminal end of the enzyme. The CDPK-related kinases (CRKs) and phosphoenolpyruvate carboxylase kinase-related kinases (PEPRKs) are also unique to plants. In addition, plants contain a large group of kinases related to the classical SNF1-type kinases from yeast, although the majority of these enzymes in Arabidopsis have a different primary structure compared with their yeast homologs. Halford and Hardie (1998) proposed the name SNF1-related kinase (SnRK) for this group and recognized three subgroups: SnRK1, SnRK2, and SnRK3.
In this report, we identify all of the kinases predicted to belong to the CDPK-SnRK superfamily based on the completed Arabidopsis genome sequence (Arabidopsis Genome Initiative, 2000) and examine their evolutionary origins. We also present suggested nomenclature for all of the kinases in this superfamily. Further details about individual kinases, including corrected sequence annotations, links to sequence alignments, and functional annotations, can be found at the PlantsP Web site (http://plantsp.sdsc.edu/).
RESULTS AND DISCUSSION
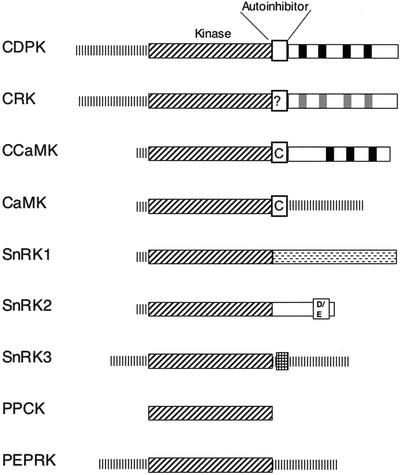
Figure 1 depicts the structural features of kinases in the CDPK-SnRK superfamily. Previous phylogenetic analyses have demonstrated that the kinases in this superfamily form a separate branch in relation to other plant protein kinases (Hardie, 1999, 2000). The common structures are discussed briefly here, and each subgroup is described in more detail in the following sections. All of these protein kinases contain a catalytic domain typical of eukaryotic Ser-Thr kinases, which was used for the initial classification of these proteins into the CDPK-SnRK superfamily (Hanks and Hunter, 1995). The proteins were assigned to particular subgroups based on the sequence and presumed function of their flanking domains. The N-terminal domains are highly variable in both length and sequence between the subgroups and even between individual protein kinases. Little is known about the function of the N-terminal domains except for the presence of putative myristoylation and palmitoylation sites in many of the CDPKs, which may contribute to membrane localization of these kinases (Hrabak, 2000).
Figure 1.
Diagrammatic representation of the domain structures of protein kinases in the CDPK-SnRK superfamily. The domains are described in more detail in the text. Broken lines at N or C termini indicate regions of variable length and function. Calcium-binding EF hands are indicated as black boxes, whereas corresponding regions in related protein kinases that lack consensus EF hand sequences are depicted in gray. The C in the autoinhibitor domain of CaMK and CCaMK indicates that the autoinhibitor domain of this kinase overlaps with a calmodulin-binding domain. The SnRK1 C-terminal domain regulatory domain is indicated by dashed lines. D/E is the acidic patch found in the SnRK2 group while the autoinhibitor (NAF/FISL) domain of the SnRK3 group is hatched.
Many CDPK-SnRK superfamily members have an autoregulatory region immediately C-terminal to the kinase domain (Fig. 1). In CDPKs, this region contains an autoinhibitor domain, which functions as a pseudosubstrate site and is involved in the intramolecular activation of the kinase (Harmon et al., 1994; Harper et al., 1994; Huang et al., 1996). In animal CaMKs (for reviews, see Hook and Means, 2001; Soderling and Stull, 2001) and calcium and calmodulin-dependent protein kinase (CCaMKs; Ramachandiran et al., 1997), this region binds a calcium-calmodulin complex required for activating the enzyme. In SnRK3s, the autoinhibitor domain (Guo et al., 2001) binds one of a family of calcium-binding proteins that includes SOS3, SOS3-like calcium binding protein (SCaBP), or calcineurin-B-like (CBL) calcium sensors (Liu and Zhu, 1998; Kudla et al., 1999; Guo et al., 2001), resulting in activation of the kinases (Guo et al., 2001; Gong et al., 2002b). It is interesting to note that the CDPKs (Yoo and Harmon, 1996; Huang and Huber, 2001), CCaMKs (Ramachandiran et al., 1997), CaMKs (Hook and Means, 2001; Soderling and Stull, 2001), and SnRK3s (Shi et al., 1999; Kim et al., 2000; Albrecht et al., 2001; Guo et al., 2001) are regulated by the interaction of their autoinhibitor domains with a calcium-binding domain or protein. No specific autoinhibitor domain has been identified for kinases in the CRK, PEP carboxylase kinase (PPCK), PEPRK, SnRK1, and SnRK2 groups.
Most members of the CDPK-SnRK superfamily have a C terminus that functions to regulate kinase activity or to mediate interactions with other proteins. The C-terminal domain of CDPKs and CCaMKs contains calcium-binding EF-hands resembling those found in either calmodulin (CDPKs; Harper et al., 1991) or visinin (CCaMKs; Patil et al., 1995). Calcium binding to the EF hands increases kinase activity. CRKs contain a C-terminal domain with some sequence similarity to the CaM-like domains of CDPKs but with poorly conserved EF-hands that do not bind calcium (Furumoto et al., 1996). The C-terminal domains of SnRKs are highly variable but in many cases are thought to function in protein-protein interactions.
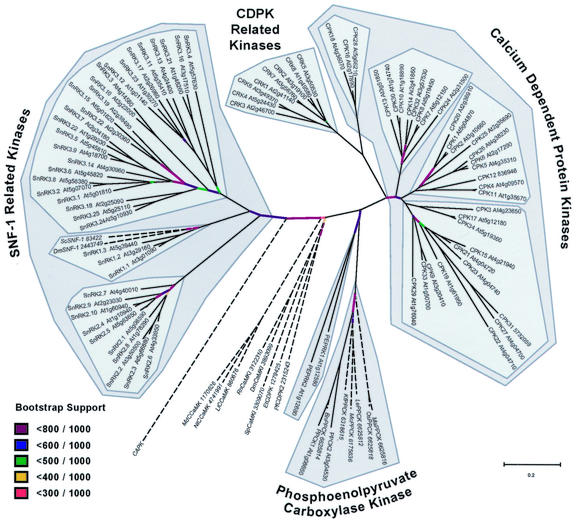
Figure 2 shows the unrooted sequence tree for the Arabidopsis CDPK-SnRK superfamily, based on comparisons of catalytic domains only. To show the relationship to other kinase groups, representative plant, animal, or fungal protein kinases from the CaMKI, CCaMK, and PPCK groups are included, and bovine cAMP-dependent protein kinase is used as an outgroup. SNF1 kinases from Brewer's yeast and fruitfly appear in the SnRK1 group of Arabidopsis kinases. There are no animal or fungal representatives of the SnRK2, SnRK3, CDPK, CRK, PPCK, or PEPRK groups. CDPK orthologs identified from vascular and nonvascular plants and green algae have been shown previously to be interspersed throughout the CDPK subgroup (Harmon et al., 2001). This distribution indicates that the diversity of CDPK sequences observed for Arabidopsis is typical for CDPK sequences in general. The apicomplexan CDPKs are distantly related to CDPKs from plants, and two apicomplexan sequences are included in Figure 2 for comparison.
Figure 2.
Sequence tree for the Arabidopsis CDPK/SnRK superfamily based on alignment of kinase catalytic domains. The five kinase groups (CDPK, CRK, PPCK, PEPRK, and SnRK) are delimited by dark gray blocks. Subgroups within the CDPK and SnRK groups are shown as lighter shaded blocks. Non-Arabidopsis sequences included as outgroups are followed by sequence identifiers and are shown in italics and with dashed lines. CAPK denotes bovine cAMP-dependent protein kinase. Abbreviations for other organisms are: Sc, Brewer's yeast; Dm, fruitfly (Drosophila melanogaster); Md, apple (Malus domestica); Nt, tobacco (Nicotiana tabacum); Ll, Lilium longiflorum; Rn, Rattus norvegicus; Sp, fission yeast (Schizosac-charomyces pombe); Et, Eimeria tenella (apicomplexan); Pf, Plasmodium falciparum (apicomplexan); Bn, canola (Brassica napus); Kf, Kalanchoe fedtschenkoi; Mc, common ice plant (Mesembryanthemum crystallinum); Le, tomato (Lycopersicon esculentum); Os, rice (Oryza sativa); and Ma, Musa acuminata. Bootstrap supports for specific branches are shown in color. Noncolored branches are supported by greater than 800 trees in 1,000 bootstrap trials.
Tables I, II, III, and IV list all of the Arabidopsis CDPKs, CRKs, PPCKs, PEPRKs, and SnRKs that we have identified to be members of the CDPK-SnRK superfamily. For the CDPKs and CRKs (Tables I and II), one nomenclature has predominated in the literature, and this is listed as the gene name. In cases where gene names were not previously assigned or where the genes have been referred to by a variety of names (Tables III and IV), we have proposed a gene name that can be used in the future for identification. For example, in the case of the SnRK3s, different labs have developed nomenclatures independently. Some names are based on functional characteristics of the proteins, whereas others are generic. We propose a new nomenclature for the SnRK group in Table IV. Although it may not be reasonable to expect researchers to change the nomenclature that they are currently using, Table IV is useful for deciphering which names are synonymous. The tables list additional important characteristics of these protein kinases that are discussed below.
