Abstract
Cellulose synthase catalytic subunits (CesAs) have been implicated in catalyzing the biosynthesis of cellulose, the major component of plant cell walls. Interactions between CesA subunits are thought to be required for normal cellulose synthesis, which suggests that incorporation of defective CesA subunits into cellulose synthase complex could potentially cause a dominant effect on cellulose synthesis. However, all CesA mutants so far reported have been shown to be recessive in terms of cellulose synthesis. In the course of studying the molecular mechanisms regulating secondary wall formation in fibers, we have found that a mutant allele of AtCesA7 gene in the fra5 (fragile fiber 5) mutant causes a semidominant phenotype in the reduction of fiber cell wall thickness and cellulose content. The fra5 missense mutation occurred in a conserved amino acid located in the second cytoplasmic domain of AtCesA7. Overexpression of the fra5 mutant cDNA in wild-type plants not only reduced secondary wall thickness and cellulose content but also decreased primary wall thickness and cell elongation. In contrast, overexpression of the fra6 mutant form of AtCesA8 did not cause any reduction in cell wall thickness and cellulose content. These results suggest that the fra5 mutant protein may interfere with the function of endogenous wild-type CesA proteins, thus resulting in a dominant negative effect on cellulose biosynthesis.
Cellulose is the most abundant biopolymer produced by plants. It is the major component of plant cell walls and, in particular, is synthesized in large quantities during secondary wall formation of tracheary elements and fibers in wood. Cellulose synthase, the enzyme responsible for cellulose biosynthesis, is located in the plasma membrane. It is imaged by transmission electron microscopy as a rosette consisting of six particles, which is termed the terminal rosette complex (Brown and Montezinos, 1976). The identification of the rosette complexes as the sites of cellulose synthesis was confirmed by the immunolocation of putative cellulose synthase catalytic subunits (CesAs) in the rosette subunits (Kimura et al., 1999). It is proposed that as many as 36 CesA protein molecules together with other putative associated proteins form one rosette complex, which produces 36 (1,4)-β-glucan chains that polymerize and crystallize into a microfibril with an estimated diameter of 8 to10 nm (Ha et al., 1998). The monosaccharide donor substrate used by CesAs, UDP-Glc, has been postulated to be provided by membrane-associated form(s) of Suc synthase (Amor et al., 1995). Recent in vitro studies suggest that the initiation of β-1,4-glucan synthesis starts with the addition of Glc units to a primer, sitosterol lipid, to form lipid-linked oligosaccharides called sitosterol cellodextrin (Peng et al., 2002). Cellodextrins are then thought to be cleaved from the sitosterol primer probably by a KOR cellulase (Nicol et al., 1998) and further elongated by addition of more Glc molecules to form a long chain of β-1,4-glucan (Peng et al., 2002). It is not known whether each CesA protein is involved in both initiation and elongation of the sugar chains, or different CesA proteins play different roles in this process (Read and Bacic, 2002).
Genes encoding CesA proteins in plants were first identified in cotton (Gossypium hirsutum) fibers (Pear et al., 1996), and their essential roles in crystalline cellulose synthesis were genetically confirmed in the Arabidopsis rsw1 mutant (Arioli et al., 1998). Since then, several additional Arabidopsis mutants with mutations in CesA genes have been isolated. It has been shown that at least four CesA genes, namely AtCesA1 (RSW1), AtCesA2, AtCesA3 (IXR1/CEV1), and AtCesA6 (PRC1/IXR2), are involved in primary wall synthesis, and mutations or antisense repression of these genes cause a reduction in cellulose synthesis (Arioli et al., 1998; Fagard et al., 2000; Scheible et al., 2001; Burn et al., 2002; Desprez et al., 2002; Ellis et al., 2002). The reduced cellulose synthesis in primary walls is associated with a decrease in cell elongation, which was shown in some of these mutants to be correlated with an increased production of jasmonate and ethylene (Ellis et al., 2002). Two other CesA genes, AtCesA7 (IRX3) and AtCesA8 (IRX1), have been shown to be essential for cellulose biosynthesis in secondary walls (Taylor et al., 1999, 2000). Mutations in these gene as seen in the irx mutants cause dramatic reductions in cellulose content and secondary wall thickness, leading to a collapsed xylem phenotype (Turner and Somerville, 1997). An additional gene, AtCesA4, has also been shown to be expressed in cells undergoing secondary wall thickening (Holland et al., 2000). The available evidence indicates that, although multiple CesA genes are expressed in the same cell types, mutation of one of them is sufficient to cause cellulose reduction (Fagard et al., 2000; Taylor et al., 2000).
Biochemical studies showed that AtCesA7 and AtCesA8 proteins co-immunoprecipitate, suggesting that different CesA proteins might interact with each other in the rosette complexes (Taylor et al., 2000). It has been hypothesized that different CesA proteins may be required to form a functional rosette complex because the CesA genes in Arabidopsis are functionally nonredundant (Scheible et al., 2001). If this is the case, incorporation of defective CesA polypeptides into a rosette complex might interfere with the function of the entire complex. In this scenario, mutation of one of the CesA genes could potentially cause a dominant or semidominant effect on cellulose synthesis. However, analyses of the irx1 and irx3 mutants showed that both mutants are recessive (Turner and Somerville, 1997). In fact, all reported CesA mutants with defects in cellulose synthesis have been shown to be recessive except two herbicide-resistant CesA mutants exhibiting semidominant resistance to herbicides (Scheible et al., 2001; Desprez et al., 2002).
