Abstract
The genome of Arabidopsis has been searched for sequences of genes involved in acyl lipid metabolism. Over 600 encoded proteins have been identified, cataloged, and classified according to predicted function, subcellular location, and alternative splicing. At least one-third of these proteins were previously annotated as “unknown function” or with functions unrelated to acyl lipid metabolism; therefore, this study has improved the annotation of over 200 genes. In particular, annotation of the lipolytic enzyme group (at least 110 members total) has been improved by the critical examination of the biochemical literature and the sequences of the numerous proteins annotated as “lipases.” In addition, expressed sequence tag (EST) data have been surveyed, and more than 3,700 ESTs associated with the genes were cataloged. Statistical analysis of the number of ESTs associated with specific cDNA libraries has allowed calculation of probabilities of differential expression between different organs. More than 130 genes have been identified with a statistical probability > 0.95 of preferential expression in seed, leaf, root, or flower. All the data are available as a Web-based database, the Arabidopsis Lipid Gene database (http://www.plantbiology.msu.edu/lipids/genesurvey/index.htm). The combination of the data of the Lipid Gene Catalog and the EST analysis can be used to gain insights into differential expression of gene family members and sets of pathway-specific genes, which in turn will guide studies to understand specific functions of individual genes.
Acyl lipids can be defined as fatty acids and their naturally occurring ester, ether, or amide derivatives. In plants, these include acylglycerols such as triacylglycerols (TAGs), phospholipids, galactolipids, and sulfolipids, plus sphingolipids, acylated steryl glycosides, oxylipins, cutins, suberins, estolides and wax, and sterol esters. The list may be extended if we consider molecules immediately derived from acyl groups, such as the epicuticular wax components (hydrocarbons, alcohols, ketones, and so on) or natural products such as anacardic acids that impart protection to predation. Polar lipids are amphipathic and as such self-associate in water to produce a variety of structures. Therefore, they provide the building blocks for biological membranes. There is substantial evidence indicating that the composition of acyl lipids in membranes influences the targeting, distribution, and functional properties of both integral and membrane-associated proteins (Sprong et al., 2001; Wallis and Browse, 2002). Furthermore, many polar lipids and the intermediates in their synthesis and degradation serve as signaling molecules. In summary, acyl lipids function in a wide range of biological processes, such as carbon and free energy storage, cell signaling, modulation of enzyme activity and protein localization, vesicle budding and fusion, waterproofing, and surface protection (Browse and Somerville, 1994).
Some acyl lipids such as TAGs, the major constituent of vegetable oils, are a primary agricultural or industrial commodity. Attempts to modify the quantity and the quality of acyl lipids in crops by metabolic engineering are underway but are hampered by the lack of knowledge of the regulation of the reaction pathways involved in both the anabolism and the catabolism (Gunstone and Pollard, 2001; Thelen and Ohlrogge, 2002).
The recent complete sequencing of the Arabidopsis genome should provide considerable new insights into the nature and the number of the proteins involved in the biosynthesis, modification, turnover, transportation, and degradation of acyl lipids in plants. However, the identification and functional annotation of genes in databases is sometimes incorrect or misleading due to the fact they are mostly performed automatically. More complete and valuable information about each gene can be obtained if sequences are examined more thoroughly by performing multiple alignments, conserved motif searches, and by carefully considering the biochemical information available in the literature. To improve the quality of the annotation of the Arabidopsis genes related to acyl lipid metabolism, a database providing data that are processed, annotated, and updated by researchers who have knowledge of the biology underlying a putative gene function is needed. Such a database should help accelerate the progress of plant lipid research by providing a common baseline of knowledge for the community, defining gaps in our knowledge, and highlighting where additional work is needed. The construction of databases specialized in one field of Arabidopsis biology is clearly a first step toward a functional catalog of the plant genome and a tool for the comparison of the metabolism and the biology within this field, between Arabidopsis and other plants, and between plants and non-plants. To improve the functional annotation of the genome, Munich Information Center for Protein Sequences (MIPS) and The Arabidopsis Information Resource databases have links to about 60 specialized databases that are dedicated to various gene families. However, none of these databases specifically concerns acyl lipid metabolism.
In 1999, when 70% of the Arabidopsis genome sequences were available, we produced a first catalog of plant genes involved in acyl lipid metabolism, which included mainly the biosynthetic reactions (Mekhedov et al., 2000; http://www.canr.msu.edu/lgc). Here, we present a second survey of lipid genes, which is based on the complete Arabidopsis genome and is substantially expanded to include almost all acyl lipid metabolism. Sterols and other isoprenoids have not been included. The number of cellular activities (i.e. reactions/type of proteins) covered by the survey has been increased more than 2-fold. Among the new genes, many encode poorly studied plant proteins such as lipolytic enzymes and lipid transfer proteins (LTPs).
Many efforts to understand the function of plant genes have used insertional mutants or other gene knockout or silencing strategies. Analysis of several hundred of such mutants indicates less than 5% show any visible phenotype (Bouché and Bouchez, 2001). Information about organ-specific expression, subcellular location, and possible biochemical activity can allow formulation of more specific hypotheses in the search for gene function. Therefore, in view of future studies on the function and organ specificity of gene family members, this survey also includes data on the number of expressed sequence tags (ESTs) associated with each gene and a statistical analysis of their distribution across organs. Also included are examples, comparisons, conclusions, and comments that can be made for specific lipid genes or, at the genome scale, for some lipid pathways, using both the Lipid Gene Catalog and the analysis of the organ distribution of ESTs. All of the gene and EST data for the 600 genes and 210 cellular activities surveyed here are available as a Web-based database, the Arabidopsis LipidGene(ALG)database(http://www.plantbiology.msu.edu/lipids/genesurvey/index.htm). We encourage readers to contact the authors with updates or corrections.
RESULTS AND DISCUSSION
The Lipid Gene Catalog
Nomenclature and Content
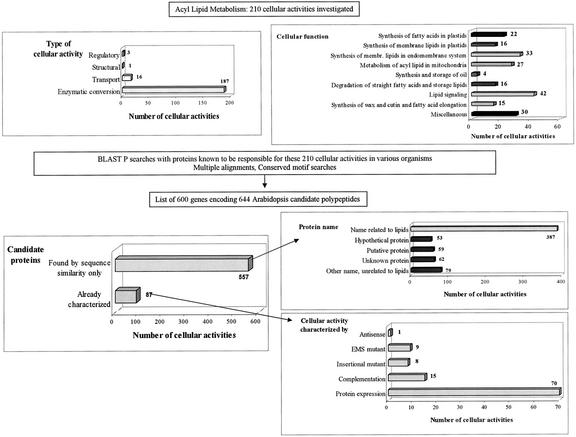
The census of genes involved in acyl lipid metabolism presented here is primarily based on sequence similarity searches using as queries only proteins whose cellular activity has been demonstrated (at least in vitro). Query proteins were from all organisms, but known plant proteins were preferentially used when available. This sequence similarity-based search was refined by multiple alignments and searches for conserved motifs when possible (see “Materials and Methods”). An overview of the gene survey and its results is shown in Figure 1.
Figure 1.
An overview of the Lipid Gene Catalog: construction and global final content. The complete content of the Lipid Gene Catalog is available in the ALG database (http://www.plantbiology.msu.edu/lipids/genesurvey/index.htm). For a summary of the type of information contained in the catalog, see Figure 2. For a summary of the candidates, see Table I.
We have surveyed 210 cellular activities that have been reported to exist in plants, in vitro at least (Table I). A cellular activity is defined here as the molecular task performed at the subcellular level by the individual product(s) of a gene or a group of genes. Therefore, plastidial lipoate synthase and mitochondrial lipoate synthase, for example, will be considered as two different cellular activities (Table I, F12 and M5). Cellular activities are grouped by the main cellular functions of acyl lipid metabolism. Some of these cellular functions might be restricted to one organelle (e.g. fatty acid synthesis in plastids), but others involve multiple cell compartments (e.g. lipid signaling). As would be expected for any list of components in a complex metabolic network, some of the classifications are arbitrary or debatable. This is particularly true for lipases and transacylases. For example, there are two cytosolic phospholipase A2 genes noted, which are placed under the heading “Lipid Signaling” (S11). However, the primary function of these gene products might alternatively fall within group E, “Synthesis of membrane lipids in the endomembrane system,” due to a role in membrane homeostasis (acyl group recycling). Eight additional cellular activities that have not been described in plants are also listed (code X) because we propose there might be some homologous proteins in plants (see next paragraph). Most of the 210 cellular activities are enzymatic conversions (187 of 210), but transport of acyl lipids (across membranes or within a cellular compartment or an extracellular fluid) and a few other functions (structural, like oleosins of oil bodies, or regulatory, like transcription factors) are also represented (Fig. 1). Six hundred Arabidopsis genes coding for the proteins that are known or thought to be responsible for one of the 210 cellular activities surveyed were found. For comparison, the number of Arabidopsis genes initially annotated as encoding proteins involved in metabolism is around 4,000 (Arabidopsis Genome Initiative, 2000). About 900 Arabidopsis proteins have been recently classified as enzymes involved in carbohydrate metabolism, which includes the enzymes of cell wall synthesis and degradation (Henrissat et al., 2001). The number of candidate genes and the corresponding number of ESTs found for each cellular activity of the Lipid Gene Catalog are listed in Table I. The identity and description of each gene and its encoded protein are given on the Web-based version of this survey (ALG database) as well as sequences, literature references, and other information where available. The structure and the content of the database are summarized in Figure 2.
Table I.
