Abstract
Xyloglucan (XyG) is a load-bearing primary wall component in dicotyledonous and non-graminaceous monocotyledonous plants. XyG fucosyltransferase (FUTase), encoded by the Arabidopsis gene AtFUT1, directs addition of fucose (Fuc) residues to terminal galactose residues on XyG side chains. Reverse transcription-polymerase chain reaction and analysis of promoter-β-glucuronidase transgenic plants indicated highest expression of AtFUT1 in the upper portion of elongating inflorescence stems of Arabidopsis. XyG FUTase activity was highest in Golgi vesicles prepared from growing Arabidopsis tissues and low in those isolated from mature tissues. There was no discernible correlation between the Fuc contents of XyG oligosaccharides derived from different Arabidopsis organs and the level of AtFUT1 expression in the organs. Thus, organ-specific variations in AtFUT1 expression and enzyme activity probably reflect differential rates of cell wall biosynthesis, rather than differences in levels of XyG fucosylation. The effects of manipulating AtFUT1 expression were examined using an Arabidopsis mutant (atfut1) containing a T-DNA insertion in the AtFUT1 locus and transgenic plants with strong constitutive expression of AtFUT1. No Fuc was detected in XyG derived from leaves or roots of atfut1. Plants overexpressing AtFUT1 had higher XyG FUTase activity than wild-type plants, but the XyG oligosaccharides derived from the transgenic and wild-type plants contained comparable amounts of Fuc, indicating that suitable acceptor substrates are limiting. Galactosyl residues had slightly higher levels of O-acetylation in XyG from plants that overexpressed AtFUT1 than in XyG from wild-type plants. O-Acetylation of galactose residues was considerably reduced in Fuc-deficient mutants (atfut1, mur1, and mur2) that synthesize XyG containing little or no Fuc. These results suggest that fucosylated XyG is a suitable substrate for at least one O-acetyltransferase in Arabidopsis.
Plant growth is largely delimited by cell wall biosynthesis. In the absence of cell wall biosynthesis, growing plant cells would eventually rupture because their walls would become so thin that they could no longer resist their internal turgor pressure (Cosgrove, 2000). Several genes encoding enzymes that synthesize cell wall polysaccharides have been identified (Arioli et al., 1998; Edwards et al., 1999; Perrin et al., 1999; Richmond and Somerville, 2000; Faik et al., 2002), but few of these genes have been characterized at the levels of gene expression, the activity of the enzyme they encode, or the quantity and structure of the synthesized glycan. Genes encoding cell wall biosynthetic enzymes are difficult to identify, because the proteins they encode do not have high sequence similarity to polysaccharide-synthesizing enzymes characterized in non-plant organisms (Perrin et al., 2001). Developing suitable enzymatic assays is challenging, because many plant glycosyltransferases and glycan synthases are unstable and often require polysaccharide acceptors that are not readily available.
Xyloglucan (XyG) fucosylation is one process in which analyses of gene expression, enzyme activity, and product structure are all achievable tasks. XyG is a quantitatively major hemicellulosic polysaccharide present in the primary walls of dicots and nongraminaceous monocots. The 1,4-linked β-d-glucan backbone of XyG is substituted at C6 with α-d-Xyl, β-d-Gal-(1,2)-α-d-Xyl or α-l-Fuc-(1,2)-β-d-Gal-(1,2)-α-d-Xyl. In the primary wall, XyGs are believed to cross-link cellulose microfibrils non-covalently, thereby forming the major load-bearing network (Hayashi, 1989; Pauly et al., 1999). Many plants including Arabidopsis synthesize XyGs that contain a fucosylated trisaccharide side chain. The gene encoding XyG fucosyltransferase (FUTase), AtFUT1 (previously referred to as AtFT1), has been identified in Arabidopsis (Perrin et al., 1999; Faik et al., 2000; Sarria et al., 2001). An enzyme assay has been developed to quantify XyG FUTase activity in plant tissues (Farkas and Maclachlan, 1988). 1H-NMR spectroscopic methods have been developed to quantify Fuc-containing XyG side chains (York et al., 1988, 1993; Zablackis et al., 1996). We describe the combined use of genetic, biochemical, and chemical methods to study XyG fucosylation in the tissue of Arabidopsis plants from different genetic backgrounds.
RESULTS
Expression of AtFUT1 in Arabidopsis Tissues
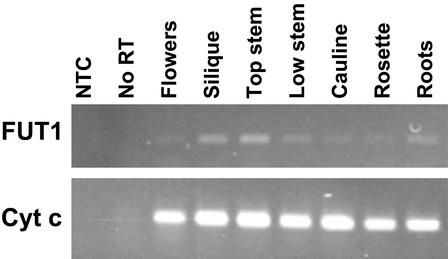
The steady-state levels of AtFUT1 transcript abundance were determined using reverse transcription (RT)-PCR and RNA prepared from various tissues of mature, hydroponically grown Arabidopsis (Columbia-0 [Col-0]) plants. These tissues were also used to determine XyG FUTase activity and the extent of XyG fucosylation. Primers specific to AtFUT1 or the “housekeeping” gene cytochrome c were used to determine gene expression levels (Fig. 1). Amplification was halted at a cycle number (22–24 cycles for AtFUT1; 28 cycles for cytochrome c) within the linear range of product accumulation (data not shown). The highest level of AtFUT1 expression occurred in the apical, youngest region of the inflorescence stem, in siliques, and in roots. Lower levels of transcript were detected in the basal, oldest region of the stem and in rosette and cauline leaves.
Figure 1.
Expression of AtFUT1 analyzed by RT-PCR. RNA isolated from Arabidopsis tissues and used for RT reactions primed with oligo(dT). An equal volume of the RT reactions was used for PCR with primers specific for either AtFUT1 (top) or cytochrome c as a standard gene control (bottom). PCR was performed for 24 (AtFUT1) or 28 (cytochrome c) cycles. A representative gel is shown from two technical replicates each of two independent biological replicates.
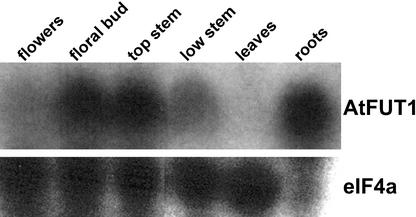
A similar pattern of expression was observed in RNA samples analyzed by northern blot (Fig. 2). In this case, the expression of AtFUT1 was markedly higher in a floral bud sample, which was derived from clusters of unopened flowers, petioles, and uppermost top inflorescence stem, than a sample derived from individual fully opened flowers and associated petioles.
Figure 2.
Expression of AtFUT1 analyzed by northern blot. Total RNA was isolated from organs of hydroponically grown Arabidopsis plants. Flowers, harvested as completely opened single flowers and associated petioles; floral buds, unopened flower clusters, associated petioles, and 2 mm of uppermost stem; top stem, the uppermost 0.5-cm segment of the inflorescence stem after removal of flowers; low stem, the segment of the inflorescence stem from a point that was 1.5 cm from its top to the base; leaves, mature rosette leaves; roots, the entire root mass. Fifteen micrograms of total RNA was analyzed for expression of AtFUT1, using a gene-specific hybridization probe (Sarria et al., 2001) or a probe for eIF4a as a loading control.