In this paper, we report that a missense mutation in the second cytoplasmic domain of AtCesA7 in the fra5 mutant results in a semidominant phenotype in the reduction of fiber wall thickness and cellulose biosynthesis. The second cytoplasmic domain is located between the second and third transmembrane helices in the predicted topology of CesA proteins (Delmer, 1999). We further demonstrate that overexpression of the fra5 mutant cDNA in wild-type plants causes a dominant negative effect on cellulose biosynthesis. Because missense mutations in all other reported CesA mutants do not cause a semidominant effect on cellulose synthesis, our results suggest that mutations in the specific region around the fra5 mutation site in the second cytoplasmic domain may be critical for the semidominant phenotype.
RESULTS
The Semidominant fra5 Mutation Occurs in a Conserved Region of the Second Cytoplasmic Domain of AtCesA7
To investigate the mechanisms controlling fiber cell wall formation, we isolated two fragile fiber mutants in Arabidopsis, fra5 and fra6, that showed a dramatic reduction in the mechanical strength of mature inflorescence stems. Anatomical analysis showed that both homozygous and heterozygous fra5 mutants exhibited reduced fiber wall thickness (Fig. 1, A–C; Table I). The reduced wall thickness in the fra5 mutants was directly correlated with a decrease in cellulose amount and Glc content (Fig. 2, A–C; Tables I and II). This indicates that the fra5 mutation causes a semidominant effect on cellulose biosynthesis. Analysis of the fra6 mutant showed that only the homozygous fra6 mutant had reductions in fiber wall thickness and cellulose amount (data not shown), suggesting that the fra6 mutation is recessive.
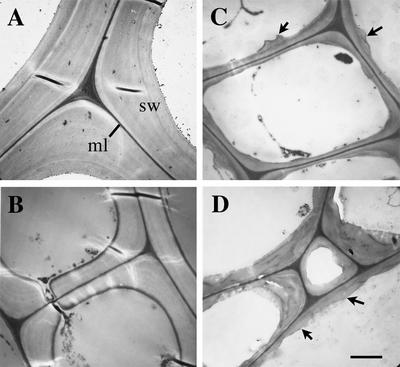
Figure 1.
Transmission electron microscopy of the walls of interfascicular fiber cells. Mature inflorescence stems were cross-sectioned for imaging of interfascicular fiber cell walls. A, Wild-type fiber cells showing thick walls. B, Heterozygous fra5 fiber cells showing markedly reduced wall thickness. C, Homozygous fra5 fiber cells showing thin walls and undulated wall surface (arrows). D, Fiber cells of the fra5-overexpressing plants showing thin walls and uneven wall surface (arrows). ml, Middle lamella; sw, secondary wall. Bar in D = 1.8 μm for A to D.
Table I.
Measurement of fiber wall thickness and cellulose
| Cell Wall Pyrolysis Mass Spectra Ion Current (× 106)c
|
||||||
|---|---|---|---|---|---|---|
| Cellulose mass-to-charge ratio (m/z) 126 + 144
|
||||||
| Sample | Fiber Wall Thicknessa | Crystalline Celluloseb | Total count | Count | Total | Wild type |
| μm | mg g-1 | % | % | |||
| Wild type | 2.75 ± 0.22 | 273 ± 11 | 5.45 | 0.182 | 3.34 | 100 |
| +/fra5 | 1.35 ± 0.25 | 208 ± 7 | 4.58 | 0.101 | 2.20 | 66 |
| fra5/fra5 | 0.55 ± 0.11 | 123 ± 14 | 6.62 | 0.126 | 1.90 | 57 |
| 35S::fra5d | 0.65 ± 0.30 | 67 ± 7 | 11.02 | 0.122 | 1.10 | 33 |
| 35S::fra6e | 2.71 ± 0.34 | 281 ± 16 | 9.42 | 0.302 | 3.21 | 96 |
Wall thickness was measured from transmission electron micrographs of fiber cells. Data are means ± SE from 20 cells. bCellulose content was calculated as milligrams per gram of dry cell walls isolated from stems. Data are means ± SE of three assays. cIon currents were calculated from in-source pyrolysis mass spectra of stem cell walls. Cellulose mass peaks at m/z 126 and 144 were used for quantitation of relative amount of cellulose. dTransgenic plants with overexpression of the fra5 mutant cDNA driven by the cauliflower mosaic virus (CaMV) 35S promoter. eTransgenic plants with overexpression of the fra6 mutant cDNA driven by the CaMV35S promoter.