Summary of the lipid gene catalog
Bold font indicates cellular activities for which, to our knowledge, no Arabidopsis candidate genes were described or suggested in the literature so far.
| Cellular Function | Cellular Activity | No. of Genes | No. of ESTs |
|---|---|---|---|
| Synthesis of fatty acids in plastids | F1. Plastidial homomeric acetyl-CoA carboxylase (ACCase; EC 6.4.1.2) | 1 | 0 |
| F2a. α-Carboxyltransferase of heteromeric ACCase | 1 | 19 | |
| F2b. β-Carboxyltransferase of heteromeric ACCase (plastid encoded) | 1 | 0 | |
| F2c. Biotin carboxyl carrier protein of heteromeric ACCase | 2 | 68 | |
| F2d. Biotin carboxylase of heteromeric ACCase | 1 | 21 | |
| F3. Malonyl-CoA: acyl-carrier protein (ACP) malonyltransferase (EC 2.3.1.39) | 1 | 3 | |
| F4a. Ketoacyl-ACP synthase I (KAS I; EC 2.3.1.41) | 1 | 39 | |
| F4b. KAS II (EC 2.3.1.41) | 1 | 2 | |
| F4c. KAS III (EC 2.3.1.41) | 1 | 6 | |
| F5. Plastidial ketoacyl-ACP reductase (EC 1.1.1.100) | 5 | 35 | |
| F6. Plastidial hydroxyacyl-ACP dehydrase (EC 4.2.1.*) | 2 | 7 | |
| F7. Plastidial enoyl-ACP reductase (EC 1.3.1.9) | 1 | 11 | |
| F8. Stearoyl-ACP desaturase (EC 1.14.19.2) | 7 | 43 | |
| F9. Plastidial ACP | 5 | 42 | |
| F10a. Acyl-ACP thioesterase FatA (EC 3.1.2.14) | 2 | 5 | |
| F10b. Acyl-ACP thioesterase FatB (EC 3.1.2.*) | 1 | 35 | |
| F11a. Plastidial pyruvate dehydrogenase E1α of pyruvate DH complex (EC 1.2.4.1) | 2 | 46 | |
| F11b. Plastidial pyruvate dehydrogenase E1β of pyruvate DH complex (EC 1.2.4.1) | 2 | 36 | |
| F11c. Plastidial dihydrolipoamide acetyltransferase of PDH complex (EC 2.3.1.12) | 4 | 33 | |
| F11d. Plastidial dihydrolipoamide dehydrogenase of PDH complex (EC 1.8.1.4) | 2 | 20 | |
| F12. Plastidial lipoate synthase | 1 | 1 | |
| F13. Plastidial lipoyltransferase | 1 | 0 | |
| Synthesis of membrane lipids in plastids | P1. Plastidial dihydroxyacetone-phosphate reductase | 1 | 10 |
| P2. Plastidial glycerol-phosphate acyltransferase (GPAT; EC 2.3.1.15) | 1 | 2 | |
| P3. Plastidial acylglycerol-phosphate acyltransferase (LPAAT; EC 2.3.1.51) | 1 | 7 | |
| P4. Plastidial CDP-diacylglycerol synthetase (EC 2.7.7.41) | 3 | 4 | |
| P5. Plastidial phosphatidylglycerol-phosphate synthase (EC 2.7.8.5) | 1 | 3 | |
| P6. Plastidial phosphatidylglycerol-phosphate phosphatase (EC 3.1.3.27) | No candidate | ||
| P7. Phosphatidylglycerol desaturase (palmitate specific; FAD4; EC 1.14.99.*) | No candidate | ||
| P8. Plastidial oleate desaturase (FAD6; EC 1.14.99.*) | 1 | 9 | |
| P9. Plastidial linoleate desaturase (FAD7/FAD8; EC 1.14.99.*) | 2 | 18 | |
| P10. Plastidial phosphatidate phosphatase (EC 3.1.3.4) | 1 | 3 | |
| P11. Monogalactosyldiacylglycerol synthase (EC 2.4.1.46) | 3 | 5 | |
| P12. Monogalactosyldiacylglycerol desaturase (palmitate-specific, FAD5; EC 1.14.99.*) | 1 | 6 | |
| P13. Digalactosyldiacylglycerol synthase (EC 2.4.1.184) | 2 | 8 | |
| P14. UDP-sulfoquinovose synthase | 1 | 5 | |
| P15. Sulfolipid synthase | 1 | 4 | |
| P16. Plastidial 1-acylglycerophosphorylcholine acyltransferase (EC 2.3.1.23) | See E3 | ||
| Synthesis of membrane lipids in the endomembrane system | E1. Endoplasmic reticulum (ER) dihydroxyacetone-phosphate reductase | 4 | 13 |
| E2. ER GPAT (EC 2.3.1.15) | 2 | 2 | |
| E3. ER LPAAT (EC 2.3.1.51) | 11 | 5 | |
| E4. ER phosphatidate phosphatase (EC 3.1.3.4) | 2 | 2 | |
| E5. ER diacylglycerol cholinephosphotransferase (EC 2.7.8.2) | 1 | 11 | |
| E6. ER oleate desaturase (FAD2; EC 1.14.99.*) | 1 | 131 | |
| E7. ER linoleate desaturase (FAD3; EC 1.14.99.*) | 1 | 31 | |
| E8. ER CDP-diacylglycerol synthetase (EC 2.7.7.41) | 2 | 9 | |
| E9. ER phosphatidylglycerol-phosphate synthetase (EC 2.7.8.5) | 1 | 7 | |
| E10. ER phosphatidylglycerol-phosphate phosphatase (EC 3.1.3.27) | No candidate | ||
| E11. Phosphatidylinositol synthase (EC 2.7.8.11) | 2 | 6 | |
| E12. Phosphatidylserine synthase (EC 2.7.8.8) | No candidate | ||
| E13a. Choline kinase (EC 2.7.1.32) | 4 | 13 | |
| E13b. Ethanolamine kinase (EC 2.7.1.82) | 1 | 0 | |
| E14a. CDP-choline synthase (EC 2.7.7.15) | 2 | 8 | |
| E14b. CDP-ethanolamine synthase (EC 2.7.7.14) | 1 | 4 | |
| E15. Phosphatidylserine decarboxylase (EC 4.1.1.65) | 3 | 10 | |
| E16. Phospholipid base exchange | 1 | 1 | |
| E17a. LCB1 subunit of Ser palmitoyltransferase (EC 2.3.1.50) | 1 | 3 | |
| E17b. LCB2 subunit of Ser palmitoyltransferase (EC 2.3.1.50) | 2 | 10 | |
| E18. Ketosphinganine reductase (EC 1.1.1.102) | 2 | 18 | |
| E19. AcylCoA: sphinganine acyltransferase (EC 2.3.1.24) | No candidate | ||
| E20. Acyl-CoA-independent ceramide synthase | No candidate | ||
| E21. Sphingolipid hydroxylase | 2 | 5 | |
| E22. Sphingolipid Δ8 desaturase (EC 1.14.99.*) | 2 | 36 | |
| E23. Ceramide glucosyltransferase | No candidate | ||
| E24. Sphingolipid fatty acid hydroxylase | 2 | 13 | |
| E25. ER 1-acylglycerophosphorylcholinecholine acyltransferase (EC 2.3.1.23) | See E3 | ||
| E26. ER 2-acylglycerophosphorylcholine acyltransferase (EC 2.3.1.62) | See E3 | ||
| E27. ER 2-acylglycerol-phosphate acyltransferase (EC 2.3.1.52) | See E3 | ||
| E28. Phosphoethanolamine N-methyltransferase (EC 2.1.1.103) | 3 | 25 | |
| E29. Sphingolipid Δ4 desaturase (EC 1.14.99.*) | 1 | 0 | |
| X1. Acyl-ceramide synthase | 2 | 6 | |
| Metabolism of acyl lipids in mito chondria | M1. Mitochondrial KAS (EC 2.3.1.41) | 1 | 0 |
| M2. Mitochondrial ketoacyl-ACP reductase (EC 1.1.1.100) | 1 | 0 | |
| M3. Mitochondrial hydroxyacyl-ACP dehydrase (EC 4.2.1.17) | No candidate | ||
| M4. Mitochondrial enoyl-ACP reductase (EC 1.3.1.9) | No candidate | ||
| M5. Mitochondrial lipoate synthase | 1 | 18 | |
| M6. Mitochondrial lipoyltransferase | 2 | 0 | |
| M7. Mitochondrial phosphatidylglycerol-phosphate synthase | 1 | 3 | |
| M8. Mitochondrial phosphatidylglycerol-phosphate phosphatase | No candidate | ||
| M9. Cardiolipin synthase (EC 2.7.8*) | No candidate | ||
| M10a. α-Ketoacid decarboxylase E1α of BC ketoacid DH complex (EC 1.2.4.4) | 2 | 11 | |
| M10b. α-Ketoacid decarboxylase E1β of BC ketoacid DH complex (EC 1.2.4.4) | 2 | 8 | |
| M10c. Dihydrolipoamide transacylase of branched chain ketoacid DH complex | 1 | 23 | |
| M10d. Mitochondrial dihydrolipoamide dehydrogenase of BCKDH complex (EC 1.8.1.4) | 2 | 15 | |
| M11. Isovaleryl-CoA dehydrogenase (EC 1.3.99.10) | 1 | 2 | |
| M12a. Methylcrotonyl-CoA carboxylase (EC 6.4.1.4), biotinylated subunit | 1 | 4 | |
| M12b. Methylcrotonyl-CoA carboxylase (EC 6.4.1.4), non-biotinylated subunit | 1 | 8 | |
| M13. Mitochondrial enoyl-CoA hydratase (EC 4.2.1.17) | 2 | 1 | |
| M14. Mitochondrial GPAT (EC 2.3.1.15) | 3 | 11 | |
| M15. Mitochondrial LPAAT (EC 2.3.1.51) | See P3 and M14 | ||
| M16. Mitochondrial phosphatidate phosphatase (EC 3.1.3.4) | 1 | 0 | |
| M17. Mitochondrial ACP | 3 | 12 | |
| M18. Malonyl-CoA synthase | No candidate | ||
| M19. Mitochondrial malonyl-CoA: ACP malonyltransferase | No candidate | ||
| M20. Malonyl-ACP Synthase | No candidate | ||
| M21. Mitochondrial CDP-diacylgycerol synthetase (EC 2.7.7.41) | See P4 | ||
| M22. Mitochondrial 1-acylglycerophosphorylcholine acyltransferase (EC 2.3.1.23) | See E3 and M14 | ||
| X2. Mitochondrial diacylglycerol cholinephosphotransferase (EC 2.7.8.2) | 1 | 4 | |
| Synthesis and storage of oil | O1. Acyl-CoA: diacylglycerol acyltransferase (DAGAT; EC 2.3.1.20) | 2 | 7 |
| O2. Oil body oleosin | 8 | 132 | |
| O3. Caleosin | 7 | 42 | |
| O4. Phospholipid: diacylglycerol acyltransferase (PDAT; EC 2.3.1.158) | 3 | 9 | |