Analysis of β-Glucuronidase (GUS) Activity in AtFUT1::GUS Plants
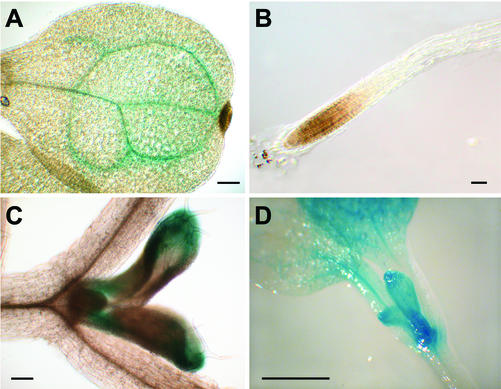
The spatial patterns of AtFUT1 expression in planta were investigated using an approximately 2-kb-long region upstream of the AtFUT1 coding region that was amplified by PCR and then used to direct expression of the GUS reporter gene. GUS activity was then determined in homozygous T3 plants carrying a single insertion of the transgene at various stages of plant development (Figs. 3 and 4) together with positive controls expressing GUS under control of the 35S reporter (data not shown). The cotyledon veins and emerging true leaves were stained in young seedlings (3–10 d old; Fig. 3). The venation-staining pattern observed in the cotyledons was not repeated in emerging true leaves, in which staining occurred first at the true leaf base and tip (Fig. 3), followed by staining only at the base of the midvein in mature leaves (Fig. 4). No staining was observed in roots, even though northern-blot and RT-PCR analyses had shown that AtFUT1 is expressed in roots. Staining patterns in cauline leaves, rosette leaves and old and young regions of the inflorescence stem generally corresponded to results obtained using RT-PCR and hybridization techniques (Figs. 1 and 2). In mature plants, a gradient of reporter activity was observed in the inflorescent stem, with most intense staining occurring in the youngest regions of the inflorescence (Fig. 4). No staining was observed in floral tissue itself, although petiole and inflorescence tissue directly below flowers did show reporter activity.
Figure 3.
GUS reporter activity in AtFUT1::GUS transgenic seedlings. Representative 3-d-old (A and B) or 10-d-old (C and D) seedlings are shown. Staining occurred in the veins of emerging cotyledons (A) and was largely absent from roots (B). C and D, In 10-d-old plants, staining occurred in base and tips of emerging leaf buds. Bar = 100 μm in A through C or 1 mm in D.
Figure 4.
GUS reporter activity in mature AtFUT1::GUS transgenic plants. Representative images are shown from 6-week-old soil-grown plants. A, Left to right, Mature rosette leaf, upper inflorescence stem (just below flower buds), lower inflorescence stem (just above rosette), flowers, silique, and cauline leaf. B, Inflorescence stem. C, Close-up of flowers.
XyG FUTase Activity Levels in Arabidopsis Tissues
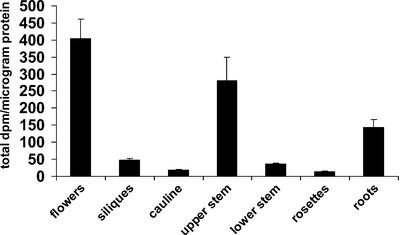
XyG FUTase is a Golgi-localized enzyme, and fucosylated XyG accumulates primarily in the trans-Golgi and trans-Golgi network (Brummell et al., 1990). Thus, Golgi vesicles have been used as an enriched source of XyG FUTase enzyme activity (Wulff et al., 2000). This procedure was developed using etiolated pea (Pisum sativum) epicotyls and was modified for use with Arabidopsis. The protein content of Arabidopsis (Col-0) Golgi vesicle-enriched preparations was quantified, and the samples were assayed for XyG FUTase activity in the presence of detergent (1% [v/v] Triton X-100). XyG FUTase activity was highest in flowers and the upper portion of the inflorescence stem (Fig. 5). Over 5-fold higher activity was observed in the upper region of the inflorescence stem compared with the lower-most region, which at the time of collection had lignified, indicating the presence of secondary cell wall material. Somewhat lower levels of activity were present in the roots (Fig. 5), whereas little or no activity was detected in fully expanded rosette leaves.
Figure 5.
XyG FUTase activity of Golgi vesicle samples. Golgi vesicles were isolated using the method of Muñoz et al. (1996). XyG FUTase assays were performed as described previously (Perrin et al., 1999). Data are averages of three replicates. An independent biological replicate resulted in a comparable profile (data not shown). Results are expressed as total dpm per microgram of protein. Error bars indicate 1 SD.
Fuc Content of XyG Oligosaccharides from Arabidopsis Organs
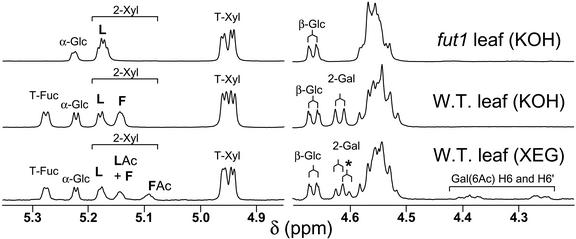
A structural reporter approach was used to determine Fuc content of Arabidopsis XyG oligosaccharides. Alcohol insoluble residues (AIR) were prepared from the remaining Arabidopsis (Col-0) samples. The enzymatically depectinated AIR was treated with a XyG-specific endoglucanse (XEG) and then with 4 m KOH. XEG treatment of the AIR hydrolyzes the “enzyme-accessible domain” of the XyG, generating a mixture of O-acetylated XyG oligosaccharides. Subsequent treatment with 4 m KOH solubilizes much of the remaining XyG as a high Mr polysaccharide with no O-acetyl substituents, which are hydrolyzed under the alkaline extraction conditions. XEG treatment of the alkali-soluble XyG generates a mixture of oligosaccharides. Figure 6 illustrates the application of this structural reporter method, showing examples of 1H-NMR spectra of KOH- and XEG-extracted XyG oligosaccharides from 4.2 to 5.3 ppm. The amount of Fuc present per unit of oligosaccharide in the Arabidopsis organ AIR samples was determined by 1H-NMR spectroscopy (Table I). XyG oligosaccharide structures are abbreviated using the nomenclature described by Fry et al. (1993). The amount of Fuc present on XyG was similar in all organs tested (Table I). Moreover, there were no discernible differences in the Fuc content of XyG from leaves ranging in age from 1 to 5 weeks (data not shown). In contrast, the Gal content of the XyG was somewhat organ dependent. For example, there was more Gal present in oligosaccharides derived from rosette leaves than other organs. O-Acetylation of Gal residues in the XEG-released oligosaccharides was also quantitated by 1H-NMR spectroscopy (Table I). In the tissues examined, less than one-half of the Gal residues in the XEG-released oligosaccharides were O-acetylated.
Figure 6.
Diagnostic resonances in the 1H-NMR spectra of XyG oligosaccharides. Example spectra of KOH- or XEG-extracted XyG oligosaccharides from wild-type or atfut1 (which contains a T-DNA insertion in AtFUT1) are shown from 4.2 to 5.3 ppm. All labeled signals are assigned as H1 (anomeric) resonances, except H6 and H6′ of 6-O-acetyl β-D-Galp. The signals labeled α-Glc and β-Glc are assigned to the reducing glucopyranose residue. Anomeric resonances of the two-linked α-D-Xylp residues (labeled 2-Xyl) are also labeled according to their substituent at O2. That is, L indicates a β-D-Gal at O2, F indicates an α-L-Fucp-(1→2)-β-D-Galp moiety at O2, LAc indicates a 6-O-acetyl-β-D-Galp at O2, and FAc indicates an α-L-Fucp-(1→2)-[6-O-acetyl]-β-D-Galp moiety at O2. The anomeric proton resonances of β-D-Galp residues with a α-L-Fucp substituent at O2 (i.e. 2-Gal) are shifted slightly up field (from δ 4.617 to δ 4.606) by O-acetylation at O6, as indicated by the asterisk. The presence or absence of the diagnostic signals, together with the methyl resonance of the O-acetyl substituents (δ 2.13, not shown) at O-6 of β-D-Galp residues, is consistent with the conclusion that fut1 oligosaccharides lack fucosyl residues, and that KOH-extracted oligosaccharides have no O-acetyl substituents.