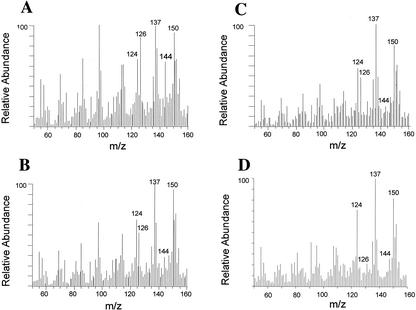
Figure 2.
In-source pyrolysis mass spectrometry of cell walls. Cell walls isolated from mature inflorescence stems were analyzed for the relative amount of different wall components by in-source pyrolysis mass spectrometry. Mass peaks of lignin had an m/z of 124, 137, 138, 150, 152, and 154. Mass peaks of cellulose had an m/z of 57, 60, 73, 85, 86, 96, 98, 100, 102, 110, 112, 126, and 144. Mass peaks of hemicellulose had an m/z of 58, 85, 86, and 114 (Niemann et al., 1992). Note that compared with the wild type (A), there is a significant reduction in the relative intensity of mass peaks for cellulose in the fra5 heterozygotes (B), the fra5 homozygotes (C), and the fra5-overexpressing plants (D).
Table II.
Monosaccharide composition of the cell walls of inflorescence stems (milligrams per gram of dry cell wall)
| Sample | Glc | Rha | Ara | Xyl | Man | Gal |
|---|---|---|---|---|---|---|
| Wild type | 266 ± 21 | 3.3 ± 1.3 | 7.1 ± 3.0 | 101 ± 8 | 15.3 ± 2.6 | 9.9 ± 0.6 |
| +/fra5 | 165 ± 7 | 3.4 ± 0.1 | 9.2 ± 0.7 | 104 ± 5 | 15 ± 1 | 9.2 ± 0.7 |
| fra5/fra5 | 138 ± 21 | 3.3 ± 0.3 | 12.7 ± 0.8 | 123 ± 10 | 15.7 ± 0.9 | 13.1 ± 0.5 |
| 35S::fra5 | 119 ± 9 | 5.7 ± 0.5 | 14.4 ± 3.4 | 190 ± 11 | 15.4 ± 1.2 | 15.1 ± 0.8 |
To investigate the molecular nature of the fra5 and fra6 mutations, we isolated their corresponding genes by a map-based approach. Using codominant amplified polymorphic sequences (CAPS) markers, we mapped the fra5 locus to a 35-kb region near the CAPS marker MKP-A on chromosome 5 and the fra6 locus to a 42-kb region near the CAPS marker AG on chromosome 4. Sequencing of putative genes led to the findings that the fra5 mutant carries a point mutation (C changed to A) in the AtCesA7 gene, and the fra6 mutant has a point mutation (G changed to A) in the AtCesA8 gene. Transformation of the wild-type AtCesA7 gene into the homozygous fra5 plants partially rescued the fra5 mutant phenotypes including stem strength, fiber wall thickness, and cellulose content (data not shown), indicating that the semidominant fra5 mutation occurs in the AtCesA7 gene. Transformation of the wild-type AtCesA8 gene into the fra6 plants completely restored the stem strength, fiber wall thickness, and cellulose amount (data not shown), confirming that the fra6 mutation occurs in the AtCesA8 gene.
Sequence analysis showed that the semidominant fra5 mutation causes a missense amino acid substitution (P557T) in the second cytoplasmic domain of AtCesA7 (Fig. 3). The recessive fra6 mutation results in a missense amino acid change (R362K) in the second cytoplasmic domain of AtCesA8 (Fig. 3). Although both fra5 and fra6 mutants have missense mutations in the second cytoplasmic domains of CesA proteins, the mutations occur in two different conserved regions, and only the fra5 mutation causes a semidominant effect on cellulose biosynthesis.
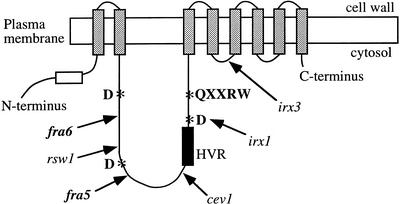
Figure 3.
A schematic diagram of the predicted topology of a CesA protein showing the locations of the fra5 and fra6 mutations relative to other missense CesA mutations. CesA proteins are predicted to contain eight transmembrane helices (gray bars), an N terminus-located first cytoplasmic domain that contains zinc-binding motifs (open box), and a second cytoplasmic domain located between the second and the third transmembrane helices (Delmer, 1999). The conserved residues (D,D,D,QXXRW; indicated by asterisks) that are believed to be important for substrate binding and catalysis are located in the second cytoplasmic domain. The relative locations of various missense mutations in different AtCesA proteins are marked with arrows. These mutations include fra6 (R362K), rsw1 (A549V; Arioli et al., 1998), fra5 (P557T), cev1 (G617E; Ellis et al., 2002), irx1-1 (D683N; Taylor et al., 2000), and irx1-2 (S679L; Taylor et al., 2000). Also shown is the irx3 mutation that causes a premature stop codon between the fourth and the fifth transmembrane helices (Taylor et al., 1999). HVR, Hypervariable region.