| Degradation of storage lipids and straight fatty acids | D1. Triacylglycerol lipase (EC 3.1.1.3) | 4 | 9 |
| D2. Monoacylglycerol lipase (EC 3.1.1.23) | 13 | 32 | |
| D3. Acyl-CoA oxidase (EC 1.3.3.6) | 6 | 61 | |
| D4. Peroxisomal enoyl-CoA hydratase (EC 4.2.1.17) | 2 | 6 | |
| D5. Hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35) | 1 | 10 | |
| D6. Ketoacyl-CoA thiolase (EC 2.3.1.16) | 3 | 65 | |
| D7a. Peroxisomal enoyl-CoA hydratase of multifunctional protein (EC 4.2.1.17) | 2 | 21 | |
| D7b. Hydroxyacyl-CoA dehydrogenase of multifunctional protein (EC 1.1.135) | |||
| D7c. Enoyl-CoA isomerase of multifunctional protein (EC 5.1.2.3) | |||
| D7d. Hydroxyacyl-CoA epimerase of multifunctional protein (EC 5.3.3.8) | |||
| D8. Dienoyl-CoA reductase (EC 1.3.1.34) | 1 | 4 | |
| D9. Hydroxyisobutyryl-CoA hydrolase (EC 3.1.2.4) | 3 | 3 | |
| D10. Fatty acid alcohol oxidase (EC 1.1.3.20) | 4 | 6 | |
| D11. Peroxisomal long-chain acyl-CoA synthetase | 2 | 7 | |
| D12. Peroxisomal fatty acid/acyl-CoA transporter | 1 | 8 | |
| X3. Acyl-CoA dehydrogenase (EC 1.3.99.2-3) | 1 | 5 | |
| Lipid signaling | S1. Diacylglycerol kinase (EC 2.7.1.107) | 9 | 33 |
| S2. Phosphatidylinositol-3-kinase (EC 2.7.1.137) | 1 | 2 | |
| S3a. Phosphatidylinositol-4-kinase-α (EC 2.7.1.67) | 2 | 6 | |
| S3b. Phosphatidylinositol-4-kinase-β (EC 2.7.1.67) | 2 | 0 | |
| S3c. Phosphatidylinositol-4-kinase-γ (EC 2.7.1.67) | 8 | 52 | |
| S4a. Phosphatidylinositol-phosphate kinase type IA (EC 2.7.1.68, 2.7.1.*) | 2 | 0 | |
| S4b. Phosphatidylinositol-phosphate kinase type IB (EC 2.7.1.68, 2.7.1.*) | 9 | 13 | |
| S4c. Phosphatidylinositol-phosphate kinase type III (EC 2.7.1.68, 2.7.1.*) | 4 | 38 | |
| S5. Phosphoinositide-specific phospholipase C (PI-PLC) (EC 3.1.4.11) | 9 | 24 | |
| S6. Nonspecific phospholipase C | 6 | 11 | |
| S7. Glycosylphosphatidylinositol-specific phospholipase C | 3 | 21 | |
| S8a. Phospholipase D-α (PLD-α; EC 3.1.4.4) | 4 | 35 | |
| S8b. PLD-β (EC 3.1.4.4) | 2 | 4 | |
| S8c. PLD-γ (EC 3.1.4.4) | 3 | 13 | |
| S8d. PLD-δ (EC 3.1.4.4) | 1 | 26 | |
| S8e. PLD-ζ (EC 3.1.4.4) | 2 | 10 | |
| S9. Phospholipase A1 (EC 3.1.1.32) | 1 | 2 | |
| S10. Secretory phospholipase A2 (PLA2; EC 3.1.1.4) | 4 | 2 | |
| S11. Cytosolic phospholipase A2 | 2 | 0 | |
| S12. Lysophospholipase (LysoPLA; EC 3.1.1.5) | 9 | 28 | |
| S13. Galactolipase (EC 3.1.1.26) | See S14 and S26 | ||
| S14. DAD1-like acylhydrolase | 12 | 54 | |
| S15a. Cytosolic lipoxygenase (EC 1.13.11.12) | 2 | 24 | |
| S15b. Plastidial lipoxygenase (EC 1.13.11.12) | 4 | 177 | |
| S16. Allene oxide synthase (EC 4.2.1.92) | 1 | 8 | |
| S17. Allene oxide cyclase (EC 5.3.99.6) | 4 | 16 | |
| S18. Oxo-phytodienoic acid reductase (OPR; EC 1.3.1.42) | 5 | 41 | |
| S19. Hydroperoxide lyase | 1 | 5 | |
| S20. Fatty acid amide hydrolase | 3 | 18 | |
| S21. N-acylphosphatidylethanolamine synthase | No candidate | ||
| S22a. α-Dioxygenase-peroxidase (involved in fatty acid α-oxidation) | 2 | 30 | |
| S22b. NAD+ oxidoreductase (involved in fatty acid α-oxidation) | 1 | 28 | |
| S23a. PTEN-like phosphoinositide 3-phosphatase | 3 | 5 | |
| S23b. Myotubularin-like phosphoinositide 3-phosphatase | 2 | 3 | |
| S24a. Type II phosphoinositide 5-phosphatase | 4 | 11 | |
| S24b. Sac domain-containing phosphoinositide phosphatase | 9 | 26 | |
| S25. Phosphoinositide 4-phosphatase | No candidate | ||
| S26. Patatin-like acyl-hydrolase | 12 | 44 | |
| S27. Sulfolipase | See S26 | ||
| S28. Peroxygenase | No candidate | ||
| S29. Hydroperoxide reductase | No candidate | ||
| S30. Epoxy alcohol synthase | No candidate | ||
| X4. Phospholipase A2-activating protein | 1 | 7 | |
| Fatty acid elongation and wax and cutin metabolism | W1. Ketoacyl-CoA synthase (KCS) | 20 | 183 |
| W2. Ketoacyl-CoA reductase | 2 | 47 | |
| W3. Hydroxyacyl-CoA dehydrase (EC 4.2.1.17) | No candidate | ||
| W4. Enoyl-CoA reductase (EC 1.3.1.44) | 1 | 31 | |
| W5. Fatty acyl-CoA reductase (EC 1.1.1.*) | 8 | 8 | |
| W6. Wax synthase (EC 2.3.1.75) | 12 | 0 | |
| W7. Aldehyde decarbonylase CER1 (EC 4.1.99.5) | 5 | 36 | |
| W8. Putative transcription factor CER2 | 3 | 28 | |
| W9. CER3 protein | 1 | 7 | |
| W10. Wax ester hydrolase (EC 3.1.1.50) | No candidate | ||
| W11. Fatty acid ω-hydroxylase | 8 | 51 | |
| W12. Aldehyde-forming fatty acyl-CoA reductase | No candidate | ||
| W13. Secondary alcohol-forming hydroxylase | No candidate | ||
| W14. Ketone-forming oxidase | No candidate | ||
| X5. ELO-like elongase | 4 | 10 | |
| Miscellaneous | Z1a. LTP1 | 14 | 362 |
| Z1b. LTP2 | 8 | 28 | |
| Z1c. LTP3 | 14 | 17 | |
| Z1d. LTP4 | 2 | 9 | |
| Z1e. LTP5 | 29 | 94 | |
| Z1f. LTP6 | 2 | 2 | |
| Z1g. LTP7 | 1 | 3 | |
| Z1h. LTP8 | 1 | 0 | |
| Z2. Acyl-CoA desaturase like (EC 1.14.99.*) | 8 | 48 | |
| Z3. ATP citrate lyase (EC 4.1.3.8) | 5 | 94 | |
| Z4. Pollen surface oleosin | 8 | 53 | |
| Z5. Acyl-CoA thioesterase (EC 3.1.2.2, EC 3.1.2.18-20) | 4 | 7 | |
| Z6. Malonyl-CoA decarboxylase (EC 4.1.1.9) | 1 | 3 | |
| Z7. Translocase | 12 | 28 | |
| Z8. Acyl-CoA-binding protein | 5 | 32 | |
| Z9. Sec14-like protein | 2 | 18 | |
| Z10. Plastid lipid-associated protein | 3 | 24 | |
| Z11a. Plastidial long-chain acyl-CoA synthetase (EC 6.2.1.3) | 3 | 10 | |
| Z11b. Long-chain acyl-CoA synthetase (other than plastidial or peroxisomal) | 6 | 53 | |
| Z12. Plastidial ABC acyl transporter | 1 | 1 | |
| Z13. Epoxide hydrolase (EC 3.3.2.3) | 6 | 46 | |
| Z14. Acetyl-CoA synthetase (AMP forming; EC 6.2.1.1) | 3 | 15 | |
| Z15. Holo-ACP synthase (EC 2.7.8.7) | 2 | 2 | |
| Z16. Cytosolic homomeric ACCase | 1 | 8 | |
| Z17. Sphingosine transfer protein | 2 | 9 | |
| Z18. Protein N-myristoyltransferase | 2 | 13 | |
| Z19. Lipid acylhydrolase like (EC 3.1.1.*) | 21 | 25 | |
| X6. Cyclopropane fatty acid synthase (EC 2.1.1.79) | 3 | 6 | |
| X7. PPT1-like thioesterase (EC 3.1.1.22, EC 3.1.2.*) | 7 | 44 | |
| X8. Glycerophosphoryl diester phosphodiesterase (EC 3.1.4.46) | 2 | 6 | |
| Total | 600 | 3,750 |
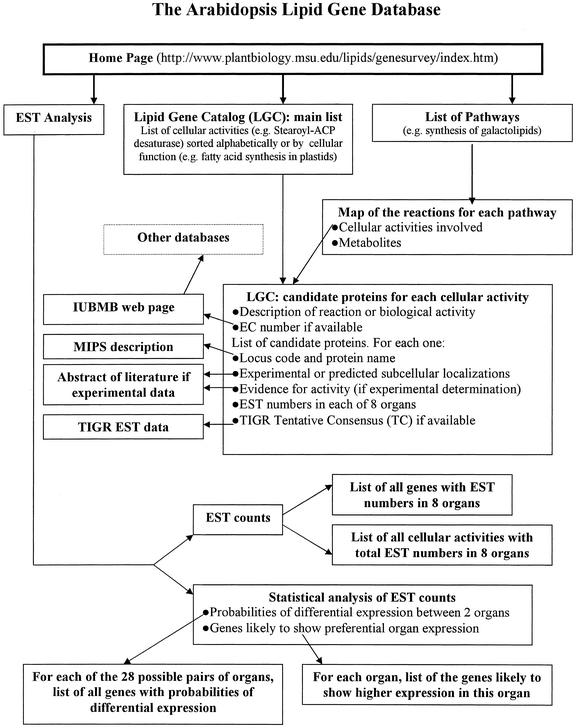
Figure 2.
Structure and content of the Web-based ALG database. Each box represents a different Web page. Arrows indicate links between pages. For each enzyme, there is a link to the corresponding Web page of the International Union of Biochemistry and Molecular Biology (http://www.chem.qmw.ac.uk/iubmb) when available. The International Union of Biochemistry and Molecular Biology Web pages have links to many other databases. TIGR and MIPS databases: see “Materials and Methods” for abbreviations and Web addresses.
Based on the Tentative Consensus data of TIGR, 42 genes (i.e. 7%) have putative alternative splice forms, including 40 genes with two forms and two genes with three forms. The total number of predicted proteins encoded by the 600 lipid genes of the catalog, therefore, is 644. The percentage of putative splice forms is also about 7% for the whole genome according to TIGR's data. Only 14% (87) of the 644 proteins have been experimentally demonstrated to have an activity related to acyl lipid metabolism (Fig. 1). Thus, 86% (557) of the gene products represent uncharacterized proteins that are identified based on sequence similarity and/or presence of conserved domains to characterized proteins from other species. The cellular activity of most (about 80%) of the characterized proteins was demonstrated by protein expression in a heterologous host, and 14 proteins were characterized by two or more methods (in most cases, characterization of an ethyl methanesulfonate mutant and functional complementation).