Table I.
Characteristics of CDPKs from Arabidopsis
| Gene Name | Synonyms | cDNAs or ESTsa | Introns | Molecular Massb | Myristoylation Motifc | PlantsP ID No.d |
|---|---|---|---|---|---|---|
| CPK1 | AK1, At5g04870 | Yes | 6 | 68.3 | Yes | 21951 |
| CPK2 | At3g10660 | Yes | 6 | 72.2 | Yes | 21670 |
| CPK3 | CDPK6, At4g23650 | Yes | 8 | 59.3 | No | 21867 |
| CPK4 | At4g09570 | Yes | 6 | 56.4 | No | 21816 |
| CPK5 | At4g35310 | Yes | 6 | 62.1 | Yes | 21913 |
| CPK6 | ATCDPK3, At2g17290 | Yes | 6 | 61.1 | Yes | 26192 |
| CPK7 | At5g12480 | Yes | 7 | 60.3 | Yes | 10200 |
| CPK8 | CDPK19, At5g19450 | Yes | 7 | 59.9 | Yes | 22074 |
| CPK9 | At3g20410 | Yes | 7 | 60.4 | Yes | 21708 |
| CPK10 | ATCDPK1, At1g18890 | Yes | 6 | 61.4 | Yes | 21443 |
| CPK11 | ATCDPK2, At1g35670 | Yes | 6 | 55.9 | No | 20998e |
| CPK12 | CDPK9, At5g23580 | Yes | 5 | 55.4 | No | 21981 |
| CPK13 | At3g51850 | Yes | 6 | 59.4 | Yes | 21791e |
| CPK14 | At2g41860 | No | 6 | 60.1 | Yes | 21512 |
| CPK15 | At4g21940 | Yes | 7 | 62.6 | Yes | 21845 |
| CPK16 | At2g17890 | Yes | 11 | 64.8 | Yes | 26190 |
| CPK17 | At5g12180 | Yes | 6 | 58.5 | Yes | 21973 |
| CPK18 | At4g36070 | Yes | 11 | 60.8 | Yes | 21917e |
| CPK19 | At1g61950 | Yes | 8 | 62.9 | No | 21152e |
| CPK20 | At2g38910 | No | 5 | 64.7 | Yes | 21594 |
| CPK21 | At4g04720 | Yes | 7 | 59.9 | Yes | 21808 |
| CPK22 | At4g04710 | No | 7 | 55.9 | Yes | 21807e |
| CPK23 | At4g04740 | Yes | 7 | 58.7 | Yes | 21809e |
| CPK24 | At2g31500 | Yes | 6 | 66.2 | Yes | 21538 |
| CPK25 | At2g35890 | Yes | 5 | 58.9 | Yes | 21601 |
| CPK26 | At4g38230 | Yes | 6 | 54.3 | No | 21919 |
| CPK27 | At4g04700 | Yes | 7 | 54.9 | Yes | 21806e |
| CPK28 | At5g66210 | Yes | 11 | 59.0 | Yes | 22061 |
| CPK29 | At1g76040 | Yes | 7 | 60.5 | No | 21002e |
| CPK30 | ATCDPK1a, At1g74740 | Yes | 6 | 61.4 | Yes | 11194 |
| CPK31 | At4g04695 | Yes | 7 | 54.7 | Yes | 11158e |
| CPK32 | At3g57530 | Yes | 7 | 60.9 | Yes | 21777e |
| CPK33 | At1g50700 | Yes | 7 | 58.6 | Yes | 21103 |
| CPK34 | At5g19360 | Yes | 6 | 58.2 | Yes | 21369 |
Sources include GenBank (http://www.ncbi.nlm.nih.gov/), The Institute for Genomic Research Arabidopsis Gene Index (http://www.tigr.org/tdb/agi/), MATDB (http://mips.gsf.de/proj/thal/db/index.html) and unpublished data. EST, Expressed sequence tag. b Molecular masses in kilodaltons were calculated at http://expasy.cbr.nrc.ca/tools/pi_tool.html. c Myristoylation motifs were predicted by PROSITE, from published myristoylation consensus sequences (Towler et al., 1988; Boutin, 1997; Hofmann et al., 1999; Falquet et al., 2002), and from unpublished data. d Additional information is available at http://plantsP.sdsc.edu/. e Annotation errors exist in some databases. See PlantsP http://plantsp.sdsd.edu for corrected amino acid sequences.
Table II.
Characteristics of CRKs from Arabidopsis
| Gene Name | Synonyms | cDNA or ESTa | Introns | Molecular Massb | Myristoylation Motifc | PlantsP ID No.d |
|---|---|---|---|---|---|---|
| CRK1 | At2g41140 | Yes | 10 | 64.3 | Yes | 21623 |
| CRK2 | At3g19100 | Yes | 9 | 67.2 | Yes | 21712 |
| CRK3 | At2g46700 | No | 10 | 66.6 | Yes | 21571 |
| CRK4 | CP4, At5g24430 | Yes | 10 | 66.5 | No | 21984 |
| CRK5 | At3g50530 | Yes | 10 | 67.0 | Yes | 21757 |
| CRK6 | At3g49370 | Yes | 10 | 66.4 | No | 21752 |
| CRK7 | At3g56760 | Yes | 10 | 64.6 | Yes | 21790 |
| CRK8 | At1g49580 | Yes | 10 | 68.0 | Yes | 21393 |
Sources include GenBank (http://www.ncbi.nlm.nih.gov/), The Institute for Genomic Research Arabidopsis Gene Index (http://www.tigr.org/tdb/agi/), and MATDB (http://mips.gsf.de/proj/thal/db/index.html). EST, Expressed sequence tag. b Molecular masses in kilodaltons were calculated at http://expasy.cbr.nrc.ca/tools/pi_tool.html. c Myristoylation motifs were predicted by PROSITE and from published myristoylation consensus sequences (Towler et al., 1988; Boutin, 1997; Hofmann et al., 1999; Falquet et al., 2002). d Additional information is available at http://plantsp.sdsc.edu.
Table III.
Characteristics of PPCKs and PEPRKs from Arabidopsis
| Gene Name | Synonyms | cDNA or ESTa | Introns | Molecular Massb | Myristoylation Motifc | PlantsP ID No.d |
|---|---|---|---|---|---|---|
| PPCK1 | At1g08650 | Yes | 1 | 31.8 | No | 21409 |
| PPCK2 | At3g04530 | Yes | 1 | 31.3 | No | 21203e |
| PEPRK1 | At1g12580 | No | 3 | 57.5 | Yes | 21496 |
| PEPRK2 | At1g12680 | Yes | 2 | 52.1 | No | 21141 |
Sources include GenBank (http://www.ncbi.nlm.nih.gov/), The Institute for Genomic Research Arabidopsis Gene Index (http://www.tigr.org/tdb/agi/), and MATDB (http://mips.gsf.de/proj/thal/db/index.html). EST, Expressed sequence tag. b Molecular masses in kilodaltons were calculated at http://expasy.cbr.nrc.ca/tools/pi_tool.html. c Myristoylation motifs were predicted by PROSITE and from published myristoylation consensus sequences (Towler et al., 1988; Boutin, 1997; Hofmann et al., 1999; Falquet et al., 2002). d Additional information is available at http://plantsp.sdsc.edu/. e Annotation errors exist in some databases. See PlantsP (http://plantsp.sdsd.edu) for corrected amino acid sequences.
Table IV.