The AtCesA7 and AtCesA8 genes have been shown previously to be responsible for the recessive irx3 and irx1 mutations, respectively, which caused a collapsed xylem phenotype (Turner and Somerville, 1997; Taylor et al., 1999, 2000). However, the fra5 and fra6 mutations did not result in a collapsed xylem phenotype (data not shown). Although both fra5 and fra6 mutations resulted in a severe reduction in fiber wall thickness, they only caused a mild decrease in vessel wall thickness (data not shown). The apparent difference between the fra and irx mutants could be due to the nature of the mutations or differences in growth conditions.
Overexpression of the fra5 Mutant cDNA Causes Dramatic Reductions in Secondary Wall Thickness and Cellulose Content
Because all other reported CesA mutants with defects in cellulose synthesis are recessive except two herbicide-resistant CesA mutants showing semidominant resistance to herbicides (Scheible et al., 2001; Desprez et al., 2002), it was intriguing to find that the fra5 mutation results in a semidominant effect on cellulose biosynthesis. There are at least two possibilities to explain the semidominant phenotype of the fra5 mutant. The reduced cellulose synthesis in the fra5 heterozygotes could be due to the reduced amount of functional AtCesA7 protein, i.e. a dosage effect. Or alternatively, it could be caused by a dominant negative effect in which the fra5 mutant protein interferes with the normal function of wild-type AtCesA7. To test these possibilities, we overexpressed the fra5 and fra6 mutant proteins in wild-type plants. We reasoned that if the fra5 semidominant phenotype were due to a dosage effect, overexpression of the nonfunctional fra5 mutant protein in wild-type plants would not be expected to affect cellulose biosynthesis if the amount of wild-type AtCesA7 protein is not altered.
To generate fra5-overexpressing plants, the full-length fra5 mutant cDNA driven by the CaMV35S promoter was introduced into wild-type plants. Fifty-two transgenic plants were produced, and 37 of them showed a weak stem strength phenotype. Three transgenic lines were chosen for further analysis. They were selfed, and the T2 generation was used for characterization. Semiquantitative reverse transcription (RT)-PCR analysis showed that all three lines expressed a high level of fra5 mutant mRNA (Fig. 4A, fra5 and fra5+CesA7). The expression level of wild-type AtCesA7 gene remained unaltered in these lines (Fig. 4A, CesA7), thus ruling out the possibility of gene silencing by cosuppression. Stem strength tests showed that like the homozygous fra5 mutant, the forces required to break mature stems of these transgenic plants, were significantly lower than those of the wild type (Fig. 5). This demonstrates that overexpression of the fra5 mutant cDNA in wild-type plants leads to a remarkable alteration in stem strength.
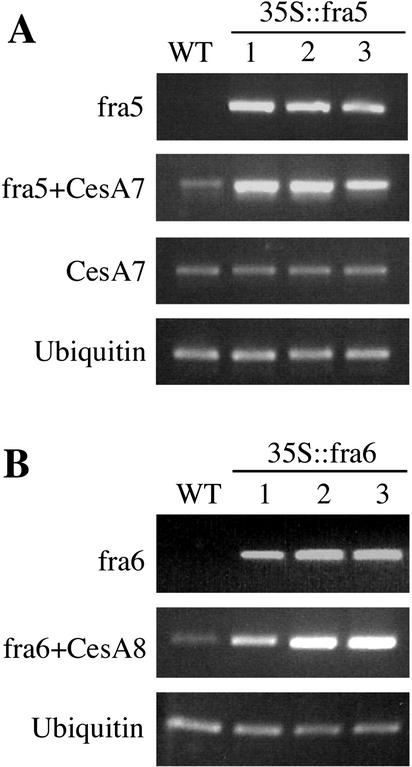
Figure 4.
Overexpression of the fra5 and fra6 mutant cDNAs in transgenic plants. Five-day-old seedlings of the wild-type (WT) and transgenic plants with overexpression of the fra5 mutant cDNA (35S::fra5) or the fra6 mutant cDNA (35S::fra6) were used for RNA isolation and subsequent RT-PCR analysis. Expression of a ubiquitin gene was used as an internal control. A, Gene-specific primers as described in “Materials and Methods” were used to determine the level of the fra5 mutant mRNA (fra5), that of the endogenous AtCesA7 mRNA (CesA7), and the combined level of the fra5 mutant mRNA and the endogenous AtCesA7 mRNA (fra5+CesA7). Note that although a high level of the fra5 mRNA was seen in the 35S::fra5 lines, the amount of the endogenous AtCesA7 mRNA was not altered. B, Gene-specific primers were used to determine the level of the fra6 mutant mRNA (fra6) and the combined level of the fra6 mutant mRNA and the endogenous AtCesA8 mRNA (fra6+CesA8).