What Is New or Improved?
The number of cellular activities covered by this survey as compared with our previous survey has been increased from 71 to 210, representing 600 genes, i.e. around 2.4% of the total number of predicted genes in Arabidopsis. To our knowledge, this is one of the most complete and extensive effort to improve the genome annotation in a specific field of Arabidopsis biology. Examples of catalogs of similar size include: about 900 carbohydrate-active enzymes (http://afmb.cnrs-mrs.fr/~cazy/CAZY/index.html; Henrissat et al., 2001), 320 receptor kinase-like proteins (http://www.cbs.umn.edu/arabidopsis/; Ward, 2001), and 260 cytochromes P450 (http://Arabidopsis-p450.biotec.uiuc.edu/).
The Lipid Gene Catalog now covers most of the known cellular activities of acyl lipid metabolism, including many that are poorly characterized (e.g. many lipolytic activities) or were recently described, such as DAD1-like acylhydrolase (Ishiguro et al., 2001). Table I gives the 11 cellular activities of acyl lipid metabolism (indicated in bold) for which, to our knowledge, no Arabidopsis candidate genes were described or suggested in the literature so far. In Table I, there are also over 40 cellular activities for which we suggest new candidates in addition to the ones that were already known, found by the genome annotation process, or mentioned in the literature. Some of these new candidates could not be identified by global sequence similarity but only by searches of specific conserved motifs (e.g. some of the 19 acyltransferases of the catalog, including the two candidates for the endoplasmic acyl-CoA:glycerol-phosphate acyltransferase and the first enzyme of the endoplasmic glycerolipid synthesis pathway).
Approximately 40% (253) of the predicted proteins found in this survey were annotated as unknown, putative, or hypothetical proteins without any indication of similarity or were given a name unrelated to acyl lipids (Fig. 1) and, therefore, could not have been related to acyl lipid metabolism solely based on the description given by MIPS or other databases. The annotation of these “anonymous” proteins was improved in this study by identifying their similarities to characterized proteins that were not available or not found at the time the annotation of the genome was performed.
An example of a group of genes for which the annotation has been much improved by this survey is the genes involved in the degradation of lipids, a field which is much less studied than the biosynthesis. In the catalog, genes encoding putative lipolytic enzymes represent about 20% of the total. Although many uncertainties remain in our classification as explained in the next section, we have clarified the annotation of this group of enzymes by a careful examination of the confusing biochemical literature describing the characterization of putative lipolytic enzymes. Only proteins clearly demonstrated as lipases (i.e. by using natural lipids as substrates) were used as queries in BLAST searches. Conserved motif searches were useful also to discard some candidate genes or refine the classification. Many sequences released in databases in recent years have been annotated as “lipases” because of sequence similarity with a group of bacterial and plant proteins originally named so (Brick et al., 1995). This group of proteins was termed “lipases” due to the fact that some of them were shown to hydrolyze artificial substrates, such as paranitrophenyl esters, that are insoluble or partially soluble in water. In fact, some other proteins, even nonenzymatic ones, can hydrolyze such artificial substrates (Beisson et al., 2000). In addition, the GXSXG motif is often considered as a signature of “lipases,” although it is encountered in various proteins that do not act on lipids (Ollis et al., 1992). Even if we take the term lipase in its broader sense (proteins catalyzing the breakdown of any covalent bond in an acyl lipid), a group of proteins bearing the GXSXG motif should not be called lipases if there are no experimental data showing that they can hydrolyze natural acyl lipids with a significant specific activity. Although some so-called “lipases” will turn out to be so, many others will likely turn out to bear a different hydrolytic activity, if any. Therefore, we have not included the many proteins showing this motif but with no biochemical activity characterized. Based on these considerations, we have been able to discard many candidates, and in our final classification, we ended up with 112 genes encoding putative lipases representing 19 different characterized lipolytic activities ranging from TAG lipase to phospholipase D.
LTPs can bind acyl lipids in vitro. It is the largest group in the catalog and represents more than 10% of all proteins. One physiological function of LTPs is thought to occur in plant defense mechanisms (Blein et al., 2002). The well-known motif of eight conserved Cys (Kader, 1996) has been used to retrieve the candidate LTPs, and the 71 putative LTPs found have been classified into eight groups (LTP1–8) based on the number of amino acids between the fourth and fifth Cys in the core of the motif. This tentative classification might turn out to group proteins with diverse functions. Nevertheless, in absence of any demonstrated cellular activities for LTPs in vivo (see next paragraph), it has the advantage of being simple and to include without modifications the two first groups of LTPs described so far (Arondel et al., 2000; Douliez et al., 2001). The LTP At5g48485 recently shown to be involved in systemic resistance signaling (Maldonado et al., 2002) belongs to a major group (LTP3) of the classification proposed here and is, to our knowledge, its only member to be characterized.
Finally, as mentioned above, we also have found in the predicted proteome of Arabidopsis some candidates (i.e. homologs of known proteins) for cellular activities that, to our knowledge, have never been reported in plants (coded X in Table I). Some of these candidates suggest, for example, the presence in plants of previously undescribed acyl-lipids (activity X1 and acyl-ceramides) or support the autonomy of organelles for the biosynthesis of some membrane lipids (activity X2 and the biosynthesis of phosphatidylcholine in mitochondria). In the case of the putative cyclopropane fatty acid synthase activity (X6), the expected product has not been detected in Arabidopsis (X. Bao, personal communication). Therefore, the candidate proteins might be responsible for a related biochemical activity yet to be discovered or might be synthesized under some special physiological conditions.
What Is Missing or Uncertain?
As can be seen in Table I, there are still 30 cellular activities of acyl lipid metabolism for which no good candidate genes could be found. For most of them, there were sequences reported in non-plant organisms but no clear homologous gene in Arabidopsis. In some cases, the missing genes for a given activity may be listed as candidates for another related activity (e.g. acylglycerophosphorylcholine acyltransferases and acylglycerol-phosphate acyltransferases) or for the same activity but in another organelle (e.g. CDP-diacylglycerol synthetase located in plastids, mitochondria, and ER). It should be pointed out that the organelle classification relies mainly on subcellular localization prediction software. When no full-length cDNA is available, these predictions also rely on the correct prediction of the first exon of the sequence by gene prediction algorithms. Although subcellular localization predictions were performed using the software targetP, which is considered as one of the best algorithms for this purpose, false positives and false negatives must be expected (Emanuelsson et al., 2000). In the ALG database, the localization predicted by targetP and the reliability of this prediction are given for each protein. Finally, it is known that some localizations are badly predicted by all algorithms, especially in the case of a dual targeting to plastids and to mitochondria, which could be a mechanism more common than previously thought (Peeters and Small, 2001). Therefore, some candidate proteins might turn out to be also targeted to mitochondria.
Another main source of uncertainty in the catalog is the fact that sequence homology alone is sometimes not sufficient to predict the correct function of the protein. There are clear examples of homologous enzymes catalyzing different reactions (Gerlt and Babbitt, 2000). However, prediction of function using a sequence similarity approach has proven to be powerful, and is now the base of COG (Clusters of Orthologous Groups of proteins), an important new method for the functional annotation of newly sequenced genomes (Tatusov et al., 1997). Sequence homology is especially useful in the case of housekeeping genes. Because many of the cellular activities involved in the metabolism of the major acyl lipids are evolutionarily conserved housekeeping functions, the annotation of these genes, therefore, is less likely to be wrong than, for example, for genes involved in secondary metabolism. Sequence similarity is also known to give its best results for species that are phylogenetically not too distant (Tatusov et al., 1997); therefore, it is in cases where no plant sequences were available that the most uncertainties remain. The group of monoacylglycerol lipases is a good example. We have listed the 13 Arabidopsis proteins showing the highest sequence similarity with the mouse (Mus musculus) protein characterized as a monoacylglycerol lipase. The Arabidopsis protein(s) responsible for this activity are probably among these 13 candidates. However, it would be surprising that there are so many monoacylglycerol lipases in Arabidopsis. Subtle amino acid changes could account for different substrate specificity, and some of these proteins might be lysophospholipases, for example. Monoacylglycerol lipases and lysophospholipases have closely related sequences (Karlsson et al., 1997), and the lack of characterized proteins in plants makes the classification difficult.
As can be seen in Table I, only three cellular activities of the catalog are classified as translocase, and many other candidates involved in the translocation of lipids between membranes are probably still to be discovered. Two of the three translocase functions (D12 and Z12 but not Z7) are ATP-binding cassette transporter genes. The ATP-binding cassette transporter gene superfamily of Arabidopsis contains 129 genes, so this class may be fertile ground for the discovery of lipid transporters. As for LTPs, it should be stressed that it is a group of proteins whose sequences are highly divergent, and their functions might be very diverse. Moreover, the LTPs characterized so far can transfer acyl lipids between vesicles in vitro (Kader, 1996), but they are extracellular (Thoma et al., 1993) and their involvement in the in vivo binding of cutin monomers or other extracellular lipids still awaits experimental confirmation. Therefore, it cannot be ruled out that none of the putative LTPs, or only a few of them, will turn out to be involved in acyl lipid binding in vivo.
About two-thirds of the 168 cellular activities for which candidate genes are available are represented by more than one gene. This apparent structural redundancy may be due to an overestimated number of candidates in those cases with many uncertainties (e.g. monoacylglycerol lipases), but it could also reflect a true functional redundancy (duplication of essential housekeeping genes) or simply be due to the expression of different members of a gene family in different parts of the plant, at different developmental stages.
Finally, we note the boundaries for this catalog. First, there are a large number of pathways supplying cofactors and non-lipidic substrates for reactions and the electron transport chain components providing reducing power for desaturation reactions, particularly the ferredoxin-ferredoxin reductase system in plastids and cytochrome b5 and cytochrome b5 reductase of the endomembrane system. Only where these are completely specific for lipids, such as in the synthesis of UDP-sulfoquinovose (Table I, P14), or where they are so central to lipid metabolism, even though they more rightly occupy a place in general metabolism, have these been included (i.e. the pathways to the synthesis of malonyl-CoA, F1, F2, F11, F12, F13, M5, M6, M10, Z3, and Z14, and key glycerolipid synthesis precursors, E1, E13, and E14). There is only one transcription factor included (W9).
Analysis of the Distribution of ESTs in Organs
Purpose
Often, information about organ expression patterns for a gene provides clues about the function of the gene. In addition, the estimation of the organ-specific or organ-preferential expression of a gene can be of help in: (a) the identification of the isoforms of lipid genes involved in metabolic pathways that occur predominantly or specifically in some organs (e.g. TAG synthesis in seeds), (b) the design of knock out strategies in the case of gene families, and (c) the finding of phenotypes in KO mutants (most of which do not show apparent phenotypes at the whole plant level in normal growth conditions; Bouché and Bouchez, 2001).