Characteristics of SnRKsa from Arabidopsis
| Gene Name | Synonyms | cDNA or ESTb | Introns | Molecular Massc | PlantsP ID No.d |
|---|---|---|---|---|---|
| SnRK1.1 | AK21, AKIN10, AKINα1, At3g01090 | Yes | 9 | 58.4 | 21659 |
| SnRK1.2 | AKIN11, AKINα 2, Akin20, At3g29160 | Yes | 9 | 58.7 | 21696 |
| SnRK1.3 | Akin30, AKINα 3, At5g39440 | No | 9 | 56.7 | 21323 |
| SnRK2.1 | ASK2, OSKL8, SRK2G, At5g08590 | Yes | 8 | 40.2 | 21293 |
| SnRK2.2 | ATHPROKINA, SPK2, OSKL3, SRK2D, At3g50500 | Yes | 8 | 41.2 | 21756 |
| SnRK2.3 | ATHPROKINB, 41K, OSKL2, SRK2I, At5g66880 | Yes | 8 | 41.1 | 22065 |
| SnRK2.4 | ASK1, OSKL7, SRK2A, At1g10940 | Yes | 8 | 41.2 | 21446 |
| SnRK2.5 | OSKL9, SRK2H, At5g63650 | Yes | 8 | 41.6 | 21362 |
| SnRK2.6 | OST1, SRK2E, At4g33950 | Yes | 9 | 40.6 | 21907 |
| SnRK2.7 | OSKL5, SRK2F, At4g40010 | No | 8 | 39.8 | 21924 |
| SnRK2.8 | OSKL4, SRK2C, At1g78290 | Yes | 5 | 38.2 | 21445 |
| SnRK2.9 | OSKL10, At2g23030 | No | 8 | 39.0 | 21604 |
| SnRK2.10 | OSKL6, SRK2B, At1g60940 | Yes | 8 | 41.0 | 21102 |
| SnRK3.1 | ATPK10, CIPK15, SIP2, PKS3, At5g01810 | Yes | 0 | 47.9 | 21940e |
| SnRK3.2 | CIPK2, PKS16, At5g07070 | Yes | 0 | 51.8 | 21956e |
| SnRK3.3 | CIPK4, PKS9, At4g14580 | Yes | 0 | 47.8 | 21928 |
| SnRK3.4 | CIPK21, PKS23, At5g57630 | Yes | 9 | 46.4 | 22037e |
| SnRK3.5 | CIPK19, PKS21, At5g45810 | Yes | 0 | 54.6 | 22004 |
| SnRK3.6 | CIPK20, PKS18, At5g45820 | Yes | 0 | 50.2 | 22005 |
| SnRK3.7 | CIPK13, PKS10, At2g34180 | Yes | 0 | 56.7 | 21627 |
| SnRK3.8 | CIPK10, SIP1, PKS2, At5g58380 | Yes | 0 | 54.6 | 17265e |
| SnRK3.9 | CIPK12, PKS8, At4g18700 | Yes | 0 | 55.0 | 21834 |
| SnRK3.10 | ATSRPK1, AtSR2, CIPK7, PKS7, At3g23000 | Yes | 0 | 48.3 | 21706 |
| SnRK3.11 | SOS2, CIPK24, At5g35410 | Yes | 12 | 50.6 | 21986 |
| SnRK3.12 | CIPK9, PKS6, At1g01140 | Yes | 13 | 50.5 | 21033e |
| SnRK3.13 | CIPK8, PKS11, At4g24400 | Yes | 13 | 50.4 | 21869 |
| SnRK3.14 | CIPK6, SIP3, PKS4, At4g30960 | Yes | 0 | 49.4 | 21894 |
| SnRK3.15 | CIPK14, PKS24, ATSR1, At5g01820 | Yes | 0 | 50.3 | 21941 |
| SnRK3.16 | CIPK1, PKS13, At3g17510 | Yes | 11 | 49.9 | 21205e |
| SnRK3.17 | CIPK3, PKS12, At2g26980 | Yes | 11 | 43.1 | 21568e |
| SnRK3.18 | CIPK16, PKS15, At2g25090 | Yes | 1 | 53.6 | 21575 |
| SnRK3.19 | CIPK22, PKS14, At2g38490 | Yes | 0 | 48.7 | 21556 |
| SnRK3.20 | CIPK18, PKS22, At1g29230 | Yes | 0 | 58.5 | 21389 |
| SnRK3.21 | CIPK17, PKS20, At1g48260 | Yes | 10 | 48.7 | 21026 |
| SnRK3.22 | CIPK11, SIP4, PKS5, At2g30360 | Yes | 0 | 49.0 | 21529 |
| SnRK3.23 | CIPK23, PKS17, At1g30270 | Yes | 14 | 53.5 | 26216e |
| SnRK3.24 | CIPK5, PKS19, At5g10930 | Yes | 0 | 50.9 | 21967 |
| SnRK3.25 | CIPK25, PKS25, At5g25110 | Yes | 0 | 55.6 | 21306e |
Subgroups were previously defined by Halford and Hardie (1998). b Sources include GenBank (http://www.ncbi.nlm.nih.gov/), The Institute for Genomic Research Arabidopsis Gene Index (http://www.tigr.org/tdb/agi/), MATDB (http://mips.gsf.de/proj/thal/db/index.html), and TAIR (http://www.arabidopsis.org/info/CBL.html#CIPK). EST, Expressed sequence tag. c Molecular masses in kilodaltons were calculated at http://expasy.cbr.nrc.ca/tools/pi_tool.html. d Additional information is available at http://plantsp.sdsc.edu. e Annotation errors exist in some databases. See PlantsP (http://plantsp.sdsc.edu) for corrected protein sequences.
CDPK
CDPKs from Arabidopsis, as well as other species, have been discussed in detail in recent reviews (Harmon et al., 2000; Hrabak, 2000; Cheng et al., 2002). Arabidopsis contains 34 genes predicted to encode CDPKs. CDPK genes and proteins are indicated by the three-letter abbreviation CPK followed by a number, according to the nomenclature of Hrabak et al. (1996). CDPKs contain a kinase catalytic domain and an autoinhibitory “junction” domain, followed by a calmodulin-like, calcium-binding regulatory domain (Fig. 1). For this reason, CDPK has occasionally been defined as calmodulin-like domain protein kinase. All CDPKs that have been characterized in detail are activated by calcium and thereby provide a mechanism to decode calcium signals (Harper et al., 1993; Urao et al., 1994; Abo-El-Saad and Wu, 1995; Hong et al., 1996; Hrabak et al., 1996; Saijo et al., 1997; Lee et al., 1998; Chico et al., 2002). Only three of the Arabidopsis CPK genes have not been shown to be expressed based on the existence of cDNA or expressed sequence tag clones. These could be nonexpressed pseudogenes, or their transcripts may have escaped detection because they are expressed only in response to specific stimuli or at specific times in development or in a limited number of cell types. The predicted CDPK proteins have molecular masses between 54.3 and 72.2 kD. All CDPK proteins have kinase, junction, and calmodulin-like domains of comparable size except CPK25, indicating that most of the variation in molecular mass is due to differences in the variable domains. All Arabidopsis CDPKs (except CPK25) have four EF hands as predicted by SMART (Schultz et al., 2000), a program for identification of protein domains (data not shown). CPK25 has a truncated C terminus that contains at most one EF hand. The majority of CDPKs have predicted myristoylation sites at their N termini (Table I), and these CDPKs also have a nearby Cys residue that could serve as a site for palmitoylation (E. Hrabak, unpublished data). Several Arabidopsis CDPKs have been shown to be myristoylated in vitro (Lu and Hrabak, 2002, S. Lu and E. Hrabak, unpublished data) including CPK5 (Table I), whose amino terminus does not fit the canonical myristoylation consensus sequence. On the basis of the myristoylation of CPK5, we predict that CPK6 and CPK13 are myristoylated also. Myristoylation appears to be an important mechanism for the membrane binding of CDPKs in Arabidopsis and other plant species (Martin and Busconi, 2000; Lu and Hrabak, 2002; Rutschmann et al., 2002).
The available information on the regulation of expression of Arabidopsis CDPK genes is limited but indicates that some genes are ubiquitously expressed (Hong et al., 1996), whereas others are induced by hormones or stress (Urao et al., 1994; Sheen, 1996). Expression of CDPKs from other plants is affected by a variety of stimuli including wounding (Chico et al., 2002), salt or drought stress (Botella et al., 1996; Patharkar and Cushman, 2000; Saijo et al., 2000), cold (Monroy and Dhindsa, 1995; Saijo et al., 2000), hormone treatment (Abo-El-Saad and Wu, 1995; Botella et al., 1996; Davletova et al., 2001), light (Frattini et al., 1999), cold (Martin and Busconi, 2001; Llop-Tous et al., 2002), and pathogens (Murillo et al., 2001; Romeis et al., 2001). Specific expression patterns have been described for CDPKs in many plant species including maize (Zea mays) pollen (Estruch et al., 1994), rice vascular tissue (Saijo et al., 2001) and seeds (Kawasaki et al., 1993; Frattini et al., 1999), and potato (Solanum tuberosum) stolons (Raices et al., 2001). Many of these CDPKs have orthologs in Arabidopsis, and it is presumed that expression of Arabidopsis CDPK genes is regulated by different stimuli and/or in specific cell types also.
The significance of the four major CDPK subgroups depicted in Figure 2 with regard to biochemical or physiological function is not known. Intriguing similarities between some CDPKs have been noted. For example, both CPK1 from Arabidopsis and a carrot (Daucus carota) CDPK are activated by phospholipids (Harper et al., 1993; Farmer and Choi, 1999), yet these two kinases are not in the same subgroup. The calcium-binding properties and the concentration of calcium required for activation was studied in depth for three soybean (Glycine max) CDPKs (Lee et al., 1998). Two of these CDPKs are very closely related and are found in the same subgroup, yet there is no consistent correlation between their kinetic properties when compared with the third soybean CDPK. More experiments are needed on all facets of CDPK function, including requirements for enzyme activation, substrate specificity, in planta expression, and subcellular localization.
CRK
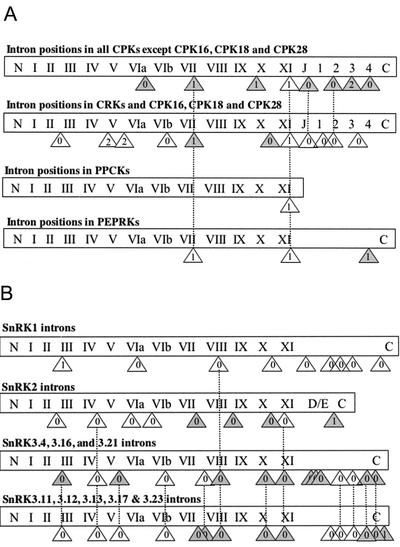
Arabidopsis contains eight CRK genes. The CRK nomenclature was proposed by Lindzen and Choi (1995). These kinases have so far only been identified in angiosperms. In the sequence tree (Fig. 2), they are located on a branch that is most closely related to a subgroup containing three CDPKs, indicating that there is a high degree of similarity between their catalytic domains. This arrangement, and the presence of apparently degenerate EF-hands in the CRKs, also suggests that the CRKs may have arisen relatively recently in evolution from a distinct subgroup of CDPKs. In support of this hypothesis, it appears that all of the CDPKs and CRKs on this branch have a pattern of intron placement that is distinct from the rest of the CDPKs (Fig. 3A; Harmon et al., 2001; Zhang and Choi, 2001).