Table I.
Side-chain analysis of xyloglucan derived from various Arabidopsis tissues
XyG oligosaccharides from the various tissues were characterized by 1H-NMR spectroscopy (see “Materials and Methods”). Data were normalized by setting the total number of side chains per oligosaccharide subunit to 3, consistent with the “XXXG-type” structure (Vincken et al., 1997) of Arabidopsis XyG. O-Acetyl substituents are hydrolyzed by 4 N KOH treatment.
| Tissue
|
Side Chains Per Subunit (Percentage of Side Chains with Acetatylated Gal Residue)
|
||
|---|---|---|---|
| Fuc→Gal→Xyl | Gal→Xyl | Xyl | |
| XEG-extracted xyoglucan | |||
| Flowers | 0.51a | 0.31a | 2.08 |
| Rosettes | 0.40a | 0.60a | 2.00 |
| Top stem | 0.43 (22) | 0.40 (14) | 2.17 |
| Lower stem | 0.43 (29) | 0.50 (18) | 2.07 |
| Siliques | 0.44a | 0.52a | 2.04 |
| Roots | 0.47a | 0.48a | 2.05 |
| KOH-extracted xyoglucan | |||
| Flowers | 0.46 | 0.41 | 2.13 |
| Rosettes | 0.53 | 0.51 | 1.96 |
| Top stem | 0.48 | 0.39 | 2.13 |
| Lower stem | 0.55 | 0.41 | 2.04 |
| Siliques | 0.50 | 0.47 | 2.03 |
| Roots | 0.49 | 0.27 | 2.24 |
| Cauline | 0.50 | 0.50 | 2.00 |
The indicated XEG-extracted samples did not contain sufficient material to accurately quantitate the levels of O-acetylation. However, the data clearly indicated that for all XEG-extracted samples in this table, less than 45% of the Fuc→Gal→Xyl side chains and less than 20% of the Gal→Xyl side chains contained an O-acetylated Gal residue. The highest levels of O-acetylation were found in rosettes, lower stem, and siliques.
Arabidopsis Plants Carrying a T-DNA Insertion in AtFUT1 Synthesize XyG That Lacks Fuc
Primers specific to AtFUT1 were used to screen T-DNA pools from the Arabidopsis Knockout Facility (Madison, WI), and plants homozygous for an insertion in the middle of the second exon (at Val-378) were identified. No full-length AtFUT1 mRNA transcripts were detected in the T-DNA mutant by RT-PCR, although transcripts of nonnative size were amplified from the atfut1 mutant using primers 5′ to the insertion site. Arabidopsis plants containing the T-DNA insertion had no visible phenotype under laboratory growth conditions. Such a result is consistent with previously published data showing that the Arabidopsis loss-of-function mutant mur2-1, in which there is a point mutation in the AtFUT1 gene, also grows normally under laboratory conditions (Vanzin et al., 2002).
Glycosyl composition analysis indicated that the cell walls prepared from mature rosettes, expanding leaves, and flowers of the atfut1 mutant contained 50% to 75% less Fuc than wild-type (Wassilewskija [WS] ecotype) walls (data not shown). Comparable amounts of Fuc were present in rhamnogalacturonans I and II isolated from the AIR of wild-type (WS2 ecotype) and atfut1 plants (data not shown). However, no Fuc was detected by 1H-NMR spectroscopic analyses of the oligosaccharides generated by XEG treatment of the 4 m KOH-soluble XyG from the shoots and roots of 7-d-old seedlings and from mature rosette leaves (Table II). These results confirm the observation that that AtFUT1 specifically fucosylates XyG (Faik et al., 2000) and indicate that the T-DNA insertion at the AtFUT1 locus causes a null allele.
Table II.
Side-chain analysis of xyloglucans from genetically modified Arabidopsis
XyG was extracted from depectinated cell wall preparations of wild-type (WS2), atfut1 (fut1), mur2, and mur1-2 (grown in the absence or presence of Fuc) rosette leaves, and the relative proportions of the various XyG side chains in each extract were determined by 1H-NMR spectroscopy.
| Sample
|
Side Chains Per Subunit (Percentage of Side Chains with Acetatylated Gal Residue)
|
||
|---|---|---|---|
| Fuc→Gal→Xyl | Gal→Xyl | Xyl | |
| XEG-extracted xyloglucan | |||
| WS2 Leaf | 0.45 (60) | 0.49 (4) | 2.07 |
| fut1 Leaf | n.d.a (0) | 1.10 (0) | 1.90 |
| mur2 Leaf | 0.03 (94) | 1.02 (0) | 1.95 |
| mur1-2b Leaf | n.d.a (—)c | 0.95 (0) | 1.97 |
| mur1-2b Leaf + Fuc | 0.19 (79)c | 0.75 (13) | 2.06 |
| KOH-extracted xyloglucan | |||
| ws2 Shootd | 0.45 | 0.42 | 2.13 |
| ws2 Rootd | 0.54 | 0.35 | 2.11 |
| ws2 Leaf | 0.46 | 0.52 | 2.03 |
| fut1 Shootd | n.d.a | 0.97 | 2.03 |
| fut1 Rootd | n.d.a | 0.92 | 2.08 |
| fut1 Leaf | n.d.a | 1.06 | 1.94 |
Not detected. bThe XEG-extracted mur1-2 xyloglucan samples also contain [α -L-galp-(1→2)-B -D-galp-(1→2)-A -D-Xylp-] trisaccharide side chains (0.10 per subunit). cO-Acetates attached to the different (Gal-Gal-Xyl or Fuc-Gal-Xyl) trisaccharide side chains were not distinguished by the method used. However, in both XEG-extracted mur1-2 leaf samples, 68% to 79% of the combined trisaccharide side chains bear an O-acetate. dDue to small sample size, the indicated samples were analyzed without purification on SEC on SD-75 and contain contaminants that reduce the accuracy of the quantitation.
Analysis of Biological and Biochemical Impact of AtFUT1 Overexpression
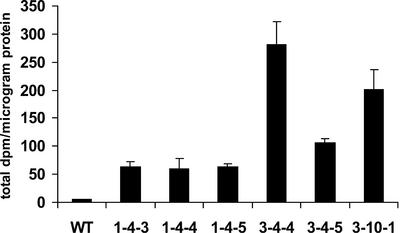
The effects of overexpression of AtFUT1 were determined by generating transgenic Arabidopsis (Col-0) plants containing a construct with cDNA encoding full-length AtFUT1 regulated by the cauliflower mosaic virus (CaMV) 35S strong constitutive promoter. The presence of the transgene in hygromycin-resistant plants was confirmed by PCR amplification using primers that span the intron of AtFUT1, allowing differentiation between the transgene and the endogenous genomic version of the gene (data not shown). Multiple lines were identified by RT-PCR with elevated expression of AtFUT1 in leaves as compared with wild-type plants, and six were chosen for further analysis. Golgi-enriched vesicle fractions were prepared from leaves of these plants, and their FUTase activities determined (Fig. 7). The transgenic lines all showed elevated levels of XyG FUTase activity compared with Golgi vesicles derived from wild-type leaves. Nevertheless, there were no visible phenotypic differences between 35S::AtFUT1 plants and wild-type plants grown under laboratory growth conditions.
Figure 7.
XyG FUTase activity in Golgi vesicles derived from 35S::AtFUT1 transgenic plants. Golgi vesicles were prepared from leaves of transgenic plants and were used in XyG FUTase activity assays. Assays were conducted at room temperature for 1 h. Duplicate samples were analyzed. Results are expressed as total dpm per microgram of protein. Error bars indicate 1 SD.