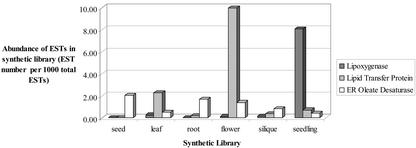
Because relative levels of gene expression can be estimated by using the frequency of gene transcripts in unbiased cDNA libraries (Okubo et al., 1992), the analysis of EST abundance in libraries made from a single organ is likely to provide some insights into the differential expression of lipid genes. This is illustrated in Figure 3 with the three proteins showing the highest numbers of total ESTs (147, 146, and 131) in the Lipid Gene Catalog. The distribution of ESTs in libraries made from the same organ shows that the lipoxygenase and the LTP are not likely to be expressed in all organs, unlike the ER oleate desaturase, a well-known housekeeping gene of lipid metabolism. It seems that their high EST number is due to a preferential expression in flowers (and maybe leaves) for the LTP and in seedling for the lipoxygenase. In these two organs, the EST frequency of the genes is at least 20 times higher than in most other organs, whereas the relative EST abundance of the ER oleate desaturase varies at most 4 to 5 times between organs. This example shows that a systematic analysis of EST abundance might be useful as a first approximation to evaluate the spatiotemporal differential expression of lipid genes and reveal a possible preferential expression. However, due to random fluctuation in EST numbers and differences in the size of libraries, EST counts in synthetic libraries (or even frequencies like in Fig. 3) cannot be rigorously compared directly. A statistical analysis is required. Therefore, the numbers of ESTs in different organs for each gene of the catalog and the results of the statistical analysis are given in the ALG database. In the following section, a few examples will illustrate how the analysis was performed and what can be expected from it.
Figure 3.
EST frequencies in organs (synthetic libraries) for the three proteins of acyl lipid metabolism showing the highest number of total ESTs. Synthetic libraries group cDNA libraries made from the same organ. Total numbers of ESTs in the synthetic libraries are: seed, 11,155; leaf, 3,944; root, 21,515; flower, 7,935; silique, 13,382; and seedling, 7,050. The proteins are: lipoxygenase At3g45140 (147 total ESTs), LTP1 At2g38540a (146 total ESTs), and endoplasmic oleate desaturase At3g12120 (131 total ESTs).
Some Examples of the Methods and the Results
For each of the 600 genes of the Lipid Gene Catalog, the TIGR Tentative Consensus data were retrieved as described in “Materials and Methods” and recorded in the catalog. Table I gives only the total number of ESTs for the cellular activities, but the complete EST data for each gene are available in the ALG database. In particular, for all genes, the number of ESTs associated with cDNA libraries from specific organs was determined. Because Arabidopsis ESTs have been derived from over 50 separate cDNA libraries, we have grouped these into eight virtual synthetic libraries that group the libraries made from the same organ (see “Material and Methods”). These synthetic libraries have recently been used to investigate the possible organ preferential expression of Arabidopsis plastid inner envelope proteins (Koo and Ohlrogge, 2002; http://www.plantbiology.msu.edu/PlastidEnvelope/index.htm). The cDNA libraries used to make the virtual “synthetic libraries” are also described in the ALG database. Of approximately 110,000 ESTs used, more than 3,700 of them (3.4%) are associated with the lipid genes of the catalog.
The EST counts of a lipid gene in each pair of synthetic libraries (organs) have been statistically compared using the version of the method developed by Audic and Claverie (1997) that is applied to libraries of different size. This statistical method was found to be the most efficient in pair-wise comparisons of EST abundance (Romualdi et al., 2001). It allows the calculation of the probability of differential expression of a given gene between two libraries, taking into account the number of ESTs in each library and the size of the two libraries. As an example, Table II presents a comparison of seeds versus leaves. The probability calculated can be used to rank genes, without discarding any gene a priori. The end user can then decide how many genes can be retained (e.g. only genes with a probability greater than 0.99, etc.). Table II displays all the genes showing a probability > 0.9 and a few others showing a lower probability. It can be seen clearly from this table that the statistical estimation of the likelihood of differential expression between two organs can avoid drawing hazardous conclusions from the direct comparison of EST counts or EST frequencies. For example, four counts in the 11,155 seed ESTs and zero in the 3,944 leaf ESTs could equally reflect a true differential expression between these two organs or a random fluctuation in EST counts. Therefore, in absence of any statistical method, the values of EST counts (or even frequencies) should not be taken as an estimate of levels of gene expression and used directly to compare the expression of different genes or to estimate how EST data compare with other quantitative or semiquantitative methods such as northern blots (O'Hara et al., 2002), especially when dealing with small EST numbers. Finally, it should be stressed that the probability of the statistical method used here is influenced not only by the EST number but also by the size of the library (for example, see Table II At2g19450 and At3g18280, two genes with the same number of ESTs but a different probability).
Table II.
EST counts and probabilities of differential expression between seeds and leaves
All genes showing a high probability of differential expression between seeds and leaves (P > 0.9), but only a few genes with a low probability (P < 0.9), are shown here. Bold font, Up-regulation in seeds. Underlined font, Up-regulation in leaves. Microarray ratios of genes upregulated in seeds (P > 0.9) were calculated from data by Girke et al. (2000; http://www.bpp.msu.edu/Seed/SeedArray.htm). Ratios based on data from a single spot on the array are marked with an asterisk. Other ratios were calculated from data of two to seven spots.
| Putative Cellular Activity | Protein | No. of Seed ESTs | No. of Leaf ESTs | Probability | Microarray Average Ratio (Seed to Leaf) |
|---|---|---|---|---|---|
| Oil-body oleosin | At3g27660 | 47 | 1 | 0.999 | 41.2 |
| Oil-body oleosin | At5g40420 | 39 | 0 | 0.999 | 45.9 |
| Caleosin | At4g26740 | 21 | 0 | 0.997 | 25.1 |
| KCS | At4g34520 | 18 | 0 | 0.993 | 7.2* |
| Plastidial pyruvate dehydrogenase E1α | At1g01090 | 16 | 0 | 0.98 | 1.9 |
| ATP citrate lyase | At1g10670 | 14 | 0 | 0.97 | 1* |
| KCS | At2g26250 | 14 | 0 | 0.97 | 0.7 |
| Oil-body oleosin | At3g01570 | 14 | 0 | 0.97 | Not available |
| ER oleate desaturase | At3g12120 | 23 | 2 | 0.96 | 2.0 |
| KAS I | At5g46290 | 11 | 0 | 0.94 | 2.4 |
| Lipoxygenase | At3g22400b | 11 | 0 | 0.94 | 1.9 |
| α-Dioxygenase-peroxidase | At1g73680 | 10 | 0 | 0.92 | 0.7 |
| ER linoleate desaturase | At2g29980 | 9 | 0 | 0.9 | 2.5 |
| Long-chain acyl-CoA synthetase | At2g47240 | 9 | 0 | 0.9 | 1.3* |
| Oil-body oleosin | At4g25140 | 9 | 0 | 0.9 | 56.5* |
| Stearoyl-ACP desaturase | At2g43710 | 9 | 0 | 0.9 | 1.1* |
| LTP1 | At2g38540a | 0 | 9 | 0.999 | |
| LTP1 | At5g59310 | 0 | 6 | 0.999 | |
| LTP1 | At5g59320 | 0 | 8 | 0.999 | |
| LTP2 | At3g18280 | 0 | 3 | 0.99 | |
| Lysophospholipase | At3g15650 | 0 | 2 | 0.96 | |
| Phosphatidylinositol-4-kinase γ | At1g26270 | 0 | 2 | 0.96 | |
| Stearoyl-ACP desaturase | At3g02630 | 6 | 0 | 0.7 | |
| Plastidial ACP | At1g54580 | 1 | 1 | 0.6 | |
| DAD1-like acylhydrolase | At4g16820 | 4 | 0 | 0.5 | |
| Ketoacyl-CoA thiolase | At2g33150 | 14 | 3 | 0.4 | |
| Acyl-CoA: DAGAT | At2g19450 | 3 | 0 | 0.4 | |
| Plastidial ACP | At3g05020 | 2 | 0 | 0.1 |
A cutoff value of 0.9 was used for the probabilities in Table II, but a different one could have been chosen, depending on the aims and the intuition of the user. The lower the threshold probability is, more false positives (genes not truly differentially expressed) and the lower the number of false negatives (genes truly differentially expressed but discarded) will occur in the list (Audic and Claverie, 1997). In the group of genes likely to be differentially expressed in seeds and leaves (Table II), some well-known seed-specific genes are present as expected (e.g. oleosins), but some other uncharacterized genes are revealed by the analysis (e.g. the lipoxygenase At3g22400). Furthermore, most of the genes likely to be up-regulated in seeds as compared with leaves (P > 0.9) also show a significant up-regulation (ratio > 1.9) in microarray experiments. Ratios marked with an asterisk are based on data from a single spot on the array and should be taken cautiously. Thus, there are only two genes showing clear discrepancies with the EST statistical analysis: KCS (At4g26740) and α-dioxygenase-peroxidase (At1g73680). These genes could be false positive in the statistical analysis (Audic and Claverie, 1997). An alternative hypothesis is that, due to cDNA cross hybridizations, the microarray ratios of these genes are an average value of intensities from different homologous genes. Interestingly, the only study involving a detailed comparison of EST abundance analysis versus cDNA arrays has shown that both methods gave qualitatively similar patterns of gene expression during maize (Zea mays) embryo development (Lee et al., 2001). For discussion about the limitations of microarray analysis, see the Arabidopsis seed array Web site (http://www.bpp.msu.edu/Seed/SeedArray.htm).
To include in a list of candidates (such as the one of Table II) more of the false negatives that have been discarded, it may be useful to perform the same analysis for the other combinations of organs (e.g. seed versus other organs than leaves). It is unlikely that genes preferentially expressed in seeds do not show a high probability (e.g. >0.95) in at least one of the five combinations of organs. This is illustrated in Table III, which gives a list of genes that show a probability > 0.95 of being more expressed in seeds than in at least one other organ. More genes known to show differential expression in seeds versus leaves are now listed, but an increase in the number of false positives is also expected.
Table III.