Figure 3.
Intron positions in Arabidopsis CDPKs, CRKs, PPCKs, PEPRKs, and SnRKs. Kinases are diagrammed as rectangles; N, N terminus; C, C terminus; Roman numerals, conserved kinase domains (Hanks and Hunter, 1995); J, junction domain; 1–4, EF hands; D/E, acidic patch. Introns are represented by triangles that contain a number indicating the phase of the intron: 0 = intron inserted between intact codons; 1 = intron inserted after first nucleotide of a codon; 2 = intron inserted after second nucleotide of a codon. White triangles are introns conserved in all members of a group, whereas gray-filled introns are only found in some members of the group. Dotted lines connect introns at comparable positions in different kinase groups. A, Intron positions in CPKs, CRK, PPCKs, and PEPRKs. B, Intron positions in SnRKs.
The CRKs range in molecular mass from 64.3 to 68 kD (Table II) and have variable domains of similar length. All but one of the CRK genes are known to be expressed. As with CDPKs, the N termini of most CRKs are predicted to be myristoylated and palmitoylated, although no CRKs have been demonstrated to be acylated. The C-terminal domains of CRKs contain apparently degenerate calcium-binding sites that are predicted to be nonfunctional. Biochemical studies with recombinant CRKs from carrot (Farmer and Choi, 1999) and maize (Furumoto et al., 1996) have shown that these representative enzymes do not bind, and are not activated by, calcium. In addition, it does not appear that CRKs contain functional autoinhibitory domains, because a maize CRK was equally active in the presence of calcium or EGTA (Furumoto et al., 1996). A rice CRK binds calmodulin in a calcium-dependent manner, although its enzymatic activity is independent of calcium and calmodulin (Zhang et al., 2002). Thus, CRKs may be constitutively active. No data is available about CRK function in vivo or potential physiological substrates.
PPCK and PEPRK
Phosphoenolpyruvate carboxylase kinases (PPCKs) have been defined biochemically as calcium-independent protein kinases with molecular mass values of 30 to 32 kD and 37 to 39 kD that phosphorylate PEP carboxylase (Vidal and Chollet, 1997, and refs. therein). The function of PPCKs is most clearly understood in crassulacean acid metabolism and C4 plants (such as K. fedtschenkoi and maize, respectively) and these enzymes have been reviewed recently (Vidal and Chollet, 1997; Nimmo, 2000; Nimmo et al., 2001). Phosphorylation of PEPCase desensitizes the enzyme to feedback inhibition by malate and serves to activate the enzyme during times of active fixation of CO2 into oxaloacetic acid. PPCK activity appears to be regulated at the level of transcription and is under the control of light in C4 plants and a circadian rhythm in crassulacean acid metabolism plants (Jiao et al., 1991; Vidal and Chollet, 1997; Hartwell et al., 1999a, 1999b; Nimmo, 2000).
All PPCK genes cloned to date encode protein kinase catalytic domains with essentially no N- or C-terminal extensions (Fig. 1). The enzymes seem to be constitutively active and are among the smallest ATP-dependent protein kinases yet described, with molecular masses in the range of 30 to 32 kD (Table III). Arabidopsis contains two such PPCK genes, and the function of the PPCK proteins has been confirmed in vitro (Hartwell et al., 1999a; Fontaine et al., 2002). Both genes contain one intron close to the end of the catalytic domain (Fig. 3A), as does the K. fedtschenkoi PPCK gene.
Arabidopsis contains two additional protein kinase genes whose catalytic domains are most related to those of the PPCKs. Only one of these genes is known to be expressed. The predicted proteins contain both N- and C-terminal extensions with no similarity to the non-catalytic domains of any of the other kinases in this superfamily (Fig. 1). Because these extensions do not contain any apparent regulatory domains, these genes may encode constitutively active kinases, although their functions are still unknown. The molecular masses of these enzymes are 57.5 and 52.1 kD (Table III), so it seems unlikely that they encode the 37- to 39-kD form of PPCKs described above. We suggest that these PPCK-related enzymes be termed PEPRKs until their functions have been established.
CaMK and CCaMK
Arabidopsis contains no representatives of the CaMKs. The distinguishing biochemical feature of CaMKs is that they are activated by the binding of calmodulin to an autoinhibitory domain located immediately C-terminal to the kinase domain (Fig. 1). A large family of these enzymes is known in animals and yeast, but only one potential CaMK, from apple, has been identified in plants (Watillon et al., 1995). Protein sequence comparisons reveal that the kinase from apple is actually most closely related to plant CCaMKs (Fig. 2).
CCaMKs have been cloned from tobacco and lily (Patil et al., 1995; Liu et al., 1998), but they are apparently absent from Arabidopsis. The arrangement of functional domains in these enzymes is similar to that of the CDPKs, but the structure of their calcium-binding regulatory domain is more similar to visinin than to calmodulin and contains only three EF hands (Harmon et al., 1987; Patil et al., 1995; Yuasa et al., 1995; Pandey and Sopory, 1998). CCaMKs contain a calmodulin-binding site that overlaps with the autoregulatory domain (Ramachandiran et al., 1997). Regulation of CCaMK by calcium and calmodulin is complex. Autophosphorylation occurs in response to the binding of calcium to the EF hands in the visinin-like domain, which increases the affinity of the enzyme for binding the calcium/calmodulin complex and enhances substrate phosphorylation (Takezawa et al., 1996; Pandey and Sopory, 1998; Sathyanarayanan et al., 2000).
SnRK
Arabidopsis contains 38 protein kinases that are related to SNF1 from yeast (Table IV). The SnRKs form three subgroups based on sequence similarity and domain structure (Fig. 1). The SnRK1 subgroup is the most closely related to SNF1 from yeast and to AMP-activated protein kinases (AMPK) from animals (Fig. 2). Only three Arabidopsis sequences belong to the SnRK1 subgroup. On average, these are the largest proteins in the SnRK group with molecular mass values from 56.7 to 58.7 kD. Studies of the biochemistry and physiology of SnRK1s from crop plants have suggested that, like SNF1 and AMPK, they may regulate metabolism in response to nutritional or environmental stress (for review, see Halford and Hardie, 1998; Halford et al., 2000, 2003; Hardie, 2000). Unlike AMPK, SnRK1s are not allosterically regulated by AMP; however, AMP indirectly regulates the activity of a spinach SnRK1 via a mechanism that involves regulation of the phosphorylation/dephosphorylation of a residue in the enzyme's activation loop (conserved subdomain VIII; Sugden et al., 1999a). In vitro phosphorylation assays with purified enzymes and synthetic peptides have shown that SnRK1s have overlapping substrate specificity with CDPKs (Bachmann et al., 1996; Huber and Huber, 1996; Sugden et al., 1999b; Huang and Huber, 2001), suggesting that some substrates such as enzymes involved in carbon metabolism may be independently regulated by separate phosphorylation pathways (Hardie, 2000).
Interactions of SnRK1s with a variety of proteins have been identified. Not unexpectedly, SnRK1s interact with plant orthologs of proteins that regulate yeast SNF1, indicating that SnRK1s are likely to form heterotrimeric complexes like their yeast and animal counterparts and may be regulated by Glc in a manner similar to yeast SNF1 (Bhalerao et al., 1999; Bouly et al., 1999; Ferrando et al., 2001). Other interactions of Arabidopsis SnRK1s with the 26S proteosome and several proteosomal subunits (Farras et al., 2001) and with a protein Tyr phosphatase (Fordham-Skelton et al., 2002) have been reported. As with its yeast and animal orthologs, these interactions are likely to occur with the C-terminal regulatory domain of SnRK1. These results indicate that SnRK1s probably have a larger role in plant metabolism than was known previously. For example, antisense expression of a barley (Hordeum vulgare) SnRK1 interfered with pollen development and caused male sterility (Zhang et al., 2001).