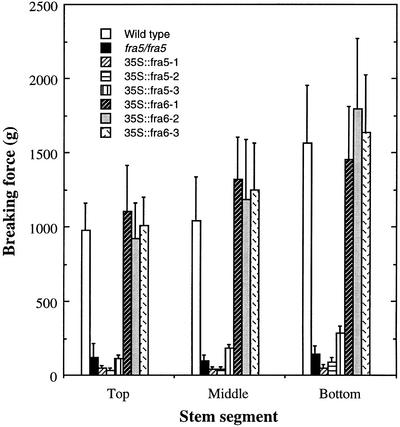
Figure 5.
Reduction of breaking strength in the inflorescence stems of the fra5-overexpressing plants. Inflorescence stems of 10-week-old plants were divided into three segments, and individual segments were used for measurement of breaking strength. Note that although the breaking strength of the stems of the 35S::fra6 lines were similar to that of the wild type, the stems of the 35S::fra5 lines were remarkably weak.
fra6-Overexpressing plants were generated by transformation of wild-type plants with a binary vector carrying the full-length fra6 mutant cDNA driven by the CaMV35S promoter. Sixty-seven transgenic plants were produced, and none of them exhibited a weak stem phenotype in the stem strength tests as shown in three representative lines (Fig. 5). RT-PCR analysis showed that the fra6 mutant mRNA was expressed in these lines (Fig. 4B).
Further anatomical and chemical analyses were performed on the three fra5-overexpressing and three fra6-overexpressing lines. Whereas the three transgenic lines showed similar results, the data from only one representative line, 35S:fra5–1 or 35S:fra6–3, are presented. To investigate whether fra5 overexpression affected secondary wall thickening, we examined the anatomy of interfascicular fibers and vessel elements. Cross sections of mature stems showed that the fiber walls in the 35S:fra5 plants were extremely thin compared with the wild type (Fig. 6, A and B). Transmission electron microscopy of fiber walls revealed that like the homozygous fra5 mutant (Fig. 1C), the surface of the thin fiber walls in the fra5-overexpressing plants was often undulated (Fig. 1D). Quantitative measurement based on transmission electron micrographs showed that the fiber wall thickness in the 35S:fra5 plants was reduced to 24% of that of the wild type (Table I). Examination of vessel elements showed that fra5 overexpression resulted in a collapsed vessel phenotype (Fig. 6D), although the anatomy of vessel elements in the homozygous fra5 mutant (Fig. 6E) did not show any apparent differences from that of the wild type (Fig. 6C). These results demonstrate that fra5 overexpression causes a severe defect in secondary wall thickening in fiber cells and to a lesser extent in vessel elements.
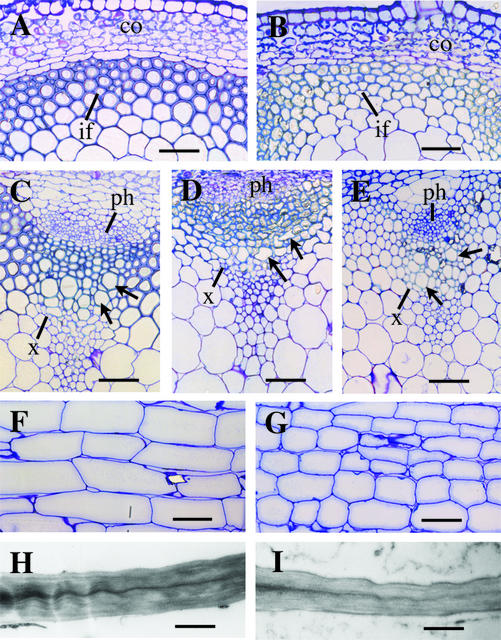
Figure 6.
Cellular morphology in the fra5-overexpressing plants. Basal parts of mature inflorescence stems were sectioned and stained with toluidine blue for observation of cellular morphology. A and B, Cross sections of stems showing interfascicular fiber cells with thin walls in the 35S::fra5 plants (B) compared with the wild type (A). C to E, Cross sections of stems showing that whereas the vessel elements (arrows) in the wild type (C) and the homozygous fra5 mutant (E) had regular shapes, those in the 35S::fra5 plants (D) were often deformed in shape. F and G, Longitudinal sections of stems showing pith cells with reduced length in the 35S::fra5 plants (G) compared with the wild type (F). H and I, Transmission electron micrographs of pith cells showing reduced wall thickness in the 35S::fra5 plants (I) compared with the wild type (H). co, Cortex; if, interfascicular fiber; ph phloem; x, xylem. Bars = 57 μm for A to E, 93 μm for F and G, and 1.14 μm for H and I.
We next analyzed the amount of cellulose in mature stems. In-source pyrolysis mass spectrometry showed that the relative intensity of cellulose-to-total cell wall mass count in the stems of the 35S:fra5 plants was reduced to 33% of that of the wild type (Fig. 2, A and D; Table I). The reduction in crystalline cellulose was confirmed by the nitric/acetic analysis (Table I). Wall composition analysis showed a 55% decrease in the amount of Glc but no reduction in other monosaccharides (Table II). In fact, there was an increase in other monosaccharides in the fra5-overexpressing plants. In addition, analysis of galacuronic acid showed a 55% increase in the fra5-overexpressing plants compared with the wild type. This observation probably is related to a compensatory change in wall composition, which was reported in cells treated with cellulose synthesis inhibitor (Shedletzky et al., 1992), in plants with virus-induced silencing of cesA genes (Burton et al., 2000) and in prc1 (Fagard et al., 2000) and kor (His et al., 2001) mutants.