Candidate genes likely to be up-regulated in seeds as compared with at least one other organ
Only genes showing probabilities of higher expression in seeds above 0.95 (bold values) are listed here.
| Cellular Functions (See Table I) | Putative Cellular Activity | Protein | Probabilities (Seed vs. Other Organ)
|
|||||
|---|---|---|---|---|---|---|---|---|
| Leaf | Root | Flower | Silique | Seedling | Mixed | |||
| FAS | Plastidial pyruvate dehydrogenase E1 alpha subunit | At1g01090 | 0.98 | 0.999 | 0.998 | 0.97 | 0.999 | 0.999 |
| Plastidial pyruvate dehydrogenase E1 beta subunit | At1g30120 | 0.5 | 0.99 | 0.1 | 0.7 | 0.97 | 0.996 | |
| Plastidial dihydrolipoamide acetyltransferase, PDH complex | At3g25860 | 0 | 0.6 | 0.1 | 0.4 | 0.4 | 0.97 | |
| Plastidial dihydrolipoamide dehydrogenase, PDH complex | At3g16950 | 0.5 | 0.8 | 0.8 | 0.2 | 0.8 | 0.97 | |
| ACCase, biotin carboxyl carrier protein | At5g15530a | 0.7 | 0.6 | 0.95 | 0.9 | 0.93 | 0.94 | |
| KAS I | At5g46290 | 0.94 | 0.8 | 0.98 | 0.96 | 0.994 | 0.997 | |
| KAS III | At1g62640b | 0.1 | 0.92 | 0.6 | 0.8 | 0.5 | 0.98 | |
| Plastidial ketoacyl-ACP reductase | At1g24360a | 0.7 | 0.91 | 0.8 | 0.2 | 0.93 | 0.98 | |
| Plastidial ketoacyl-ACP reductase | At1g24360b | 0.4 | 0.92 | 0.6 | 0.8 | 0.5 | 0.98 | |
| Stearoyl-ACP desaturase | At2g43710 | 0.9 | 0.995 | 0.95 | 0.95 | 0.92 | 0.996 | |
| Stearoyl-ACP desaturase | At3g02630 | 0.7 | 0.998 | 0.95 | 0.96 | 0.93 | 0.999 | |
| Plastidial ACP | At3g05020 | 0.1 | 0.5 | 0.6 | 0.8 | 0 | 0.98 | |
| P | Plastidial dihydroxyacetone-phosphate reductase | At5g40610 | 0.6 | 0.98 | 0.7 | 0.98 | 0.8 | 0.993 |
| Digalactosyldiacylglycerol synthase | At3g11670 | 0.1 | 0.92 | 0.6 | 0.8 | 0.5 | 0.98 | |
| ER | ER dihydroxyacetone-phosphate reductase | At2g41540a | 0.4 | 0.97 | 0.3 | 0.91 | 0.7 | 0.996 |
| ER dihydroxyacetone-phosphate reductase | At2g41540b | 0.4 | 0.97 | 0.7 | 0.91 | 0.7 | 0.996 | |
| ER oleate desaturase | At3g12120 | 0.96 | 0.5 | 0.7 | 0.99 | 0.996 | 0.993 | |
| ER linoleate desaturase | At2g29980 | 0.9 | 0.995 | 0.95 | 0.6 | 0.8 | 0.996 | |
| ER CDP-diacylglycerol synthase | At1g62430 | 0.5 | 0.99 | 0.8 | 0.8 | 0.8 | 0.97 | |
| Mito | α-Ketoacid decarboxylase E1a subunit, BCKDH Complex | At5g09300 | 0 | 0.99 | 0.8 | 0.8 | 0.8 | 0.999 |
| Dihydrolipoamide transacylase, BCKDH complex | At3g06850 | 0.6 | 0.95 | 0.4 | 0.4 | 0.8 | 0.8 | |
| Mitochondrial GPAT | At1g06520 | 0.1 | 0.5 | 0.6 | 0.8 | 0.5 | 0.98 | |
| Mitochondrial lipoate synthase | At2g20860 | 0 | 0.99 | 0.8 | 0.2 | 0.8 | 0.7 | |
| Oil Synth. and Stor. | Acyl-CoA: DAGAT | At2g19450 | 0.4 | 0.97 | 0.7 | 0.91 | 0.7 | 0.996 |
| Oil body oleosin | At3g01570 | 0.97 | 0.999 | 0.999 | 0.999 | 0.998 | 0.999 | |
| Oil body oleosin | At3g18570 | 0.1 | 0.92 | 0.6 | 0.8 | 0.5 | 0.98 | |
| Oil body oleosin | At3g27660 | 0.999 | 0.999 | 0.999 | 0.999 | 0.999 | 0.999 | |
| Oil body oleosin | At4g25140 | 0.9 | 0.999 | 0.99 | 0.9 | 0.98 | 0.999 | |
| Oil body oleosin | At5g40420 | 0.999 | 0.999 | 0.999 | 0.999 | 0.999 | 0.999 | |
| Caleosin | At1g70670 | 0.4 | 0.5 | 0.6 | 0.1 | 0.5 | 0.98 | |
| Caleosin | At4g26740 | 0.997 | 0.999 | 0.999 | 0.999 | 0.999 | 0.999 | |
| Caleosin | At5g55240 | 0.7 | 0.998 | 0.95 | 0.991 | 0.93 | 0.999 | |
| Degradation | Fatty acid alcohol oxidase | At4g19380 | 0.1 | 0.92 | 0.6 | 0.8 | 0.5 | 0.98 |
| Monoacylglycerol lipase | At2g39400 | 0.1 | 0.92 | 0.6 | 0.1 | 0.4 | 0.98 | |
| Acyl-CoA oxidase | At2g35690 | 0.1 | 0.92 | 0.6 | 0.8 | 0.5 | 0.98 | |
| Ketoacyl-CoA thiolase | At1g04710 | 0.4 | 0.97 | 0.7 | 0.91 | 0.7 | 0.98 | |
| Ketoacyl-CoA thiolase | At2g33150 | 0.4 | 0.999 | 0.995 | 0.7 | 0.8 | 0.998 | |
| Dienoyl-CoA reductase | At3g12790 | 0.4 | 0.92 | 0.6 | 0.5 | 0.5 | 0.98 | |
| Lipid signaling | Secretory phospholipase A2 | At4g29070 | 0.1 | 0.92 | 0.6 | 0.8 | 0.5 | 0.98 |
| DAD1-like acylhydrolase | At4g16820 | 0.5 | 0.5 | 0.5 | 0.96 | 0.8 | 0.8 | |
| Lipoxygenase | At1g17420 | 0.4 | 0.9 | 0.7 | 0.91 | 0.1 | 0.3 | |
| Lipoxygenase | At3g22400a | 0.6 | 0.996 | 0.92 | 0.98 | 0.8 | 0.999 | |
| Lipoxygenase | At3g22400b | 0.94 | 0.999 | 0.996 | 0.999 | 0.994 | 0.999 | |
| α-Dioxygenase-peroxidase (involved in FA α-oxidation) | At1g73680 | 0.92 | 0.999 | 0.994 | 0.997 | 0.99 | 0.999 | |
| Type II phosphoinositide 5-phosphatase | At1g65580b | 0.4 | 0.97 | 0.7 | 0.91 | 0.7 | 0.93 | |
| Patatin-like acyl-hydrolase | At4g37050 | 0.4 | 0.97 | 0.7 | 0.4 | 0.7 | 0.996 | |
| Nonspecific phospholipase C | At3g03530 | 0.1 | 0.92 | 0.6 | 0.8 | 0.5 | 0.98 | |
| Phospholipase A2-activating protein | At3g18860 | 0.1 | 0.5 | 0.6 | 0.1 | 0 | 0.98 | |
| Wax/cutin | KCS | At1g68530 | 0.4 | 0.999 | 0.8 | 0.2 | 0.3 | 0.9 |
| KCS | At2g26250 | 0.97 | 0.999 | 0.98 | 0.2 | 0.8 | 0.999 | |
| KCS | At2g26640 | 0.4 | 0.9 | 0.7 | 0.91 | 0.7 | 0.98 | |
| KCS | At4g34250 | 0.6 | 0.996 | 0.92 | 0.98 | 0.3 | 0.999 | |
| KCS | At4g34520 | 0.993 | 0.999 | 0.999 | 0.999 | 0.999 | 0.999 | |
| Fatty acid omega-hydroxylase | At4g00360 | 0.5 | 0.99 | 0.5 | 0.2 | 0.1 | 0.3 | |
| Ketoacyl-CoA reductase | At1g67730a | 0.2 | 0.995 | 0.99 | 0.995 | 0.98 | 0.999 | |
| Acyl-CoA reductase (NADPH dependent) | At3g44540 | 0.1 | 0.7 | 0.6 | 0.8 | 0.5 | 0.98 | |
| Putative transcription factor CER2 | At4g13840 | 0.6 | 0.996 | 0.92 | 0.6 | 0.8 | 0.993 | |
| Miscellaneous | Long-chain acyl-CoA synthetase (not plastidial, not peroxisomal) | At2g47240 | 0.9 | 0.998 | 0.8 | 0.4 | 0.98 | 0.999 |
| Epoxide hydrolase | At4g02340 | 0.8 | 0.996 | 0.92 | 0.92 | 0.97 | 0.999 | |
| Protein N-myristoyltransferase | At5g57020 | 0.5 | 0.96 | 0.8 | 0 | 0.8 | 0.97 | |
| LTP1 | At2g38530 | 0.8 | 0.999 | 0.1 | 0.997 | 0.993 | 0.98 | |
| LTP5 | At3g43720 | 0.2 | 0.97 | 0.3 | 0.1 | 0.1 | 0.5 | |
| Acyl-CoA desaturase like | At2g31360 | 0.7 | 0.6 | 0.95 | 0.8 | 0.7 | 0.92 | |
| ATP citrate lyase | At1g10670 | 0.97 | 0.999 | 0.98 | 0.1 | 0.96 | 0.999 | |
For each of the 28 possible pairs of organs, probabilities of differential expression between the two organs have been calculated for all lipid genes. The 28 corresponding tables and the tables of the multiple organ analysis and some supplemental tables are available in the ALG database. The lipid genes have been ranked by decreasing probability of differential expression, and over 130 genes have been identified with a statistical probability > 0.95 of preferential expression in seed, leaf, root, or flower.
Limitations
The results of the statistical analysis might be biased by the fact that some cDNA libraries have been normalized and/or subtracted. However, among the libraries used here, most were not normalized/subtracted or were only processed by removal of very highly abundant transcripts (e.g. the PLR2 library containing approximately 27,600 ESTs from mixed organs [Newman et al., 1994] and the developing seed with 10,500 [White et al., 2000]). Moreover, in the normalized libraries used here (mainly silique, flower, and some root libraries), it can be seen easily that many transcripts are in fact approximately as abundant as in standard libraries (e.g. LTPs and also many other non-lipid genes such as translation factors, chaperones, etc.). Therefore, the normalization/subtraction procedures may not be very efficient, and it seems unlikely that the results of the EST analysis were strongly biased.
This statistical analysis provides a more rigorous strategy for the analysis of EST data, and we believe it will help users to spot genes with interesting patterns among the hundreds of the Lipid Gene Catalog. However, it eliminates neither the role of intuition and chance nor the need for experimental confirmation of gene expression by methods such as quantitative reverse transcriptase-PCR. For example, because the “virtual synthetic libraries” (just like real ones) are from whole organs, a fairly uniform distribution of ESTs across these libraries does not necessarily imply constitutive gene expression because most of the organs will contain, for example, meristems or epidermal cell layers. Also, specific expression patterns from a very small region of the organ, say the pericycle in roots or stigmas in flowers, may be missed because of dilution from the remainder of the tissue in the organ.