The SnRK2 and SnRK3 groups appear to be unique to plants (Halford et al., 2000). The SnRK2s are about 140 to 160 amino acids shorter than the SnRK1s, averaging about 40 kD in size, and have a characteristic patch of acidic amino acids in their C-terminal domains (Halford et al., 2000). There are 10 SnRK2 genes in Arabidopsis. One of these, SnRK2.6, has been characterized recently (Mustilli et al., 2002; Yoshida et al., 2002). SnRK2.6 is expressed in guard cells and in the vascular system, and SnRK2.6 mutants are affected in stomatal closure in response to abscisic acid (ABA). SnRK2.6 activity is stimulated by ABA but gene expression is not regulated by ABA (Mustilli et al., 2002; Yoshida et al., 2002). Other SnRK2s include PKABA1 from wheat (Triticum aestivum; Anderberg and Walker-Simmons, 1992) and REK from rice (Hotta et al., 1998). In contrast to SnRK2.6 of Arabidopsis, transcription of PKABA1 is induced by ABA and occurs in embryos and seedlings, sites of relatively high ABA concentrations. PKABA1 has been implicated as a component in the pathway by which ABA treatment antagonizes the effects of GA3 via suppression of GA3-inducible genes (Gomez-Cadenas et al., 1999, 2001; Shen et al., 2001). An ABA-response element binding factor (TaABF) has been shown by two-hybrid screening to specifically bind to PKABA1. TaABF is seed specific, suggesting the TaABF may serve as a physiological substrate for PKABA1 in ABA-signaling processes in wheat seeds (Johnson et al., 2002). Expression of PKABA1 in Escherichia coli or Pichia pastoris yields inactive enzyme, however, the active form can be obtained by expression in insect cells (S.D. Verhey, L.D. Holappa, and M.K. Walker-Simmons, personal communication). This implies that PKABA1 requires posttranslational modification and/or regulatory subunits for activity and that these are available in insect cells but not in bacterial or yeast cells. In contrast, REK, a SnRK2 from rice, autophosphorylates when expressed in bacterial cells, and the addition of 10 mm calcium stimulates this activity (Hotta et al., 1998). Although 10 mm calcium is much higher than physiological concentrations, this observation raises the question of whether the activity of some SnRK2s may be regulated by calcium.
Twenty-five of the Arabidopsis SnRKs fall into the SnRK3 group. This group is represented by kinases previously published as CBL-interacting protein kinase (CIPKs; Kudla et al., 1999; Shi et al., 1999; Kim et al., 2000; Albrecht et al., 2001), salt overly sensitive 2 (SOS2; Zhu et al., 1998; Halfter et al., 2000; Liu et al., 2000), SOS3-interacting proteins (SIPs) (Halfter et al., 2000) and protein kinase S (PKS) (Guo et al., 2001; Table IV). These protein kinases interact with calcium-binding proteins such as SOS3, SCaBPs, and CBL proteins, which are related to animal neuronal calcium sensors and to the regulatory B subunit of the protein phosphatase calcineurin (Liu and Zhu, 1998; Kudla et al., 1999; Kim et al., 2000; Albrecht et al., 2001; Guo et al., 2001). These kinases are involved in responses to salt stress and in sugar and ABA signaling (Guo et al., 2001, 2002; Gong et al., 2002a; Gong et al., 2002c).
The research groups of Kudla, Luan, and Zhu independently discovered the SnRK3 group of kinases, and these groups have developed different nomenclatures for these enzymes. In a search for calcineurin regulatory subunits from plants, Kudla et al. (1999) isolated a family of genes encoding CBL calcium-binding proteins. When AtCBL1, a stress-responsive member of the CBL family, was subsequently used as bait in a yeast two-hybrid analysis, Shi et al. (1999) identified CIPKs as target proteins for CBL1. Further work showed that different CBLs and CIPKs interact with some degree of isoform specificity (Kim et al., 2000; Albrecht et al., 2001). Independently, Zhu's group isolated mutant Arabidopsis plants having a “salt overly sensitive” phenotype in a genetic screen (Zhu et al., 1998) and later showed that two of the mutant lines carried mutations in either a calcium-binding protein named SOS3 (Liu and Zhu, 1998) or a protein kinase named SOS2 (Liu et al., 2000). These results implicate SOS2 and SOS3 in salt resistance in Arabidopsis. SOS3 interacts with SOS2 and other related protein kinases called SIPs (Halfter et al., 2000). SOS2 is the only SnRK3 subgroup member whose function has been identified by genetic analysis.
One feature that differentiates the SnRKs from the CDPK/CRK/PPCK/PEPRK group is that all of the SnRKs contain a Thr in their activation loops (conserved subdomain VIII). Most CDPKs and all CRKs have Asp or Glu at this position: Two CDPKs have basic amino acids, whereas PPCKs and PEPRKs have Gly. This is especially interesting because of the effect of this residue on enzyme activity (Guo et al., 2001; Gong et al., 2002b). Replacement of the Thr residue in the activation loop of SOS2 with an Asp residue confers significantly higher activity on the enzyme, which does not require either SOS3 or calcium (Guo et al., 2001; Gong et al., 2002b). Similar activation after replacement of this Thr residue has been reported for other SnRK3s (Gong et al., 2002a; Gong et al., 2002c). A potential requirement of this Thr residue as a site for phosphorylation by another kinase could explain the lack of activity of some SnRK3s when expressed in bacteria.
The activity of SnRK3s expressed in bacteria is stimulated by the addition of SOS3 or a SOS3-like calcium-binding protein in the presence of calcium. These calcium-binding proteins interact with SnRK3s via a 21- to 24-residue region called the NAF or FISL domain (Shi et al., 1999; Albrecht et al., 2001; Guo et al., 2001), which also serves as an autoinhibitory domain for kinase activity (Guo et al., 2001; Gong et al., 2002b). These observations indicate that the regulation of SnRK3s in vivo is complex and is only beginning to be understood.
Unlike CDPKs and CRKs, almost none of the SnRK1, SnRK2, or SnRK3 kinases contain a putative N-terminal myristoylation sequence and thus are unlikely to be membrane associated due to this hydrophobic modification. Interestingly, SnRK3.11 (also known as SOS2) regulates the activity of SOS1, a plasma membrane Na+/H+ exchanger (Qiu et al., 2002), even though SOS2 is not by itself a membrane-associated protein. SOS2 may be membrane localized through its interaction with SOS3 (Halfter et al., 2000).
Evolutionary Origins
A phylogenetic analysis of kinase domains representative of all Ser/Thr kinases from Arabidopsis indicated that the CDPK and SnRK subfamilies cluster next to each other on a branch distinct from other kinases (Hardie, 2000) and we have termed this group of kinases the CDPK-SnRK superfamily. The evolutionary relationships of the CDPK, CRK, PPCK, and PEPRK groups have been analyzed recently (Harmon et al., 2001; Zhang and Choi, 2001; Cheng et al., 2002). CDPKs and CRKs have four highly conserved intron placements: two phase 1 introns in the catalytic domain, a phase 0 intron at the border of the junction and calmodulin-like domains, and another phase 0 intron in the calmodulin-like domain (Fig. 3A). One of the phase 1 introns is also conserved in both PPCK and PEPRK genes, whereas the other phase 1 intron is only found in PEPRKs. The sequence tree (Fig. 2) supports the hypothesis that CDPKs, CRKs, PPCKs, and PEPRKs have a common evolutionary origin. However, a number of questions about these genes still remain. For example, we do not know whether there were independent gene fusion events that gave rise to the plant and protistan CDPKs or whether the visinin-like regulatory domain of CCaMKs arose from the divergence of a calmodulin-like domain of a CDPK or from a separate gene fusion event.
Although the kinase domain of the SnRK subfamily is most closely related to the CDPK subfamily, based on our analyses, there are no conserved intron positions between these two groups of kinases. Within the SnRK subfamily, few intron positions are conserved between all SnRK groups. The SnRK3 group is particularly diverse with two major branches. One branch contains genes with zero or one intron, whereas the genes in the other branch have nine or more introns (Fig. 2; Table IV). On the former branch, a single SnRK3 gene contains one intron, whereas the remainder have none. This single intron is a phase two intron in kinase subdomain X and is not similar to any other SnRK intron position (data not shown). The SnRK3 branch with nine or more introns is composed of two smaller groups with related but distinct intron structures (Fig. 3B). The differences in intron structure between the two major SnRK3 branches raises the possibility that the genes on the branch with zero or one intron arose after reverse transcription of a mRNA from the group with nine or more introns, followed by reinsertion into the genome and subsequent divergence. SnRK1s share one intron position with the branch of SnRK3s that contains many introns, whereas SnRK2s share two intron positions with these SnRK3s (Fig. 3B). SnRK1.1 and SnRK1.2 are apparently the only members of the CDPK-SnRK superfamily to have an intron in their 5′-untranslated region which may play a role in regulation of gene expression (data not shown; J. Bouly and M. Thomas, unpublished data).
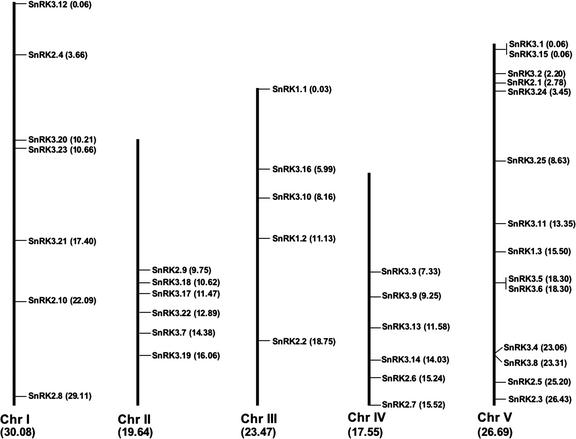
Chromosome Positions
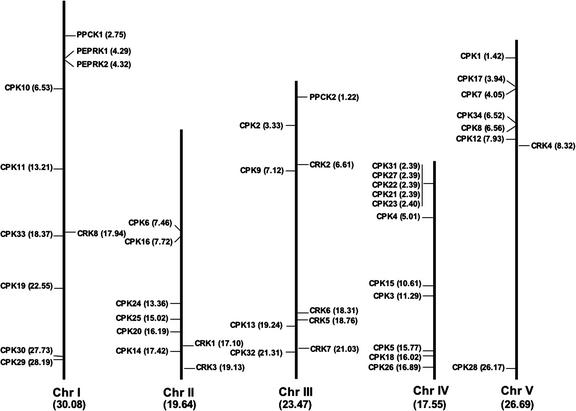
CDPKs and SnRKs are found on all chromosomes (Figs. 4 and 5). No CRKs are located on chromosome IV, whereas PPCKs and PEPRKs are only found on chromosomes I and III. One group of CDPKs (CPK31, CPK27, CPK22, CPK21, and CPK23) is arranged in a cluster of five adjacent genes on the top of chromosome IV. These five genes are transcribed in the same direction (data not shown) and are closely related on the sequence tree (Fig. 2), indicating that they may have arisen by a relatively recent gene duplication. The extremely close linkage for some isoforms will make it difficult for researchers to obtain the double and triple knockout mutants that could be helpful in determining CDPK function.