In contrast, overexpression of the fra6 mutant cDNA did not cause any reduction in secondary wall thickness and cellulose amount (Table I). These results indicate that expression of the defective fra6 protein in wild-type plants appears not to affect the activity of endogenous CesA proteins, which is consistent with the recessive phenotype of the fra6 mutant. On the other hand, the dramatic reduction in cellulose synthesis in the fra5-overexpressing plants suggests that the fra5 mutant protein most likely interferes with the activity of endogenous wild-type CesA proteins, thus causing a dominant negative effect on cellulose synthesis.
Overexpression of the fra5 Mutant cDNA Alters Plant Growth
Three genes (AtCesA4, AtCesA7, and AtCesA8) are believed to be associated with cellulose synthesis in secondary cell wall formation (Taylor et al., 1999; Holland et al., 2000) and mutations of AtCesA7 as seen in irx3 (Turner and Somerville, 1997) and fra5 plants (data not shown) do not affect plant growth except for the pendent stem phenotype due to weakened stems. However, fra5 overexpression not only caused a pendent stem phenotype (Fig. 7A) but also resulted in altered plant growth. The inflorescence stems of the fra5-overexpressing plants were much shorter compared with the wild type (Fig. 7A; Table III). Longitudinal sections of stems showed that the length of pith cells in the 35S:fra5 plants was reduced to 52% of that of the wild type (Fig. 6, F and G; Table III), indicating that the shortened stem length was due to a reduced cell length. Transmission electron microscopy revealed that the thickness of pith cell walls in the 35S:fra5 plants was decreased by 27% compared with the wild type (Fig. 6, H and I; Table III). fra5 mutants did not exhibit any alterations in the length and wall thickness in pith cells (data not shown). These results suggest that fra5 overexpression not only interferes with cellulose synthesis during secondary wall thickening but also affects cellulose synthesis during primary wall formation.
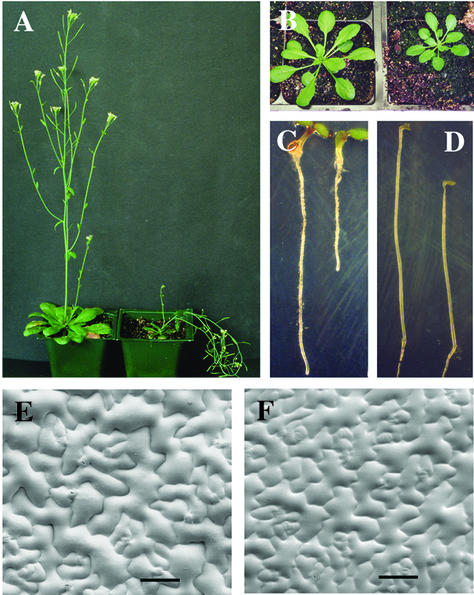
Figure 7.
Alteration of plant morphology by fra5 overexpression. A, Six-week-old 35S::fra5 plant (right) showing short pendent stem phenotype compared with the long erect stems of the wild type (left). B, Four-week-old 35S::fra5 plant (right) showing smaller rosette leaves compared with the wild type (left). C, Four-day old light-grown 35S::fra5 seedling (right) showing the reduced root length compared with the wild type (left). D, Four-day-old etiolated 35S::fra5 seedling (right) showing the reduced hypocotyl length compared with the wild type (left). E and F, Scanning electron micrographs of leaf epidermal cells showing the reduced cell size in the 35S::fra5 plants (F) compared with the wild type (E). Bars = 81 μm for E and F.
Table III.
Length and width of organs of wild-type and the 35S::fra5 plants
Data are means ± SE from 15 to 20 samples.
| Organ | Wild Type | 35S::fra5 |
|---|---|---|
| Stem height (cm)a | 34.9 ± 4.1 | 15.8 ± 2.3 |
| Pith cell length (μm)b | 201 ± 38 | 104 ± 15 |
| Pith wall thickness (μm)c | 0.67 ± 0.08 | 0.49 ± 0.07 |
| Leafd | ||
| Blade length (mm) | 23.8 ± 1.4 | 17.7 ± 1.9 |
| Blade width (mm) | 13.4 ± 1.0 | 10.0 ± 0.7 |
| Petiole length (mm) | 18.2 ± 2.3 | 10.6 ± 1.2 |
| Root length (mm)e | 9.4 ± 2.1 | 5.7 ± 1.2 |
| Hypocotyl length (mm)f | 15.4 ± 1.5 | 12.0 ± 1.5 |
Inflorescence stems of 6-week-old plants were measured for their height. bPith cell length was measured from longitudinal sections of inflorescence stems. cWall thickness was measured from transmission electron micrographs of stem pith cells. dFifth leaves of 4-week-old plants were used for measurement. eRoots of 4-day-old light-grown seedlings were used for measurement. fHypocotyls of 4-day-old dark-grown seedlings were used for measurement.