AIR were prepared from 11 different AtFUT1-overexpressing plants, and the Fuc contents of their XyGs were then determined (Table III). No increase in fucosylation was detected by 1H-NMR spectroscopic analysis of the XyG oligosaccharides derived from any of the 35S::AtFUT1 plants. However, there was a consistent increase in the O-acetylation of Gal residues in the Fuc-Gal-Xyl side chains of the XyG from 35S::AtFUT1 plants (Table III).
Table III.
Analysis of XyG from 35S::AtFUT1 transgenic plants
XyG was extracted from depectinated cell wall preparations from the rosette leaves of wild-type and overexpressing transgenic lines, and the relative proportions of the various XyG side chains in each extract were determined by 1H-NMR spectroscopy.
| Line | Side Chains Per Subunit (Percentage of Side Chains with Acetatylated Gal Residue) | ||
|---|---|---|---|
| Fuc→Gal→Xyl | Gal→Xyl | Xyl | |
| XEG-extracted xyloglucan | |||
| Wild type | 0.44 (45) | 0.52 (9) | 2.04 |
| 1-4-3 | 0.48 (72) | 0.50 (9) | 2.02 |
| 1-4-4 | 0.46 (72) | 0.50 (9) | 2.04 |
| 1-4-5 | 0.46 (76) | 0.50 (9) | 2.03 |
| 3-4-3 | 0.49 (61) | 0.47 (6) | 2.04 |
| 3-4-4 | 0.47 (65) | 0.46 (6) | 2.07 |
| 3-4-5 | 0.48 (69) | 0.51 (9) | 2.01 |
| 3-10-1 | 0.45 (74) | 0.57 (13) | 1.98 |
| KOH-extracted xyloglucan | |||
| Wild type | 0.49 | 0.48 | 2.03 |
| 1-4-3 | 0.49 | 0.44 | 2.06 |
| 1-4-4 | 0.48 | 0.48 | 2.04 |
| 1-4-5 | 0.50 | 0.47 | 2.04 |
| 3-4-3 | 0.48 | 0.48 | 2.05 |
| 3-4-4 | 0.50 | 0.45 | 2.06 |
| 3-4-5 | 0.49 | 0.47 | 2.04 |
| 3-10-1 | 0.47 | 0.50 | 2.03 |
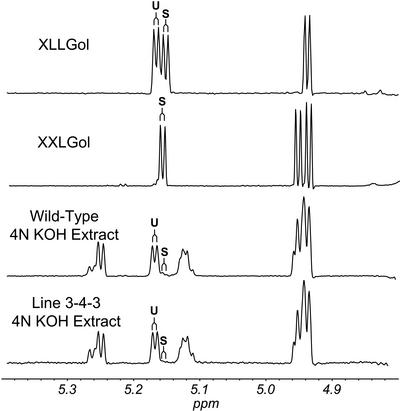
Side chains terminated by a d-Gal residue are potential acceptor substrates for the AtFUT1 enzyme. Therefore, 1H-NMR spectroscopy was used to determine the number and position of terminal d-Gal residues in XyG from wild-type and AtFUT1-overexpressing plants. XyG oligosaccharides were treated with sodium borohydride, converting them to oligosaccharide alditols. 1H-NMR analysis of these oligoglycosyl alditols allows the galactosyl residues of “suitable” AtFUT1 acceptor substrates (e.g. XXLG) to be distinguished from galactosyl residues that are “unsuitable” AtFUT1 acceptor substrates (e.g. XLLG and XLXG; see Fig. 8). The observed distinctions between suitable and unsuitable acceptors correspond with observations by Faik et al. (2000), who found that fucosylation of tamarind XyG by purified pea FUTase resulted in only XXFG or XLFG, but not XFXG or XFLG. Low levels of suitable acceptors were found in both wild-type and overexpressing lines, suggesting that acceptor substrate availability is a limiting factor for XyG fucosylation.
Figure 8.
Partial 1H-NMR spectrum of reduced XyG oligosaccharides. Oligosaccharides XLLG and XXLG from tamarind seed XyG (York et al., 1993) were purified and reduced to obtain the products (XLLGol and XXLGol). Oligosaccharides prepared from the 4 N KOH extract from rosette leaves of wild-type (Col-0) Arabidopsis and the line 3-3-4 (overexpressing AtFUT1) were also reduced. 1H-NMR spectra of all reduced products were recorded at 500 MHz. H1 resonances for α-D-Xylp residues that bear a terminal β-D-Galp residue at O2 and that are adjacent to the glucitol, which indicate the presence of a Gal-Xyl side chains that are suitable AtFUT1 acceptor substrates, are indicated by the letter S. H1 resonances of α-D-Xylp residues that bear a terminal β-D-Galp residue at O2 but are not adjacent to the glucitol, which indicate the presence of Gal-Xyl side chains that are unsuitable AtFUT1 acceptor substrates, are indicated by the letter U.
Determination of XyG Acetylation Levels in mur1-2, mur2, and atfut1
Our data (Table III) when taken together with the results of a previous study (Pauly et al., 2001a) suggest that the extent of O-acetylation of Arabidopsis XyG is correlated with the extent of fucosylation of this polysaccharide and/or abundance of AtFUT1. Additional support for this hypothesis was obtained by structural characterization of the XyG oligosaccharides generated by XEG treatment of the AIR from the rosette leaves of wild-type (WS2 ecotype), atfut1, mur2, mur1-2 plants, and mur1-2 plants that were grown in the presence of 10 mm l-Fuc (O'Neill et al., 2001). The mur2 mutant is defective in the AtFUT1 gene encoding XyG FUTase, whereas the aerial portions of mur1 plants are defective in an isoform of GDP-d-Man-4,6-dehydratase, an enzyme required for the formation of GDP-Fuc (Bonin et al., 1997; Vanzin et al., 2002). Virtually no Fuc and O-acetylated Gal residues were detected in the XyG oligosaccharides from atfut1 and mur2 leaves (Table II). No Fuc was detected in the XyG oligosaccharides from mur1-2 leaves, although low amounts of O-acetylated β-d-Gal residues were present. The O-acetylation of mur1-2 XyG may be due to the presence of α-l-Gal residues, which partially replace the missing α-l-Fuc residues in mur1-2 XyG (Zablackis et al., 1996). However, mur1-2 plants grown in the presence of Fuc synthesize XyG that contains α-l-Fuc residues and O-acetylated β-d-Gal residues (Table II). Thus, the presence of α-l-Fuc or α-l-Gal residues probably determines whether the XyG is a suitable acceptor substrate for at least one O-acetyltransferase in Arabidopsis.
DISCUSSION
Cell wall biosynthesis is believed to be correlated with plant growth, but few studies have examined the relationship between these two processes at the molecular level. This study demonstrates that fucosylation of XyG can be investigated at the levels of gene regulation, enzyme activity, and product composition.
A mutant referred to as mur2-1 has a point mutation in AtFUT1 and synthesizes XyG that contains less than 2% of the amounts of Fuc than the XyG derived from wild-type plants in various tissues including leaves, stems, flowers, and roots (Vanzin et al., 2002). Disruption of AtFUT1 by a T-DNA insertion in the first exon results in plants whose leaves and roots synthesize XyG with no detectable Fuc residues. Thus, AtFUT1 is likely to be the only gene in Arabidopsis encoding a XyG-specific FUTase. Although nine different genes are present in Arabidopsis that have 47% to 62% sequence identity with AtFUT1, of those tested, none encodes enzymes that catalyze the transfer of Fuc to XyG (Sarria et al., 2001).