Combining the Lipid Gene Catalog and the EST Analysis to Gain New Insights into Lipid Metabolism
Organ Expression of Gene Family Members
An analysis of EST expression as described above can be performed for all organs to further investigate the organ expression profiles. For example, results are summarized in Table IV for the KCS isoform At1g68530. No preferential expression clearly appears; thus, the gene may be expressed at about the same level in most organs, except in roots where it seems to be down-regulated or not expressed. In fact, the mutation of this locus in the Arabidopsis CUT1 mutant results in waxless stems and siliques and conditional male sterility (Millar et al., 1999). In addition, this gene was found to be expressed in wild-type shoots but not in roots (Hooker et al., 2002). For purpose of comparison, probabilities of differential expression are also shown in Table V for another KCS isoform (At2g26250) that seems to show a preferential expression. This gene has ESTs in flowers and seedlings that are consistent with its proposed role in epidermal cuticle formation in flowers and ovule primordia (Pruitt et al., 2000) and in young leaves (Yephremov et al., 1999). However, according to the probabilities of differential expression, it seems reasonable to hypothesize that this isoform is preferentially expressed in seeds and, thus, also has an important role in seed development (e.g. in cuticle formation in outer integument cells at the seed coat).
Table IV.
Probabilities of differential expression between organs for At1g68530 ′all but root′ isoform (CUT1) KCS isoform
Probabilities are indicated in bold when they are >0.95 and correspond to an up-regulation in the organs also indicated in bold (first row). Nos. of ESTs are indicated between brackets.
| Seed (7) | Leaf (1) | Root (0) | Flower (1) | Silique (7) | |
|---|---|---|---|---|---|
| Seed | - | 0.4 | 0.999 | 0.8 | 0.2 |
| Leaf | 0.4 | - | 0.7 | 0.4 | 0.2 |
| Root | 0.999 | 0.7 | - | 0.8 | 0.998 |
| Flower | 0.8 | 0.4 | 0.8 | - | 0.8 |
| Silique | 0.2 | 0.2 | 0.998 | 0.8 | - |
Table V.
Probabilities of differential expression between organs for At2g26250 ′seed/silique′ KCS isoform
Probabilities are indicated in bold when they are >0.95 and correspond to an up-regulation in the organs also indicated in bold (first row). Nos. of ESTs are indicated between brackets.
| Seed (14) | Leaf (0) | Root (0) | Flower (2) | Silique (15) | |
|---|---|---|---|---|---|
| Seed | - | 0.97 | 0.999 | 0.98 | 0.2 |
| Leaf | 0.97 | - | 0.6 | 0.4 | 0.96 |
| Root | 0.999 | 0.6 | - | 0.94 | 0.999 |
| Flower | 0.98 | 0.4 | 0.94 | - | 0.96 |
| Silique | 0.2 | 0.96 | 0.999 | 0.96 | - |
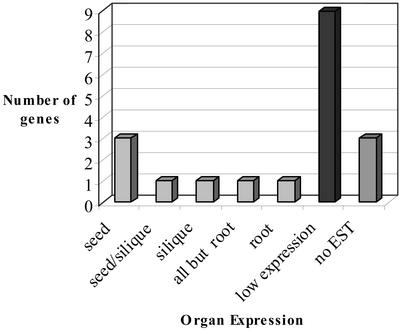
If all members of a gene family are analyzed using such an approach, a general picture of the possible organ expression profile of a gene family can be tentatively drawn. Such a summary is shown in the case of the KCS family in Figure 4. In the complete Arabidopsis genome, there are 21 Arabidopsis genes related to known KCSs. Three of the 21 KCS do not have ESTs and may not be expressed. Because one of the 18 expressed KCS was discarded in the new catalog due to an amino acid change in the conserved motif for the putative catalytic triad (Ghanevati and Jaworski, 2001), there are, therefore, 17 expressed putative KCS genes that could be involved in the synthesis of wax, sphingolipids, and storage lipids. Among the four isoforms that seem to be more specific for seeds (Tables IV and V), some of them could be specialized in the synthesis of storage TAGs, and some others could be specialized in the wax/cutin of the seed coat. One or two isoforms showing preferential expression in silique are good candidates for the synthesis of the waxes of the silique coat. Nine KCS isoforms could not be classified because they have a low number of ESTs. These genes could be involved in the housekeeping synthesis of membrane sphingolipids or could be expressed at a low level in some restricted organs like flowers. The three isoforms that do not have ESTs might not be functional or might be inducible isoforms (by a pathogen attack, for example).
Figure 4.
Possible preferential organ expressions in the KCS family based on EST data. Each of the 19 KCS genes was classified in one category only based on the probabilities of differential expression between organs (see Tables IV and V for an example of the data used).
The complete EST analysis available in the ALG database suggests that organ-preferential expression is likely to account, at least partly, for the existence of multiple members in a few other families than the KCS family. For example, the seven stearoyl-ACP desaturases present in the Lipid Gene Catalog seem to include two seed preferentially expressed isoforms (At2g43710 and At3g02630) and a leaf isoform (At1g43800). In fact, in most cases, only one isoform of a gene family is clearly found to be preferentially expressed in an organ. However, this limited information might be very valuable when bearing some specific biological questions in mind.
Finally, it should be stressed that the expression of organ-specific protein isoforms can also be achieved by alternative splice forms in addition to gene families. As an example, it can be seen in Table III that genes with alternative splice forms (locus codes with “a” or “b” at the end) represented 15% of the genes likely to be seed specific (but only 7% of the genes in the catalog and the genome).
Insights into Metabolites and Pathways
In several cases, the EST analysis provided a confirmation of previous observations and hypotheses. For example, the presence of pyruvate dehydrogenase and ATP:citrate lyase in the list of Table III is consistent with suggestions that up-regulation of these two enzymes (but not of acetyl-CoA synthase) may be the main source of plastidial and cytosolic acetyl-CoA, respectively, for the synthesis of fatty acids and cuticular waxes in seeds (Fatland et al., 2000; Ke et al., 2000; Schwender and Ohlrogge, 2002).
In other cases, several unexpected enzymes involved in the same pathway or reaction may be found in the lists of genes likely to be up-regulated in a organ. This may help in revealing pathways or metabolites not described previously or not thought as important in some organs. For example, the occurrence of the thioesterase fatB, a ketosphinganine reductase and a sphingolipid hydroxylase in the list of genes that may be preferentially expressed in roots, may indicate an important role of sphingolipids in root cell membranes (see EST analysis section in the ALG database). Another example in roots is the up-regulation of three enzymes producing phosphocholine: two choline kinase isoforms and a phosphoethanolamine N-methyltransferase (the latter is known to be involved in salt sensitivity; Mou et al., 2002).
Sets of Pathway-Specific Genes
The potential existence of some special isoforms catalyzing the same reactions as housekeeping isoforms but involved specifically in pathways predominant in certain organs may be of primary importance in view of engineering these pathways. For example, the synthesis of storage TAGs takes place in many organs but is highly abundant in seeds. Furthermore, the pathway has many reactions in common with the synthesis of membrane lipids, but these reactions could be performed by special isoforms. Alternatively, some particular housekeeping isoforms might be up-regulated specifically in the pathway and may indicate key control points. A statistical analysis of EST data similar to the one summarized in Table III can help by revealing sets of pathway-specific or pathway-up-regulated isoforms. For example, a detailed analysis (P > 0.9) spots three acyltransferases among the 19 candidates of the catalog. One of them is the well-known acylCoA:DAGAT At2g19450, and the two others are candidates for the two first acyltransferases of the glycerolipid biosynthesis pathway (Ohlrogge and Browse, 1995). A tentative set of genes involved in TAG synthesis in seeds that can be deduced from a statistical analysis and from data of the literature is given in Table VI. The two first acyltransferases of the pathway may be seed-specific isoforms, whereas the other candidate genes are expressed in many organs and may be up-regulated in seeds. The presence of specific acyltransferase isoforms in seeds favors the hypothesis of the existence of distinct pathways for the synthesis of membrane and storage glycerolipids in specific subdomains of the ER (Cahoon and Ohlrogge, 1994; Lacey and Hills, 1996; Vogel and Browse, 1996).
Table VI.
A list of genes potentially playing a specific role in TAG synthesis in seeds
| Putative Cellular Activity | Protein | Expression of the Gene in the Pathway |
|---|---|---|
| Plastidial pyruvate dehydrogenase E1 α | At1g01090 | Up-regulated |
| Ketoacyl-ACP Synthase I | At5g46290 | Up-regulated |
| Stearoyl-ACP desaturase | At2g43710 | Up-regulated |
| ER glycerol-phosphate acyltransferase | At3g11430 | Specific? |
| ER Acylglycerol-phosphate acyltransferase | At1g75020b | Specific? |
| ER oleate desaturase | At3g12120 | Up-regulated |
| ER linoleate desaturase | At2g29980 | Up-regulated |
| Acyl-CoA: DAGAT | At2g19450 | Up-regulated |
CONCLUSIONS
Despite some uncertainties in the catalog for some poorly characterized groups of enzymes, most notably lipases/transacylases and lipid translocators, we feel that this first genome-wide classification of 600 Arabidopsis genes involved in acyl lipid metabolism (including over 200 that were previously not annotated or were functionally misannotated) will assist in experimental works because, for any given reaction, it gives a list of the most probable candidates. Moreover, the accuracy and, thus, the utility of protein classifications based on sequence similarity obviously increases with time as new biochemical information becomes available. Therefore, the main intent of this catalog is to provide the plant biology community with a first Web-based common platform of information, where new data can be collected and organized to fully take advantage of their accumulation. Most immediately, the list will facilitate studies in systematic biology, particularly proteomics, where it will be important to recognize and distinguish different polypeptides from gene families for a particular reaction of acyl lipid metabolism. In addition, analysis of EST distribution in organs can help in revealing organ-preferential expressions and, therefore, should prove a useful guide to the designing and the interpretation of gene silencing or disruption experiments as well as providing clues of the function of lipid genes at the cell and plant levels.
MATERIALS AND METHODS
BLAST Searches
To retrieve amino acid sequences, the blastp option of the BLAST program (Altschul et al., 1990) available at The Arabidopsis Information Resource (http://www.Arabidopsis.org/Blast/) was used. BlastP queries were performed with default parameters and amino acid sequences of proteins from plant, animal, fungal, or bacterial origin demonstrated to be involved in acyl lipid metabolism. The Lipid Gene Catalog (http://www.canr.msu.edu/lgc) was a convenient source of Arabidopsis and rice (Oryza sativa) query sequences. The other query sequences were from SWISS-PROT (http://us.expasy.org/sprot/) and GenBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html) databases.