fra5 Overexpression also affected the expansion of leaves and the elongation of primary roots and dark-grown hypocotyls (Fig. 7, B–D). The length and width of leaf blades of the 35S:fra5 plants were decreased by 26% and 25%, respectively, compared with the wild type (Fig. 7B; Table III). The smaller leaf blades were accompanied with smaller leaf epidermal cells (Fig. 7, E and F), indicating that fra5 overexpression caused a reduction in the expansion of leaf epidermal cells. The length of petioles was reduced by 42% in these plants (Table III). The lengths of roots and dark-grown hypocotyls of the 35S:fra5 plants were reduced by 39% and 22%, respectively, compared with the wild type (Table III). The reduced root length was correlated with a decrease in cell length as revealed by imaging of root cells with confocal microscopy (data not shown). The reduced expansion or elongation of cells and organs was observed in all three fra5-overexpressing lines. Overexpression of the fra6 mutant cDNA did not cause any changes in the elongation of cells and organs (data not shown). These results are consistent with the fact that reduction of cellulose synthesis in primary walls affects cell elongation as seen in several other CesA mutants (Arioli et al., 1998; Fagard et al., 2000; Williamson et al., 2001; Ellis et al., 2002). However, unlike these mutants, the fra5-overexpressing lines did not exhibit any increase in the widths of cells or organs, which might be due to the less severity of cellulose reduction in the primary walls of these transgenic plants. Reduced cell length without an increase in cell width also has been reported in antisense AtCesA1 and AtCesA3 transgenic lines that exhibit a reduced cellulose content (Burn et al., 2002).
DISCUSSION
The fra5 Mutation Causes a Dominant Negative Effect on Cellulose Biosynthesis
The available evidence suggests that the semidominant fra5 phenotype is not caused by a dosage effect. If the reduced cellulose synthesis in the fra5 heterozygotes were caused by loss of one-half of the normal amount of functional AtCesA7 protein, overexpression of the fra5 mutant cDNA in wild-type plants should not result in defects in cellulose biosynthesis because the expression of endogenous AtCesA7 gene remains unaltered. However, analyses of the fra5-overexpressing plants showed that fra5 overexpression causes a severe reduction in cellulose synthesis. In contrast, overexpression of the fra6 mutant cDNA in wild-type plants had no effect on cellulose synthesis. In addition, overexpression of the wild-type AtCesA7 cDNA did not result in any reduction in cellulose content (Z.-H. Ye, unpublished data). These results indicate that the reduction in cellulose synthesis caused by fra5 overexpression is specific. They further suggest that the fra5 mutant protein most likely interferes with the activity of endogenous CesA proteins, thus leading to a defect in cellulose synthesis.
The suggestion that the semidominant fra5 mutant phenotype is not caused by loss of one-half of the normal amount of functional AtCesA7 is consistent with the previous analyses of other CesA mutants. So far, mutations occurring in five of the 10 CesA genes in Arabidopsis have been reported, and none of them have been shown to cause a dominant or semidominant phenotype in terms of cellulose synthesis (Arioli et al., 1998; Taylor et al., 1999, 2000;; Fagard et al., 2000; Ellis et al., 2002). The exceptions are the ixr1 and ixr2 mutants that show semidominant resistance to herbicide drugs, and the mutations are proposed to disrupt the binding sites of the drugs (Scheible et al., 2001; Desprez et al., 2002). The recessive phenotype caused by nonsense mutations of the AtCesA7 and AtCesA3 genes in the irx3 (Taylor et al., 1999) and the prc1 (Fagard et al., 2000) mutants, respectively, suggests that loss of one-half of the normal amount of one CesA protein does not cause any detectable effects on cellulose synthesis. In addition, disruption of the function of one type CesA protein by missense mutation of a CesA gene, as shown in rsw1 (Arioli et al., 1998), irx1 (Taylor et al., 2000), cev1 (Ellis et al., 2002), and fra6 mutants, does not necessarily result in a dominant or semidominant phenotype. The fact that the fra5 mutation causes a semidominant effect on cellulose synthesis indicates that the fra5 mutation is novel among all known CesA mutations.
It is not clear whether the fra5 mutant protein may interfere with the function of only AtCesA7 protein or also some other CesA proteins. The fact that fra5 overexpression in wild-type plants causes a more severe effect on the reduction of cellulose synthesis than does the homozygous fra5 mutant (Table I) suggests that the fra5 mutant protein might also affect the activity of other CesA proteins such as AtCesA4 and AtCesA8 in addition to AtCesA7, all of which are expressed during secondary wall formation (Taylor et al., 1999, 2000; Holland et al., 2000).