Distinct patterns of gene expression and XyG FUTase activity were observed in Arabidopsis stems, leaves, and flowers. Gene expression and enzyme activity levels were highest in rapidly growing tissues and lowest in the oldest tissues of the mature plant. Flowers showed high enzyme activity, but RT-PCR analysis of the same sample did not show a high level of gene expression. The variation in gene expression in floral tissue may be due to the amount of pedicel and inflorescence that is present, because promoter-reporter analysis indicated that expression of AtFUT1 is particularly high in those tissues. In that respect, it is interesting to note that northern analysis of gene expression indicated higher expression of AtFUT1 in floral bud clusters, which included a portion of top stem and pedicel tissue, than in isolated mature flowers and associated pedicels. AtFUT1::GUS transgenic plants did not exhibit strong reporter staining in reproductive tissue itself. No reporter activity was detected in roots of AtFUT1::GUS lines, although expression of AtFUT1 was observed in roots analyzed by RT-PCR and northern analysis. It is possible that regulatory regions (such as the single intron present in this gene or the 86-nucleotide-long 3′-untranslated region) directing expression of AtFUT1 in root tissues were not present in the construct used for the promoter-reporter analysis. The low reporter activity observed in mature rosette leaves and the oldest regions of the stem and high activity observed in the youngest inflorescence stem tissue paralleled AtFUT1 expression results obtained using RT-PCR and hybridization techniques.
No correlations between the level of AtFUT1 expression and the Fuc content of XyG oligosaccharides were observed in the different organs of wild-type plants. Notably, the amounts of fucosylated XyG did not increase in the upper inflorescence stem even though this tissue had increased levels of AtFUT1 expression and XyG FUTase activity. Such a result suggests that differing levels of AtFUT1 gene expression and enzyme activity reflect the rate and amounts of cell wall deposition occurring in planta, rather than variations in XyG fucosylation patterns.
The results of XyG side-chain analysis indicate that side chains terminating in Gal are more abundant in leaf and silique XyG than in XyG from other tissues. This is consistent with a previous analysis demonstrating that the “central” side chain of the oligosaccharide subunits is more likely to be L (i.e. terminated with β-d-Galp) in leaf XyG than in XyG from stems (Pauly et al., 2001b). Furthermore, the extent of O-acetylation is highly correlated to fucosylation of the side chain next to the reducing Glc of the oligosaccharide subunits.
Ectopic expression of a gene is often used to study the effects of gene manipulation in planta. Transgenic plants expressing AtFUT1 under control of a strong constitutive promoter show higher gene expression and XyG FUTase activity than wild-type plants. Nevertheless, the overexpressing and wild-type plants synthesized XyG with comparable structures, which suggests that FUTase is not a limiting factor for XyG fucosylation. Our data show that the overexpression of AtFUT1 could not have resulted in a significant increase in XyG fucosylation, because the acceptor substrates (XLLG and XXLG) for the enzyme are almost fully fucosylated even in wild-type plants. The enzyme transfers a fucosyl to the galactosyl residue that terminates the side chain that is adjacent to the unbranched glucosyl residue, to produce XLFG and XXFG, respectively. Virtually no XFFG or XFLG is present in Arabidopsis XyG because side chains that are not adjacent to an unbranched glucosyl residue are not efficiently fucosylated by AtFUT1. In fact, Gal-Xyl side chains that are not in an appropriate position to act as AtFUT1 substrates are found in both wild-type and AtFUT1-overexpressing plants.
Previous studies of sycamore (Acer pseudoplatanus) XyG have shown that the major site of O-acetylation is O-6 of Gal residues. O-Acetylation also occurs at the O-4 and O-3 positions, and some Gal residues may be di-O-acetylated (York et al., 1988). Acetyl Co-A has been shown to be the donor substrate for O-acetylation (Pauly and Scheller, 2000), but the enzyme(s) responsible for the O-acetylation of XyG has not been identified. The biological role of O-acetylated residues in XyG is not known, although it has been speculated to hinder enzymatic fragmentation of the polymer (Pauly and Scheller, 2000).
The amounts of O-acetylated Gal residues in Arabidopsis XyG are positively correlated with the Fuc content of the XyG or AtFUT1 enzyme abundance. The amount of O-acetylation was consistently higher in plants that overexpress the AtFUT1 gene than in wild-type plants. Conversely, the levels of O-acetylation are considerably reduced in XyG isolated from atfut1 or mur2 plants, which have lesions in XyG FUTase, or in mur1-2 plants, which have a defect in the GDP-Fuc biosynthetic pathway. The XyG of mur1 plants does contain some O-acetyl substituents, presumably due to the presence of the J side chain [α-l-Galp-(1→2)-β-d-Galp-(1→2)-α-d- Xylp-], in which l-Fuc is replaced by l-Gal. Growing mur1 plants in the presence of Fuc partially restores both fucosylation and O-acetylation of the XyG. This suggests that the preferred substrate for at least one Arabidopsis O-acetyl transferase is fucosylated (or l-galactosylated) XyG. It is possible that recognition by the O-acetyl transferase is enhanced by a change in the molecular conformation of the XyG that occurs when Fuc residues are present. However, this alone does not explain the increased O-acetylation observed in plants that overexpress AtFUT1, because the level of fucosylation is no greater in these plants than in wild-type plants. More extensive analysis would be necessary to determine the basis for the correlation and to conclusively determine whether increased O-acetylation is the direct result of overexpression of AtFUT1. Nevertheless, it is possible that this correlation could stem from a process-based relationship between fucosylation and O-acetylation. For example, increasing FUT1 abundance in the Golgi may lead to more rapid fucosylation of the nascent XyG. If the residence time of XyG in the Golgi is relatively constant, this would lead to an increase in the time that the acceptor substrate (fucosylated xyoglucan) is available to the O-acetyl transferase. Furthermore, XyG may be a substrate for several different O-acetyl transferases in Arabidopsis, and the acceptor substrate specificities of these putative enzymes may be different. This hypothesis is consistent with the observation that the most abundant oligosaccharide subunit of the XyG produced by suspension-cultured mur1 cells is O-acetylated XXLG, an oligosaccharide subunit that does not contain any l-Gal or l-Fuc residues (Pauly et al., 2001a). If the high level of O-acetylation for the XXLG subunit in these cells depends on l-fucosylation or l-galactosylation of the XyG, then the O-acetyl transferase must be recognizing the presence of l-Gal or l-Fuc residues in subunits that are distal to the O-acetylation site. The data described herein confirm that, in general, Fuc-Gal-Xyl side chains are more frequently acetylated than Gal-Xyl side chains, suggesting that O-acetyl transferases can recognize the presence of a fucosyl residue that is proximal to the acetylation site.
Despite the XyG structural changes observed in AtFUT1-overexpressing plants, atfut1 and mur2, no visible changes in phenotype were apparent. This suggests that fucosylation and O-acetylation of XyG are not required for normal growth and development under laboratory conditions. However, fucosylation and O-acetylation of XyGs are widely observed in a taxonomically diverse range of dicotyledenous plants (Hayashi, 1989; Carpita, 1997). It is possible that fucosylation and O-acetylation confer some selective advantage under conditions yet to be determined in a laboratory setting.
In conclusion, AtFUT1 expression and XyG FUTase activity were positively correlated with growth in the aerial portions of Arabidopsis. Determination of the gene expression profile of AtFUT1 provides a baseline of comparison with other genes thought to encode enzymes involved in primary cell wall biosynthesis. Disruption of the AtFUT1 locus by insertion of a T-DNA in the second exon results in XyG containing no detectable Fuc and reduced O-acetylation of Gal residues. Increasing the expression of AtFUT1 using the strong constitutive CaMV 35S promoter results in increased XyG FUTase activity and increased XyG O-acetylation. This result, when taken together with the virtual absence of O-acetylation in atfut1, mur1-2, and mur2 XyGs, provides evidence that there is a positive correlation between XyG O-acetylation and fucosylation.