Figure 4.
Chromosome positions of CDPK, CRK, PPCK, and PEPRK genes. The positions of CDPKs are shown to the left of each chromosome, whereas the CRKs, PPCKs, and PEPRKs are indicated on the right. Values in parentheses are distances in megabases.
Figure 5.
Chromosome positions of SnRK genes. Values in parentheses are distances in megabases.
In most cases, kinases that are closely related based on the sequence tree (Fig. 2) are not located near each other on the physical map of the Arabidopsis genome. However, there is evidence that the Arabidopsis genome has undergone extensive duplication and reorganization (Blanc et al., 2000). Therefore, we compared whether closely related pairs of kinases are found in known duplicated regions of the genome. Six closely related pairs of CPK genes (CPK1/2, CPK9/33, CPK5/6, CPK16/18, CPK14/32, and CPK10/30) and two pairs of CRK genes (CRK2/8 and CRK1/7) were found in duplicated genomic regions. In addition, two pairs of SnRK2s (SnRK2.2/2.3 and SnRK2.4/2.10) and five pairs of SnRK3s (SnRK3.3/3.10, SnRK3.5 and 3.6/3.9, SnRK3.7/3.20, SnRK3.16/3.21, and SnRK3.24/3.25) occurred in duplicated regions. These results support the hypothesis that these kinases are paralogs that arose by divergence after genome duplication events.
PlantsP Database
Our alignments, corrected sequence annotations, and functional annotations of the Arabidopsis sequences are available at the PlantsP database (http://plantsp.sdsc.edu). Our analysis revealed several annotation problems with the published genomic sequence. Such problems are not uncommon and are unavoidable with the current sequence analysis technology but can be confusing to researchers. To address this issue, the PlantsP database provides a community-enhanced resource to resolve annotation problems and to continuously update information on protein kinases from all plants. Input to the database from other researchers studying plant protein kinases and phosphatases is welcome through the User Registration link.
CONCLUSIONS
Our understanding of the members of the Arabidopsis CDPK-SnRK superfamily is similar to that of many other Arabidopsis gene families in that the genes and the proteins they encode have been identified, but studies to clarify their roles in plant growth and development are still under way. Multiple approaches are being used to investigate the functions of these proteins including site-directed mutagenesis or deletion of key residues or domains, reverse genetics (knockouts) to identify null mutations, protein interaction screens to identify potential substrates, biochemical analyses to characterize kinetic properties, and expression and localization studies to clarify where and when various family members are expressed. These experiments can be complicated by the existence of multiple, closely related genes that may have similar functions. For instance, knockouts in these kinase genes usually have not had an obvious phenotype, indicating either that the correct conditions to identify a phenotype were not tested or that the protein's function can be fulfilled by other related family members. In some cases, double, triple, or higher order mutants may have to be made to uncover a phenotype. Mutant combinations could be made at random, or specific mutants could be combined based on the hypothesis that kinases that are near each other on the sequence tree might have similar or redundant functions. Clearly there is much exciting research yet to come.
Presumably, the proliferation of divergent kinases has played an important role in the evolution of successful plant species. We expect that a comprehensive bioinformatics analysis of all kinases and phosphatases encoded in the Arabidopsis genome, starting here with the CDPK-SnRK superfamily, will provide an important foundation for understanding the role of phosphoregulation in plants.
MATERIALS AND METHODS
Construction of Trees
Sequences of CDPK family members from all species were retrieved from the GenBank database. The complete set of CDPK and CRK sequences were identified by repetitive BLAST analyses (Altschul et al., 1990) of the proteins predicted from the Arabidopsis genomic sequence and from GenBank. A set of known CDPK and SnRK proteins sequences were used as query sequences (the query panel). Candidate members of the CDPK-SnRK group were identified as those sequences most similar to the query sequences. Each of the identified sequences was used as a query in further searches. Candidate sequences were accepted as members of the CDPK-SnRK group when they were closer to the query panel than to any other group and when they identified the CDPK-SnRK group as their nearest relative in reciprocal searches. In cases where the GenBank sequences contained errors predicted from comparison with known CDPK-SnRK family members, manually corrected sequences were used. Most errors occurred due to incorrect intron splice site predictions. Sequences of Arabidopsis isoforms were also obtained from The Institute for Genomic Research (ftp://ftp.tigr.org/pub/data/a_thaliana/ath1/PUBLICATION_RELEASE/) and from the Munich Information Center for Protein Sequences (ftp://ftp.mips.biochem.mpg.de/pub/cress/). Sequence trees were constructed using the neighbor-joining method as implemented in the ClustalW program (Thompson et al., 1994). Alignments were constructed by a combination of automatic and manual procedures. In brief, sequences were initially aligned with ClustalW after first omitting the most divergent sequences. Alignments were used to construct an evolutionary profile (Gribskov and Veretnik, 1996). All sequences were then aligned to the profile to construct the multiple alignment. Finally, this alignment was manually edited to correct errors. Sequences were assigned to families and subfamilies based on the calculated tree and additional information such as intron/exon patterns.
Identification of Gene Duplications
Data on duplicated regions in the Arabidopsis genome were obtained from The Institute for Genomic Research (http://www.tigr.org/tdb/e2k1/ath1/arabGenomeDups.html). Duplications were confirmed by direct examination of the gene order flanking the kinases.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.011999.
Note Added in Proof
Two additional references may be of interest to readers.
Kim K-N, Cheong YH, Grant JJ, Pandey GK, Luan S (2003) CIPK3, a calcium-sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 14 411–423
Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W (2002) Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell 14 S389–S400
This work was supported by the National Science Foundation (grant nos. DBI–9975808 and MCB-9973770 to A.C.H., IBN–9728563 to M.R.S., IBN–9416038 and MCB–9723539 to J.F.H., and DBI–9975808 to E.M.H., A.C.H., J.F.H., and M.G.), by the Deutsche Forschungsgemeinschaft (grant nos. Ku 931/3–2 and Ku931/4–1 to J.K.); by the U.S. Department of Agriculture (grant nos. 95–37304-2364 to M.R.S. and 98–35304–6510 to E.M.H.), and by the National Institutes of Health (grant no. R01GM59138 to J.K.Z.).