For a given query, all sequences with P < 0.1 were recorded for further analysis. Based on multiple alignments and/or presence/absence of conserved motifs, some initial sequence “hits” were then discarded.
Multiple Alignments and Motif Searches
Protein sequences were aligned using the programs ClustalW (Thompson et al., 1994) or T-Coffee (Notredame et al., 2000) available at http://www.ch.embnet.org/index.html. Conserved protein motifs were searched by eye in the multiple alignments. Known protein motifs were searched in the Arabidopsis proteome by using the PatternFind server of the Swiss Institute of Bioinformatics (http://hits.isb-sib.ch/cgi-bin/hits_patsearch) or the pattern search of the MIPS server (http://mips.gsf.de/proj/thal/db/search/search_frame.html).
Subcellular Localization Predictions
Predictions were performed for all protein sequences using the targetP program (Emanuelsson et al., 2000) available at the Center for Biological Sequence Analysis (http://www.cbs.dtu.dk/services/TargetP).
EST Searches and Assignment to Organs
Arabidopsis ESTs were retrieved using the BLASTN program available at TIGR (http://tigrblast.tigr.org/tgi/) with cDNA or predicted open reading frame DNA sequences as queries. Data from 55 different cDNA libraries and 110,154 ESTs were analyzed, resulting in identification of 3,750 acyl lipid-related ESTs. To assign these ESTs to the organs from which they originated, the 55 different cDNA libraries were classified and grouped into eight synthetic libraries based on the organ used for construction of each cDNA library. The composition of these synthetic cDNA libraries is available on the ALG database (http://www.plantbiology.msu.edu/lipids/genesurvey/EST_Libraries.htm).
Statistical Analysis of EST Data
For each lipid gene, the numbers of ESTs in the synthetic libraries (organs) was used to compute the probability of differential expression between all pairs of organs by using the UNIX version of the statistical program of Audic and Claverie (1997; http://igs-server.cnrs-mrs.fr/~audic/significance.html).
Acknowledgments
We thank Matt Larson and Drs. Robert Halgren, Curtis Wilkerson, and Ann Jones for their help in computing the probabilities and their comments on the EST analysis. We also thank Drs. Vincent Arondel, Christoph Benning, John Browse, Edgar Cahoon, Kent Chapman, Margrit Frentzen, Ian Graham, Sergei Mekhedov, Martine Miquel, Hajime Wada, and Xuemin Wang for giving expert advice on some gene families or suggesting gene candidates.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.022988.
This work was supported by the Department of Energy (grant no. DE–FG02–87ER13729), by the National Science Foundation (grant no. MCB 98–17882), and by the Michigan Agricultural Experiment Station.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Arondel V, Vergnolle C, Cantrel C, Kader JC (2000) Lipid transfer proteins are encoded by a small multigene family in Arabidopsis thaliana. Plant Sci 157: 1–12 [DOI] [PubMed] [Google Scholar]
- Audic S, Claverie JM (1997) The significance of digital expression profiles. Genome Res 7: 986–995 [DOI] [PubMed] [Google Scholar]
- Beisson F, Tiss A, Rivière C, Verger R (2000) Methods for lipase detection and assay: a critical review. Eur J Lipid Sci Technol 2: 133–153 [Google Scholar]
- Blein J-P, Coutos-Thévenot P, Marion D, Ponchet M (2002) From elicitins to lipid-transfer proteins: a new insight in cell signalling involved in plant defence mechanisms. Trends Plant Sci 7: 293–296 [DOI] [PubMed] [Google Scholar]
- Bouché N, Bouchez D (2001) Arabidopsis gene knockout: phenotypes wanted. Curr Opin Plant Biol 4: 111–117 [DOI] [PubMed] [Google Scholar]
- Brick DJ, Brumlik MJ, Buckley JT, Cao JX, Davies PC, Misra S, Tranbarger TJ, Upton C (1995) A new family of lipolytic plant enzymes with members in rice, Arabidopsis and maize. FEBS Lett 377: 475–480 [DOI] [PubMed] [Google Scholar]
- Browse J, Somerville C (1994) Glycerolipids. In EM Meyerowitz, CR Somerville, eds, Arabidopsis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, pp 881–912
- Cahoon EB, Ohlrogge JB (1994) Apparent role of phosphatidylcholine in the metabolism of petroselinic acid in developing Umbelliferae endosperm. Plant Physiol 104: 845–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douliez JP, Pato C, Rabesona H, Molle H, Marion D (2001) Disulfide bond assignment, lipid transfer activity and secondary structure of a 7-kDa plant lipid transfer protein, LTP2. Eur J Biochem 268: 1400–1403 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Fatland B, Anderson M, Nikolau BJ, Wurtele ES (2000) Molecular biology of cytosolic acetyl-CoA generation. Biochem Soc Trans 28: 593–595 [PubMed] [Google Scholar]
- Gerlt JA, Babbitt PC (2000) Can sequence determine function? Genome Biol 1: 5.1–5.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanevati M, Jaworski JG (2001) Active-site residues of a plant membrane-bound fatty acid elongase β-ketoacyl-CoA synthase FAE1 KCS. Biochim Biophys Acta 1530: 77–85 [DOI] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J (2000) Microarray analysis of developing Arabidopsis seeds. Plant Physiol 124: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunstone FD, Pollard M (2001) Vegetable oils with fatty acid changed by plant breeding or genetic modification. In FD Gunstone, ed, Structured and Modified Lipids. Marcel Dekker, New-York, pp 155–184
- Henrissat B, Countinho PM, Davies GJ (2001) A census of carbohydrate-active enzymes in the genome of Arabidopsis thaliana. Plant Mol Biol 47: 55–72 [PubMed] [Google Scholar]
- Hooker TS, Millar AA, Kunst L (2002) Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol 129: 1568–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K (2001) The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader JC (1996) Lipid-transfer proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 627–654 [DOI] [PubMed] [Google Scholar]
- Karlsson M, Contreras JA, Hellman U, Tornqvist H, Holm C (1997) cDNA cloning, tissue distribution, and identification of the catalytic triad of monoglyceride lipase: evolutionary relationship to esterases, lysophospholipases, and haloperoxidases. J Biol Chem 272: 27218–27223 [DOI] [PubMed] [Google Scholar]
- Ke J, Behal RH, Back SL, Nikolau BJ, Wurtele ES, Oliver DJ (2000) The role of pyruvate dehydrogenase and acetyl-CoA synthetase in fatty acid synthesis in developing Arabidopsis seeds. Plant Physiol 123: 497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJK, Ohlrogge JB (2002) The predicted candidates of Arabidopsis plastid envelope membrane proteins and their expression profiles. Plant Physiol 130: 823–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey DJ, Hills MJ (1996) Heterogeneity of the endoplasmic reticulum with respect to lipid synthesis in developing seeds of Brassica napus L. Planta 199: 545–551 [Google Scholar]
- Lee JM, Williams ME, Tingey SV, Rafalski JA (2001) DNA array profiling of gene expression changes during maize embryo development. Funct Integr Genomics 2: 13–27 [DOI] [PubMed] [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK (2002) A putative lipid transfer protein involved in systemic resistance in Arabidopsis. Nature 419: 399–403 [DOI] [PubMed] [Google Scholar]
- Mekhedov S, Martínez de Llárduya, Ohlrogge JB (2000) Toward a functional catalog of the plant genome: a survey of genes for lipid biosynthesis. Plant Physiol 122: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Clemens S, Zachgo S, Giblin EM, Taylor DC, Kunst L (1999) CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11: 825–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, Wang X, Fu Z, Dai Y, Han C, Ouyang J, Bao F, Hu Y, Li J (2002) Silencing of phosphoethanolamine N-methyltransferase results in temperature-sensitive male sterility and salt hypersensitivity in Arabidopsis. Plant Cell 14: 2031–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman T, de Bruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M et al. (1994) Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol 106: 1241–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205–217 [DOI] [PubMed] [Google Scholar]
- O'Hara P, Slabas AR, Fawcett T (2002) Fatty acid and lipid biosynthetic genes are expressed at constant molar ratios but different absolute levels during embryogenesis. Plant Physiol 129: 310–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J (1995) Lipid Biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo K, Hori N, Matoba R, Niiyama T, Fukushima A, Kojima Y, Matsubara K (1992) Large scale cDNA sequencing for analysis of quantitative and qualitative aspects of gene expression. Nat Genet 2: 173–179 [DOI] [PubMed] [Google Scholar]
- Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J et al. (1992) The alpha/beta hydrolase fold. Protein Eng 5: 197–211 [DOI] [PubMed] [Google Scholar]
- Peeters N, Small I (2001) Dual targeting to mitochondria and chloroplasts. Biochim Biophys Acta 1541: 54–63 [DOI] [PubMed] [Google Scholar]
- Pruitt RE, Vielle-Calzada JP, Ploense SE, Grossniklaus U, Lolle SJ (2000) FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc Natl Acad Sci USA 1: 1311–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romualdi C, Bortoluzzi S, Danieli GA (2001) Detecting differentially expressed genes in multiple tag sampling experiments: comparative evaluation of statistical tests. Hum Mol Genet 10: 2133–2141 [DOI] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge JB (2002) Probing in vivo metabolism by stable isotope labeling of storage lipids and proteins in developing Brassica napus embryos. Plant Physiol 130: 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprong H, van der Sluijs P, van Meer G (2001) How proteins move lipids and lipids move proteins. Nat Rev Mol Cell Biol 2: 504–513 [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ (1997) A genomic perspective on protein families. Science 278: 631–637 [DOI] [PubMed] [Google Scholar]
- Thelen JJ, Ohlrogge JB (2002) Metabolic Engineering of fatty acid biosynthesis in plants. Metab Eng 4: 12–21 [DOI] [PubMed] [Google Scholar]
- Thoma S, Kaneko Y, Somerville C (1993) A non-specific lipid transfer protein from Arabidopsis is a cell wall protein. Plant J 3: 427–436 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel G, Browse J (1996) Cholinephosphhotransferase and diacylglycerol acyltransferase: substrate specificities at a key branch point in seed lipid metabolism. Plant Physiol 110: 923–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis JG, Browse J (2002) Mutants of Arabidopsis reveal many roles for membrane lipids. Prog Lipid Res 41: 254–278 [DOI] [PubMed] [Google Scholar]
- Ward JM (2001) Identification of novel families of membrane proteins from the model plant Arabidopsis thaliana. Bioinformatics 17: 560–563 [DOI] [PubMed] [Google Scholar]
- White JA, Todd J, Newman T, Focks N, Girke T, de Ilarduya OM, Jaworski JG, Ohlrogge JB, Benning C (2000) A new set of Arabidopsis expressed sequence tags from developing seeds: the metabolic pathway from carbohydrates to seed oil. Plant Physiol 124: 1582–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H (1999) Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11: 2187–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]