MATERIALS AND METHODS
Plant Growth Conditions and Tissue Collection
Arabidopsis Col-0 plants were grown hydroponically as previously described (Gibeaut et al., 1997) in growth chambers (16 h of light, 22°C). Plants were grown to maturity (8 weeks under these conditions). Soil-grown plants were grown on greenhouse-prepared soil media under the same growth chamber conditions.
Arabidopsis Col-0, mur1-2, and mur2 plants were grown in potting soil (Fafard 3B, Fafard, Inc., Anderson, SC) in a controlled environmental chamber with a 14-h-light and 10-h-dark cycle at 19°C and 15°C, respectively. Arabidopsis mur1-2 plants were also grown in the presence of 10 mm l-Fuc as previously described (O'Neill et al., 2001). The rosette leaves were harvested from 4-week-old plants and stored at –80°C. WS2 and atfut1 plants were grown on vertically oriented plates containing Murashige and Skoog media with 2% (w/v) Suc.
Flowers, the uppermost 6 cm of the inflorescence stem, the lowest 6 cm of the inflorescence stem, siliques, cauline leaves, rosette leaves, and roots were harvested from mature hydroponically grown Arabidopsis plants. Tissues were collected on ice, weighed, and divided into portions to be used for Golgi vesicle, RNA, and cell wall preparations. Golgi vesicles were prepared using fresh tissue. RNA and cell wall samples were prepared using tissue that had been frozen in liquid nitrogen and kept at –80°C.
Preparation of RNA from Arabidopsis
RNA was prepared by grinding tissue under liquid nitrogen to a fine powder and then suspending in 65°C extraction buffer (2% [w/v] hexadecyltrimethylammonium bromide, 2% [w/v] polyvinylpyrrolidone K 30, 100 mm Tris-HCl pH 8.0, 25 mm EDTA, 2.0 m NaCl, 0.5 g L–1 spermidine, and 2% [v/v] β-mercaptoethanol). The suspensions were extracted twice with an equal volume of chloroform. One-quarter volume of 10 m LiCl was added to the supernatant, and the mixture was kept at 4°C for several hours to precipitate RNA. The samples were centrifuged (7,000 rpm, 5,800g) at 4°C for 20 min, and pellets were then resuspended in Tris-buffered EDTA (0.5% [w/v] SDS, 10 mm Tris, pH 7.5 [diethyl pyrocarbonate (DEPC) treated], and 1 mm EDTA [DEPC treated]). An equal volume of phenol:chloroform: isoamyl alcohol (25:24:1, v/v) was added, and the mixtures were vortexed and centrifuged at room temperature for 15 min (1,000g). The upper phase was removed and 0.25 volume of 10 m LiCl was added. Samples were incubated at 4°C for several hours and then centrifuged (5,800g) for 20 min at 4°C to pellet RNA. The pellets were resuspended in 2 mL of DEPC-treated water, and 0.1 volume of 3 m sodium acetate (pH 5.2) and 2.5 volumes of ethanol were added. The samples were centrifuged (5,800g) for 15 min to pellet RNA. The supernatant was removed, an appropriate amount of DEPC-treated water was used to resuspend the pellet, and samples were centrifuged briefly to remove any insoluble material.
Analysis of AtFUT1 Expression by RT-PCR
RNA samples were treated with RNase-free DNase I using the DNA-free kit (Ambion, Austin, TX) according to the manufacturer's instructions. RNA (1 μg) was used in a RT reaction with 0.5 μg of oligo(dT) primer (Invitrogen, Carlsbad, CA), 1× first-strand buffer, 10 mm dithiothreitol (DTT), 5 mm dNTPs, and 200 units of Superscript II reverse transcriptase (Invitrogen). RNA, water, and the reverse primer were incubated at 70°C for 10 min, transferred to ice for 2 min, centrifuged, and mixed with first-strand buffer, DTT, and dNTPs. The samples were incubated at 42°C for 1 min and reverse transcriptase was added. The samples were kept at 42°C for 50 min and then at 70°C for 15 min. PCR was performed using equal volumes of the original RT reactions as template in 20-μL PCR reactions with 1× PCR buffer, 150 μm MgCl2, 125 μm each forward and reverse primer, and 5 units of Amplitaq (Roche Diagnostics, Mannheim, Germany). Primers used to amplify AtFUT1 were: forward primer, 5′-GAA GGG CTA CTT GCT TCT GGT TTT-3′; reverse primer, 5′-CCC GAT GAA TGT TTG GTC TCC TT-3′. These primers amplify a 578-bp fragment of AtFUT1 from nucleotides 571 to 1,149. Thermal cycling parameters for amplification of AtFUT1 were: 94°C for 1 min (hot start); 92°C for 30 s, 58°C for 1 min, 72°C for 1 min and 30 s for 22 to 24 cycles; and 72°C for 5 min. Primers used to amplify cytochrome c were: forward primer, 5′-TCG CTT ATT TGA AGG AAG TG-3′; reverse primer, 5′-CTC TTC ACA TCA ATA GCGT AAT-3′. These primers amplify a 212-bp fragment of cytochrome c (Arabidopsis thaliana Gene Index no. TC87099), located on chromosome 4. Thermal cycling parameters for the amplification of cytochrome c were: 94°C for 1 min (hot start); 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min for 28 cycles.
Analysis of AtFUT1 Expression by RNA Gel Blot
RNA was isolated from hydroponically grown wild-type (Col-0) Arabidopsis plants as described above. For the purposes of these experiments, “flower” samples refers to individual, fully opened flowers and associated petioles; “floral buds” refers to a cluster of unopened flowers, associated petioles, and approximately 2 mm of uppermost inflorescence stem; “top stem” refers to the uppermost 0.5 cm of inflorescence stem after removal of flowers; “low stem” refers to a segment of the inflorescence stem from 1.5 cm from the top of the stem (after removal of flowers) to the base of the stem; “leaves” refers to mature rosette leaves; and “roots” refers to the total root mass. Fifteen micrograms of total RNA was subjected to RNA gel-blot analysis using a hybridization probe specific for AtFUT1 (Sarria et al., 2001). The blot was then stripped and hybridized with a probe to detect eIF4a as a loading control.
Preparation of Golgi Vesicles from Arabidopsis
Golgi vesicles were prepared using the method of Muñoz et al. (1996), adapted for use in Arabidopsis. Tissue was weighed and 1 mL of 0.5 m Suc solution (0.5 m Suc, 0.1 m KH2PO4, pH 6.65, 5 mm MgCl2, 1 mm DTT, and EDTA-free complete protease inhibitor cocktail [Roche Diagnostics] was added per g of tissue. The tissue was chopped by hand with a razor blade, ground with a mortar and pestle, and homogenized with a Pyrex Dounce homogenizer. The homogenate was filtered through Miracloth and centrifuged at 1,000g for 5 min. The supernatant was removed and layered on top of a 1.3 m Suc solution cushion (1.3 m Suc, 0.1 m KH2PO4, pH 6.65, and 5 mm MgCl2) and centrifuged at 100,000g for 90 min in a swinging bucket rotor. The upper phase was removed without disturbing the interface and 1.1 m Suc solution (1.1 m Suc, 0.1 m KH2PO4, pH 6.65, and 5 mm MgCl2) was layered on top, followed by a layer of 0.25 m Suc solution (0.25 m Suc, 0.1 m KH2PO4, pH 6.65, and 5 mm MgCl2). The samples were centrifuged at 100,000g for 100 min in a swinging bucket rotor. The 0.25 m/1.1 m Suc interface fraction was collected, 1 volume of water was added, and the samples were centrifuged at 100,000g for 50 min in a fixed angle rotor. The supernatant was discarded, and the pellet was suspended in Suc-Tris-Manganese Chloride buffer (0.5 m Suc, 2 mm MgCl2, 20 mm Tris-HCl, pH 7.5, and protease inhibitor cocktail tablets). Aliquots of the preparations were reserved for quantification by the Bradford assay.