References
- Abo-El-Saad M, Wu R (1995) A rice membrane calcium-dependent protein kinase is induced by gibberellin. Plant Physiol 108 787–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht V, Ritz O, Linder S, Harter K, Kudla J (2001) The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J 20 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215 403–410 [DOI] [PubMed] [Google Scholar]
- Anderberg RJ, Walker-Simmons MK (1992) Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proc Natl Acad Sci USA 89 10183–10187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Bachmann M, Shiraishi N, Campbell WH, Yoo B-C, Harmon AC, Huber SC (1996) Identification of Ser-543 as the major regulatory phosphorylation site in spinach leaf nitrate reductase. Plant Cell 8 505–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao RP, Salchert K, Bako L, Okresz L, Szabados L, Muranaka T, Machida Y, Schell J, Koncz C (1999) Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc Natl Acad Sci USA 96 5322–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M (2000) Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella JR, Arteca JM, Somodevilla M, Arteca RN (1996) Calcium-dependent protein kinase gene expression in response to physical and chemical stimuli in mungbean (Vigna radiata). Plant Mol Biol 30 1129–1137 [DOI] [PubMed] [Google Scholar]
- Bouly J, Gissot L, Lessard P, Kreis M, Thomas M (1999) Arabidopsis thaliana proteins related to the yeast SIP and SNF4 interact with AKINalpha1, a SNF1-like protein kinase. Plant J 18 541–550 [DOI] [PubMed] [Google Scholar]
- Boutin JA (1997) Myristoylation. Cell Signal 9 15–35 [DOI] [PubMed] [Google Scholar]
- Cheng S, Willmann MR, Chen H, Sheen J (2002) Calcium signaling through protein kinases: the Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol 129 469–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico JM, Raices M, Tellez-Inon MT, Ulloa RM (2002) A calcium-dependent protein kinase is systemically induced upon wounding in tomato plants. Plant Physiol 128 256–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Meszaros T, Miskolczi P, Oberschall A, Torok K, Magyar Z, Dudits D, Deak M (2001) Auxin and heat shock activation of a novel member of the calmodulin like domain protein kinase gene family in cultured alfalfa cells. J Exp Bot 52 215–221 [PubMed] [Google Scholar]
- Estruch JJ, Kadwell S, Merlin E, Crossland L (1994) Cloning and characterization of a maize pollen-specific calcium-dependent calmodulin-independent protein kinase. Proc Natl Acad Sci USA 91 8837–8841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJ, Hofmann K, Bairoch A (2002) The PROSITE database, its status in 2002. Nucleic Acids Res 30 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer PK, Choi JH (1999) Calcium and phospholipid activation of a recombinant calcium-dependent protein kinase (DcCPK1) from carrot (Daucus carota L.). Biochim Biophys Acta 1434 6–17 [DOI] [PubMed] [Google Scholar]
- Farras R, Ferrando A, Jasik J, Kleinow T, Okresz L, Tiburcio A, Salchert K, delPozo C, Schell J, Koncz C (2001) SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J 20 2742–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando A, Koncz-Kalman Z, Farras R, Tiburcio A, Schell J, Koncz C (2001) Detection of in vivo protein interactions between Snf1-related kinase subunits with intron-tagged epitope-labelling in plant cells. Nucleic Acids Res 29 3685–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine V, Hartwell J, Jenkins GI, Nimmo HG (2002) Arabidopsis thaliana contains two phosphoenolpyruvate carboxylase kinase genes with different expression patterns. Plant Cell Environ 25 115–122 [Google Scholar]
- Fordham-Skelton AP, Chilley P, Lumbreras V, Reignoux S, Fenton TR, Dahm CC, Pages M, Gatehouse JA (2002) A novel higher plant protein tyrosine phosphatase interacts with SNF1-related protein kinases via a KIS (kinase interaction sequence) domain. Plant J 29 705–715 [DOI] [PubMed] [Google Scholar]
- Frattini M, Morello L, Breviario D (1999) Rice calcium-dependent protein kinase isoforms OsCDPK2 and OsCDPK11 show different responses to light and different expression patterns during seed development. Plant Mol Biol 41 753–764 [DOI] [PubMed] [Google Scholar]
- Furumoto T, Ogawa N, Hata S, Izui K (1996) Plant calcium-dependent protein kinase-related kinases (CRKs) do not require calcium for their activities. FEBS Lett 396 147–151 [DOI] [PubMed] [Google Scholar]
- Gomez-Cadenas A, Verhey SD, Holappa LD, Shen Q, Ho TH, Walker-Simmons MK (1999) An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proc Natl Acad Sci USA 96 1767–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cadenas A, Zentella R, Walker-Simmons MK, Ho TH (2001) Gibberellin/abscisic acid antagonism in barley aleurone cells: site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13 667–679 [PMC free article] [PubMed] [Google Scholar]
- Gong D, Gong Z, Guo Y, Chen X, Zhu J-K (2002a) Biochemical and functional characterization of PKS11, a novel Arabidopsis protein kinase. J Biol Chem 277 28340–28350 [DOI] [PubMed] [Google Scholar]
- Gong D, Gong Z, Zhu J (2002b) Expression, activation and biochemical properties of a novel Arabidopsis protein kinase. Plant Physiol 129 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D, Zhang C, Chen X, Gong Z, Zhu J-K (2002c) Constitutive activation and transgenic evaluation of the function of an Arabidopsis PKS protein kinase. J Biol Chem 277 42088–42096 [DOI] [PubMed] [Google Scholar]
- Gribskov M, Veretnik S (1996) Identification of sequence pattern with profile analysis. Methods Enzymol 266 198–212 [DOI] [PubMed] [Google Scholar]
- Guo Y, Halfter U, Ishitani M, Zhu JK (2001) Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13 1383–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Song CP, Gong D, Halfter U, J.K. Z (2002) A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev Cell 3 233–244 [DOI] [PubMed] [Google Scholar]
- Halford NG, Bouly JP, Thomas M (2000) SNF1-related protein kinases (SnRKs): regulators at the heart of the control of carbon metabolism and partitioning. Adv Bot Res 32 405–434 [Google Scholar]
- Halford NG, Hardie DG (1998) SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Mol Biol 37 735–748 [DOI] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Paul M, Zhang Y (2003) Metabolic signalling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. J Exp Bot 54 467–475 [DOI] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu JK (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97 3735–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Hunter T (1995) The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9 576–596 [PubMed] [Google Scholar]
- Hardie DG (1999) Plant protein serine/threonine kinases: classification and functions. Annu Rev Plant Physiol Plant Mol Biol 50 97–131 [DOI] [PubMed] [Google Scholar]
- Hardie DG (2000) Plant protein-serine/threonine kinases: classification into subfamilies and overview of function. Adv Bot Res 32 1–44 [Google Scholar]
- Harmon AC, Gribskov M, Gubrium E, Harper JF (2001) The CDPK superfamily of protein kinases. New Phytol 151 175–183 [DOI] [PubMed] [Google Scholar]
- Harmon AC, Gribskov M, Harper JF (2000) CDPKs: a kinase for every Ca2+ signal? Trends Plant Sci 5 154–159 [DOI] [PubMed] [Google Scholar]
- Harmon AC, Putnam-Evans C, Cormier MJ (1987) A calcium-dependent but calmodulin-independent protein kinase from soybean. Plant Physiol 83 830–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon AC, Yoo B-C, McCaffery C (1994) Pseudosubstrate inhibition of CDPK, a protein kinase with a calmodulin-like domain. Biochemistry 33 7278–7287 [DOI] [PubMed] [Google Scholar]
- Harper JF, Binder BM, Sussman MR (1993) Calcium and lipid regulation of an Arabidopsis protein kinase expressed in Escherichia coli. Biochemistry 32 3282–3290 [DOI] [PubMed] [Google Scholar]
- Harper JF, Huang J-F, Lloyd SJ (1994) Genetic identification of an autoinhibitor in CDPK, a protein kinase with a calmodulin-like domain. Biochemistry 33 7267–7277 [DOI] [PubMed] [Google Scholar]
- Harper JF, Sussman MR, Schaller GE, Putnam-Evans C, Charbonneau H, Harmon AC (1991) A calcium-dependent protein kinase with a regulatory domain similar to calmodulin. Science 252 951–954 [DOI] [PubMed] [Google Scholar]
- Hartwell J, Gill A, Nimmo GA, Wilkins MB, Jenkins GI, Nimmo HG (1999a) Phosphoenolpyruvate carboxylase kinase is a novel protein kinase regulated at the level of expression. Plant J 20 333–342 [DOI] [PubMed] [Google Scholar]
- Hartwell J, Jenkins GI, Wilkins MB, Nimmo HG (1999b) The light induction of maize phosphoenolpyruvate carboxylase kinase translatable mRNA requires transcription but not translation. Plant Cell Environ 22 883–889 [Google Scholar]
- Hofmann K, Bucher P, Falquet L, Bairoch A (1999) The PROSITE database, its status in 1999. Nucleic Acids Res 27 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Takano M, Liu C-M, Gasch A, Chye M-L, Chua N-H (1996) Expression of three members of the calcium-dependent protein kinase gene family in Arabidopsis thaliana. Plant Mol Biol 30 1259–1275 [DOI] [PubMed] [Google Scholar]
- Hook SS, Means AR (2001) Ca(2+)/CaM-dependent kinases: from activation to function. Annu Rev Pharmacol Toxicol 41 471–505 [DOI] [PubMed] [Google Scholar]
- Hotta H, Aoki N, Matsuda T, Adachi T (1998) Molecular analysis of a novel protein kinase in maturing rice seed. Gene 213 47–54 [DOI] [PubMed] [Google Scholar]
- Hrabak EM (2000) Calcium-dependent protein kinases and their relatives. Adv Bot Res 32 185–223 [Google Scholar]
- Hrabak EM, Dickmann LJ, Satterlee JS, Sussman MR (1996) Characterization of eight new members of the calmodulin-like domain protein kinase gene family of Arabidopsis thaliana. Plant Mol Biol 31 405–412 [DOI] [PubMed] [Google Scholar]
- Huang J-F, Teyton L, Harper JF (1996) Activation of a Ca(2+)-dependent protein kinase involves intramolecular binding of a calmodulin-like regulatory domain. Biochemistry 35 13222–13230 [DOI] [PubMed] [Google Scholar]
- Huang J-Z, Huber SC (2001) Phosphorylation of synthetic peptides by a CDPK and plant SNF1-related protein kinase: influence of proline and basic amino acid residues at selected positions. Plant Cell Physiol 42 1079–1087 [DOI] [PubMed] [Google Scholar]