XyG FUTase Activity Assays
XyG FUTase activity assays were conducted as previously described (Faik et al., 2000). All assays of Golgi vesicle preparations included 1% (v/v) Triton X-100 to permeabilize the vesicles and thereby allow access of tamarind XyG substrate to the enzyme.
Isolation and Enzymatic Fragmentation of XyG
Cell Wall Preparations
AIR were prepared by grinding frozen tissue to a powder under liquid nitrogen. The ground tissue was suspended in 80% (v/v) ethanol (approximately 20 mL g–1 tissue) and further disrupted using a Polytron homogenizer. The AIR was collected on nylon mesh and washed with aqueous 80% (v/v) ethanol and then with absolute ethanol. The washed AIR was suspended in methanol:chloroform (1:1, v/v), stirred for 1 h at room temperature, collected by filtration through Whatman paper, washed with acetone, and air dried.
Partial Depectination of AIR
AIR were suspended in 10 mL of 50 mm NaOAc, pH 5, containing 0.01% (w/v) thimerosal. Endo-polygalacturonase (5 units) from Aspergillus niger (provided by Dr. Carl Bergmann, CCRC), and pectin methylesterase (5 units) from Aspergillus oryzae (supplied by Novozymes A/S, Bagsvaerd, Denmark) were added. The suspension was incubated for 24 h at 24°C in a shaking incubator. The suspensions were filtered, and the solid residues were treated a second time with endo-polygalacturonase and pectin methylesterase.
XEG Treatment
The partially depectinated AIR were suspended in 10 mL of 20 mm NaOAc, pH 5, containing 0.01% (w/v) thimerosal. XEG (10 units, supplied by Novozymes A/S) was added, and the suspensions were incubated at 24°C for 24 h in a shaking incubator and then filtered. The filtrate, containing XyG oligosaccharides, was applied to a C-18 cartridge (Supelcelan LC-18 SPE tube, Supelco, Bellefonte, PA), which was then washed with water (10 mL) to remove salts. The oligosaccharides were then eluted from the cartridge with 10 mL of aqueous 25% (v/v) methanol. The eluant was concentrated under vacuum and lyophilized. The oligosaccharides were further purified on a Superdex-75 HR10/30 column (Amersham Biosciences, Uppsala) eluted with 50 mm ammonium formate, pH 5.0, at a flow rate of 0.5 mL min–1. Carbohydrate in the eluant was monitored by refractive index. Fractions containing the oligosaccharides were pooled and lyophilized several times to remove volatile ammonium formate salts.
1 N KOH and 4 N KOH Treatment
The insoluble residues remaining after XEG treatment were suspended in 10 mL of 1 n KOH containing 1% (w/v) NaBH4 and stirred at room temperature for 24 h. The suspensions were filtered, and the insoluble residues were then suspended in 10 mL of 4 n KOH containing 1% (w/v) NaBH4. The suspensions were stirred for 24 h at room temperature and then filtered. The filtrate was adjusted to pH 5 with glacial AcOH and dialyzed (3,500 Mr cutoff tubing, Spectrum Laboratories, Rancho Dominguez, CA) against six changes of deionized water over 2 d. The retentates were lyophilized to yield the 4 n KOH-solubilized XyG polysaccharides, which were digested with XEG (as described above) to generate XyG oligosaccharides, which were isolated from the reaction mixture as described above.
1H-NMR Spectroscopy
Solutions of XyG oligosaccharides in D2O (0.6 mL, 99.9%; Cambridge Isotope Laboratories, Andover, MA) were analyzed at 25°C using Inova NMR spectrometers (Varian Medical Systems, Palo Alto, CA) operating at 500 and 600 MHz. Five diagnostic regions in the anomeric region of the spectra (Fig. 6) were integrated: (a) δ 5.27, H1 of all α-Fucp residues, (b) δ 5.18, H1 of α-Xylp residues bearing a terminal β-Galp residue at O2, (c) δ 5.14, H1 of α-Xylp residues bearing either an O-acetylated terminal β-Galp residue or an a-Fucp-(1→2)-β-Galp disaccharide at O2, (d) δ 5.09, H1 of α-Xylp residues bearing an O-acetylated a-Fucp-(1→2)-β-Galp disaccharide at O2, and (e) δ 4.94–4.96, H1 of terminal α-Xylp residues. The relative amount of the unacetylated a-Fucp-(1→2)-β-Galp-(1→2)-α-Xylp side chain was calculated by subtracting the signal area at δ 5.09 (Fuc-containing O-acetylated side chains) from the area at δ 5.27 (all Fuc-containing side chains). The amount of the acetylated β-Galp-(1→2)-α-Xylp side chain was calculated by subtracting the result of the last calculation (unacetylated a-Fucp-(1→2)-β-Galp-(1→2)-α-Xylp side chain) from the area at δ 5.14 (acetylated β-Galp-(1→2)-α-Xylp side chain plus unacetylated a-Fucp-(1→2)-β-Galp-(1→2)-α-Xylp side chain). The results were verified by integration of other diagnostic signals in the spectra, including the methyl proton signal (δ 2.13) of O-acetyl substituents of the a-Fucp-(1→2)-β-Galp-(1→2)-α-Xylp side chain. The spectra of oligosaccharides from the 4 n KOH fractions do not contain resonances corresponding to O-acetylated side chains.
Generation of AtFUT1::GUS-GFP Transgenic Plants
A 1,987-bp fragment upstream of the AtFUT1 open reading frame was amplified from Arabidopsis genomic DNA. The primers used for amplification were: 5′-GGG GGA TCC CTA TAG TGG CTG TCT GCT TGA GGA-3′ and 5′-GGG CCA TGG ATT GCT CTT GAG GGA-3′. Primers included restriction sites for BamHI and NcoI digestion. Fragments were cloned in pGemT-Easy vector (Promega, Madison, WI) and then into BamHI and NcoI sites in the pCAMBIA 1303 vector. This caused a fusion of the first ATG codon originating from AtFUT1 with the gusA-mGFP5 double reporter present in the pCAMBIA 1303 vector. The constructs were introduced into Agrobacterium tumefaciens by standard transformation methods and transgenic Arabidopsis plants were generated by vacuum infiltration (Bechtold and Pelletier, 1998). Lines bearing single insertions of the transgene were identified by determining ratios of selective marker inheritance in T2 and T3 plants on hygromycin-containing media.
GUS-Staining Conditions
Whole seedlings or plant tissues were harvested and suspended in cold aqueous 90% (v/v) acetone for 20 min on ice to permeabilize the tissues. Samples were washed three times for 15 min each in working solution (100 mm NaH2PO4 containing 10 mm EDTA, 0.5 mm ferrocyanide, 0.5 mm ferricyanide, and 0.1% [v/v] Triton X-100). Samples were then kept at 37°C for 16 to 36 h in staining solution (working solution containing 2 mm X-Gluc [Rose Scientific, Edmonton, Canada]). Samples were cleared of chlorophyll by washing with aqueous 70% (v/v) ethanol and photographed using a Coolpix 995 digital camera (Nikon, Tokyo) attached to a light microscope or stereoscope.
Identification of T-DNA Allele in AtFUT1
The T-DNA pools of the Wisconsin collection were screened as recommended by the Biotechnology Center-University of Wisconsin (http://www.biotech.wisc.edu/NewServicesAndResearch/Arabidopsis/Guidelines-Index.html). Forward and reverse primers used for the mutant screen were respectively, 5′-ACAATTAAACCATGTCGTGGAGACATGTT-3′ and 5′-TCATACTAGCTTAAGTCCCCAGCTGATAT-3′. The reverse primer in combination with the T-DNA left border primer (JL202) amplified the insertion. RT-PCR of mutant and wild-type RNA was performed using 1 μg of total RNA and the One-Step RT-PCR kit (Qiagen, Germantown, MD) according to the manufacturer's directions. Primers for RT-PCR spanned the intron and are as follows: 5′-TTCTCGACGCCGGAGTTT-3′ and 5′-CCCTCAGTATCAATCACCT-3′.
Generation of 35S::AtFUT1 Transgenic Plants
A cDNA fragment encoding the open reading frame of AtFUT1 was subcloned into the NcoI/XhoI sites of pET14-b. The AtFUT1 cDNA was then cloned into the SalI/XbaI sites of pCAMBIA 1300 MCS (a derivative of pCAMBIA with additional sites in the MCS generated by Dr. Anton Sanderfoot [Michigan State University]). This generated a construct in which the 35S CaMV strong constitutive promoter controls the expression of AtFUT1. The construct was introduced into A. tumefaciens by standard transformation methods and transformed into wild-type (Col-0) Arabidopsis plants using the vacuum infiltration method (Bechtold and Pelletier, 1998). Transgenic plants were selected using hygromycin and the presence of the transgene was confirmed by PCR amplification across the AtFUT1 intron, which results in a 489-bp product for the genomic version and a 212-bp product for the transgene. RNA was prepared from rosette leaves of T2 plants using the RNeasy Mini Plant Kit (Qiagen) and used to analyze AtFUT1 expression by RT-PCR as described above.
Acknowledgments
We thank Dr. Carl Bergmann of the CCRC and Novozymes A/S for supplying enzymes used in the analysis of XyG.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.016642.
This work was supported by the Department of Energy Biosciences Program and the Plant Genome Program at the National Science Foundation, by the U.S. Department of Energy (grant no. DE–FG05–93ER20220 to Z.J., M.A.O., and W.S.Y.), and by the U.S. Department of Energy-funded Center for Plant and Microbial Complex Carbohydrates (grant no. DE–FG05–93ER20097 to Z.J., M.A.O., and W.S.Y.).
References
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazinski J, Birch R et al. (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Bonin CP, Potter I, Vanzin GF, Reiter WD (1997) The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-d-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-l-fucose. Proc Natl Acad Sci USA 94: 2085–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Camirand A, Maclachlan GA (1990) Differential distribution of xyloglucan transferases in pea Golgi dictyosomes and secretory vesicles. J Cell Sci 96: 705–710 [Google Scholar]
- Carpita NC (1997) Structure and biosynthesis of plant cell walls. In DT Dennis, DH Turpin, DD Lefebvre, DB Layzell, eds, Plant Metabolism, Ed 2. Longman Scientific & Technical, New York, pp 124–147
- Cosgrove DJ (2000) Expansive growth of plant cell walls. Plant Physiol Biochem 38: 109–124 [DOI] [PubMed] [Google Scholar]
- Edwards ME, Dickson CA, Chengappa S, Sidebottom C, Gidley MJ, Reid JS (1999) Molecular characterisation of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J 19: 691–697 [DOI] [PubMed] [Google Scholar]
- Faik A, Bar Peled M, DeRocher AE, Zeng W, Perrin RM, Wilkerson C, Raikhel NV, Keegstra K (2000) Biochemical characterization and molecular cloning of an alpha-1,2-fucosyltransferase that catalyzes the last step of cell wall xyloglucan biosynthesis in pea. J Biol Chem 275: 15082–15089 [DOI] [PubMed] [Google Scholar]
- Faik A, Price NJ, Raikhel NV, Keegstra K (2002) An Arabidopsis gene encoding an alpha-xylosyltransferase involved in xyloglucan biosynthesis. Proc Natl Acad Sci USA 99: 7797–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas V, Maclachlan G (1988) Fucosylation of exogenous xyloglucans by pea microsomal membranes. Arch Biochem Biophys 264: 48–53 [DOI] [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, Darvill A, Bayashi R, Joseleau JP, Kato Y, Lorences EP, Maclachlan GA, McNeil M et al. (1993) An unambiguous nomenclature for xylogucan-derived oligosaccharides. Physiol Plant 89: 1–3 [Google Scholar]
- Gibeaut DM, Hulett J, Cramer CR, Seemann JR (1997) Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol 115: 317–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T (1989) Xyloglucans in the primary cell wall. Annu Rev Plant Physiol Plant Mol Biol 40: 139–168 [Google Scholar]
- Muñoz P, Norambuena L, Orellana A (1996) Evidence for a UDP-glucose transporter in Golgi apparatus-derived vesicles from pea and its possible role in polysaccharide biosynthesis. Plant Physiol 112: 1585–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill M, Eberhard S, Alberscheim P, Darvill A (2001) Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294: 846–849 [DOI] [PubMed] [Google Scholar]
- Pauly M, Albersheim P, Darvill A, York WS (1999) Molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. Plant J 20: 629–639 [DOI] [PubMed] [Google Scholar]
- Pauly M, Eberhard S, Alberscheim P, Darvill A, York W (2001a) Effects of the mur1 mutation on xyloglucans produced by suspension-cultured Arabidopsis cells. Planta 214: 67–74 [DOI] [PubMed] [Google Scholar]
- Pauly M, Qin Q, Greene H, Albersheim P, Darvill A, York WS (2001b) Changes in the structure of xyloglucan during cell elongation. Planta 212: 842–850 [DOI] [PubMed] [Google Scholar]
- Pauly M, Scheller HV (2000) O-Acetylation of plant cell wall polysaccharides: identification and partial characterization of a rhamnogalacturonan O-acetyl-transferase from potato suspension-cultured cells. Planta 210: 659–667 [DOI] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K (1999) Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284: 1976–1979 [DOI] [PubMed] [Google Scholar]
- Perrin RM, Wilkerson C, Keegstra K (2001) Golgi enzymes that synthesize plant cell wall polysaccharides: finding and evaluating candidates in the genomic era. Plant Mol Biol 47: 115–130 [PubMed] [Google Scholar]
- Richmond TA, Somerville CR (2000) The cellulose synthase superfamily. Plant Physiol 124: 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarria R, Wagner TA, O'Neill MA, Faik A, Wilkerson CG, Keegstra K, Raikhel NV (2001) Characterization of a family of Arabidopsis genes related to xyloglucan fucosyltransferase1. Plant Physiol 127: 1595–1606 [PMC free article] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD (2002) The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci USA 99: 3340–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincken JP, York WS, Beldman G, Voragen AG (1997) Two general branching patterns of xyloglucan, XXXG and XXGG. Plant Physiol 114: 9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff C, Norambuena L, Orellana A (2000) GDP-fucose uptake into the Golgi apparatus during xyloglucan biosynthesis requires the activity of a transporter-like protein other than the UDP-glucose transporter. Plant Physiol 122: 867–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS, Harvey LK, Guillen R, Albersheim P, Darvill AG (1993) Structural analysis of tamarind seed xyloglucan oligosaccharides using β-galactosidase digestion and spectroscopic methods. Carbohydr Res 248: 285–301 [DOI] [PubMed] [Google Scholar]
- York WS, Oates JE, van Halbeek H, Darvill AG, Albersheim P, Tiller PR, Dell A (1988) Location of the O-acetyl substituents on a nonasaccharide repeating unit of sycamore extracellular xyloglucan. Carbohydr Res 173: 113–132 [DOI] [PubMed] [Google Scholar]
- Zablackis E, York WS, Pauly M, Hantus S, Reiter WD, Chapple CC, Albersheim P, Darvill A (1996) Substitution of l-fucose by l-galactose in cell walls of Arabidopsis mur1. Science 272: 1808–1810 [DOI] [PubMed] [Google Scholar]