- Huber SC, Huber JL (1996) Role and regulation of sucrose-phosphate synthase in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47 431–444 [DOI] [PubMed] [Google Scholar]
- Hunter T, Plowman GD (1997) The protein kinases of budding yeast: six score and more. Trends Biochem Sci 22 18–22 [DOI] [PubMed] [Google Scholar]
- Jiao J-A, Echevarria C, Vidal J, Chollet R (1991) Protein turnover as a component in the light/dark regulation of phosphoenolpyruvate carboxylase protein-serin kinase activity in C4 plants. Proc Natl Acad Sci USA 88 2712–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Wagner R, Verhey SD, Walker-Simmons MK (2002) The ABA-responsive kinase PKABA1 interacts with a seed-specific ABA response element binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol 130 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Hayashida N, Baba T, Shinozaki K, Shimada H (1993) The gene encoding a calcium-dependent protein kinase located near the sbe1 gene encoding starch branching enzyme I is specifically expressed in developing rice seeds. Gene 129 183–189 [DOI] [PubMed] [Google Scholar]
- Kim K-N, Cheong YH, Gupta R, Luan S (2000) Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases. Plant Physiol 124 1844–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Xu Q, Harter K, Gruissem W, Luan S (1999) Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc Natl Acad Sci USA 96 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-Y, Yoo B-C, Harmon AC (1998) Kinetic and calcium-binding properties of three calcium-dependent protein kinase isoenzymes from soybean. Biochemistry 37 6801–6809 [DOI] [PubMed] [Google Scholar]
- Lindzen E, Choi JH (1995) A carrot cDNA encoding an atypical protein kinase homologous to plant calcium-dependent protein kinases. Plant Mol Biol 28 785–797 [DOI] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97 3730–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu J-K (1998) A calcium sensor homolog required for plant salt tolerance. Science 280 1943–1945 [DOI] [PubMed] [Google Scholar]
- Liu Z, Xia M, Poovaiah BW (1998) Chimeric calcium/calmodulin-dependent protein kinase in tobacco: differential regulation by calmodulin isoforms. Plant Mol Biol 38 889–897 [DOI] [PubMed] [Google Scholar]
- Llop-Tous I, Dominguez-Puigjaner E, Vendrell M (2002) Characterization of a strawberry cDNA clone homologous to calcium-dependent protein kinases that is expressed during fruit ripening and affected by low temperature. J Exp Bot 53 2283–2285 [DOI] [PubMed] [Google Scholar]
- Lu SX, Hrabak EM (2002) An Arabidopsis calcium-dependent protein kinase is associated with the endoplasmic reticulum. Plant Physiol 128 1008–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ML, Busconi L (2000) Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitoylation. Plant J 24 429–435 [DOI] [PubMed] [Google Scholar]
- Martin ML, Busconi L (2001) A rice membrane-bound calcium-dependent protein kinase is activated in response to low temperature. Plant Physiol 125 1442–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroy AF, Dhindsa RS (1995) Low-temperature signal transduction: induction of cold acclimation-specific genes of alfalfa by calcium at 25 C. Plant Cell 7 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo I, Jaeck E, Cordero MJ, Segundo BS (2001) Transcriptional activation of a maize calcium-dependent protein kinase gene in response to fungal elicitors and infection. Plant Mol Biol 45 145–158 [DOI] [PubMed] [Google Scholar]
- Mustilli A-C, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kianse mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo HG (2000) The regulation of phosphoenolpyruvate carboxylase in CAM plants. Trends Plant Sci 5 75–80 [DOI] [PubMed] [Google Scholar]
- Nimmo HG, Fontaine V, Hartwell J, Jenkins GI, Nimmo GA, Wilkins MB (2001) PEP carboxylase kinase is a novel protein kinase controlled at the level of expression. New Phytol 151 91–97 [DOI] [PubMed] [Google Scholar]
- Pandey S, Sopory SK (1998) Biochemical evidence for a calmodulin-stimulate calcium-dependent protein kinase in maize. Eur J Biochem 255 718–726 [DOI] [PubMed] [Google Scholar]
- Patharkar OR, Cushman JC (2000) A stress-inducible calcium-dependent protein kinase from Mesembryanthemum crystallinum phosphorylates a two-component pseudo-response regulator. Plant J 24 679–691 [DOI] [PubMed] [Google Scholar]
- Patil S, Takezawa D, Poovaiah BW (1995) Chimeric plant calcium/calmodulin-dependent protein kinase gene with a neural visinin-like calcium-binding domain. Proc Natl Acad Sci 92 4897–4901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowman GD, Sudarsanam S, Bingham J, Whyte D, Hunter T (1999) The protein kinases of Caenorhabditis elegans: a model for signal transduction in multicellular organisms. Proc Natl Acad Sci USA 96 13603–13610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q-S, Guo Y, Dietrich MA, Schumaker KS, Zhu J-K (2002) Regulation of SOS1, a plasma membrane Na +/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raices M, Chico JM, Telez-Inon MT, Ulloa RM (2001) Molecular characterization of StCDPK1, a calcium-dependent protein kinase from Solanum tuberosum that is induced at the onset of tuber development. Plant Mol Biol 46 591–601 [DOI] [PubMed] [Google Scholar]
- Ramachandiran S, Takezawa D, Wang W, Poovaiah BW (1997) Functional domains of plant chimeric calcium/calmodulin-dependent protein kinase: regulation by autoinhibitory and visinin-like domains. J Biochem 197 984–990 [DOI] [PubMed] [Google Scholar]
- Romeis T, Ludwig AA, Martin R, Jones JDG (2001) Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J 20 5556–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutschmann F, Stalder U, Piotrowski M, Oecking C, Schaller A (2002) LeCPK1, a calcium-dependent protein kinase from tomato: plasma membrane targeting and biochemical characterization. Plant Physiol 129 156–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23 319–327 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Hata S, Sheen J, Izui K (1997) cDNA cloning and prokaryotic expression of maize calcium-dependent protein kinases. Biochim Biophys Acta 1350 109–114 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Kinoshita N, Ishiyama K, Hata S, Kyozuka J, Hayakawa T, Nakamura T, Shimamoto K, Yamaya T, Izui K (2001) A Ca2+-dependent protein kinase that endows rice plants with cold- and salt-stress tolerance functions in vascular bundles. Plant Cell Physiol 42 1228–1233 [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan PV, Cremo CR, Poovaiah BW (2000) Plant chimeric Ca2+/calmodulin-dependent protein kinase: role of the neural visinin-like domain in regulating autophosphorylation and calmodulin affinity. J Biol Chem 275 30417–30422 [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P (2000) SMART: a Web-based tool for the study of genetically mobile domains. Nucleic Acids Res 28 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J (1996) Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274 1900–1902 [DOI] [PubMed] [Google Scholar]
- Shen Q, Gomez-Cadenas A, Zhang P, Walker-Simmons MK, Sheen J, Ho TH (2001) Dissection of abscisic acid signal transduction pathways in barley aleurone layers. Plant Mol Biol 47 437–448 [DOI] [PubMed] [Google Scholar]
- Shi J, Kim K-N, Ritz O, Albrecht V, Gupta R, Harter K, Luan S, Kudla J (1999) Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11 2393–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderling TR, Stull JT (2001) Structure and regulation of calcium/calmodulin-dependent protein kinases. Chem Rev 101 2341–2352 [DOI] [PubMed] [Google Scholar]
- Sugden C, Crawford RM, Halford NG, Hardie DG (1999a) Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5′-AMP. Plant J 19 433–439 [DOI] [PubMed] [Google Scholar]
- Sugden C, Donaghy PG, Halford NG, Hardie DG (1999b) Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol 120 257–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezawa D, Ramachandiran S, Paranjape V, Poovaiah BW (1996) Dual regulation of a chimeric plant serine/threonine kinase by calcium and calcium/calmodulin. J Biol Chem 271 8126–8132 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler DA, Gordon JI, Adams SP, Glaser L (1988) The biology and enzymology of eukaryotic protein acylation. Annu Rev Biochem 57 69–99 [DOI] [PubMed] [Google Scholar]
- Urao T, Katagiri T, Mizoguchi T, Yamaguchi-Shinozaki K, Hayashida N, Shinozaki K (1994) Two genes that encode Ca2+-dependent protein kinases are induced by drought and high-salt stresses in Arabidopsis thaliana. Mol Gen Genet 244 331–340 [DOI] [PubMed] [Google Scholar]
- Vidal J, Chollet R (1997) Regulatory phosphorylation of C4 PEP carboxylase. Trends Plant Sci 2 230–237 [Google Scholar]
- Watillon B, Kettmann R, Boxus P, Burny A (1995) Structure of a calmodulin-binding protein kinase gene from apple. Plant Physiol 108 847–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo B-C, Harmon AC (1996) Intramolecular binding contributes to the activation of CDPK, a protein kinase with a calmodulin-like domain. Biochemistry 35 12029–12037 [DOI] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Alonso J, Ecker JR, Shinozaki K (2002) ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol 43 1473–1483 [DOI] [PubMed] [Google Scholar]
- Yuasa T, Takahashi K, Muto S (1995) Purification and characterization of a Ca2+-dependent protein kinase from the halotolerant green alga Dunaliella tertiolecta. Plant Cell Physiol 36 699–708 [Google Scholar]
- Zhang L, Liu BF, Liang S, Jones RL, Lu YT (2002) Molecular and biochemical characterization of a calcium/calmodulin-binding protein kinase from rice. Biochem J 368 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XS, Choi JH (2001) Molecular evolution of calmodulin-like domain protein kinases (CDPKs) in plants and protists. J Mol Evol 53 214–224 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Shewry PR, Jones H, Barcelo P, Lazzeri PA, Halford NG (2001) Expression of antisense SnRK1 protein kinase sequence causes abnormal pollen development and male sterility in transgenic barley. Plant J 28 431–441 [DOI] [PubMed] [Google Scholar]
- Zhu JK, Liu J, Xiong L (1998) Genetic analysis of salt tolerance in Arabidopsis. Evidence for a critical role of potassium nutrition. Plant Cell 10 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]