Possible Mechanisms for the Dominant Negative Effect of the fra5 Mutation
The fra5 missense mutation occurs in a conserved amino acid in the second cytoplasmic domain of AtCesA7. The second cytoplasmic domains of CesA proteins are highly conserved except for a short hypervariable region. The conserved amino acids that form the putative catalytic site are located in this domain (Fig. 3). Besides the catalytic activity, the second cytoplasmic domain of CesA proteins has been postulated to play a role in protein-protein interactions (Delmer, 1999). This hypothesis is supported by the fact that a missense mutation in the second cytoplasmic domain of AtCesA1 in the rsw1 mutant causes a disappearance of the rosettes (Arioli et al., 1998). However, the rsw1 mutation is recessive and has been shown to cause a reduction in crystalline cellulose formation without affecting (1,4)-β-glucan polymerization (Arioli et al., 1998). The fra5 mutant is distinguished from rsw1 by its semidominant phenotype and a severe reduction in both crystalline cellulose and glucan content (Tables I and II), which suggests how the fra5 mutation affects cellulose synthesis might be different from that of the rsw1 mutation.
Because the fra5 mutation causes a missense amino acid change, it is possible that the fra5 mutant protein may be able to incorporate into the rosette complex, and its presence may affect the activity of the whole rosette complex, thus leading to a dominant negative effect on cellulose biosynthesis. Although the precise mechanism of how the fra5 mutant protein causes a dominant negative effect is not clear, we speculate the following possibilities. The fra5 mutation might affect CesA-CesA interactions in the rosette complex or interactions between CesA proteins and other cellular components essential for cellulose biosynthesis. Tests of protein-protein interactions in the yeast (Saccharomyces cerevisiae) two-hybrid system did not reveal any interactions between the second cytoplasmic domains of CesA proteins, and the second cytoplasmic domains did not show any interactions with the first cytoplasmic domains (data not shown). The first cytoplasmic domains of CesA proteins have been shown recently to interact with each other and are proposed to mediate the formation of the rosette complexes (Kurek et al., 2002). Our data suggest that the second cytoplamsic domains of AtCesA proteins might not be involved in direct interactions between CesA polypeptides; rather, they might mediate interactions of CesA proteins with other cellular components in the rosette complex. We hypothesize that elimination of such an interaction in one or several CesA subunits in the rosette complex, such as by the fra5 mutation, might reduce the functional efficiency or disrupt the function of the whole complex, thus leading to a dominant negative effect on cellulose synthesis.
Several other missense mutations in the second cytoplasmic domains of CesA proteins have been reported (Fig. 3). The missense mutations in irx1 (AtCesA8) occur at or around an Asp residue essential for forming the catalytic site, presumably leading to a loss of the catalytic activity (Taylor et al., 2000). The missense mutations in cev1 (AtCesA3; Ellis et al., 2002) and fra6 (AtCesA8) occur in different conserved regions of the second cytoplasmic domains. How these mutations affect the activity of the CesA proteins and result in reduced cellulose synthesis is not clear. Nevertheless, all these mutations have been shown to be recessive, suggesting that unlike the fra5 mutant protein, these mutant proteins appear not to affect the function of wild-type CesA proteins. Further investigation on the roles of the conserved regions of the second cytoplasmic domains of CesA proteins in cellulose synthesis is crucial in our understanding of how these conserved regions exert their effects on the activities of CesA proteins.
fra5 Overexpression May Also Affect the Functions of AtCesA Proteins Involved in Cellulose Biosynthesis in Primary Walls
It has been proposed that CesA proteins are functionally nonredundant because mutations in one type of CesA protein result in a severe loss of cellulose synthesis even though other CesA proteins are still present in the same cell types (Scheible et al., 2001). In addition, it has been shown that overexpression of the AtCesA3 protein in the rsw1 mutant with a defective AtCesA1 does not rescue the mutant phenotype, indicating that different types of CesA proteins may not be functionally interchangeable (Burn et al., 2002). Despite the fact that mutations in the AtCesA7 gene only affect cells undergoing secondary wall thickening, expression of the fra5 mutant form of AtCesA7 under the CaMV 35S promoter not only causes a dominant negative effect on cellulose synthesis during secondary wall formation but also appears to affect cellulose synthesis during primary wall formation. This suggests that in the fra5-overexpressing plants, the fra5 mutant protein may also be able to incorporate into cellulose-synthesizing rosettes in elongating cells, thus affecting the activity of CesA proteins involved in primary wall synthesis. Further studies on how the fra5 mutant protein interferes with the activity of wild-type CesA proteins will likely contribute to our understanding of the complex process of cellulose biosynthesis.
MATERIALS AND METHODS
Mutant Isolation
Ethyl methanesulfonate-mutagenized M2 Arabidopsis populations (ecotype Columbia) were grown in a greenhouse and screened for mutants with reduced stem strength. The mechanical strength of inflorescence stems was determined by measuring the force needed to break stem segments (Zhong et al., 1997). Mutant lines were backcrossed with wild-type Columbia three times before further analysis.
Microscopy
Mature inflorescence stems from 10-week-old plants were fixed and embedded in Araldite resin (Electron Microscopy Sciences, Fort Washington, PA). One-micrometer-thick sections were cut and stained with toluidine blue for light microscopy. For transmission electron microscopy, 90-nm-thick sections were cut, post-stained with uranyl acetate and lead citrate, and viewed with an EM 902A electron microscope (Zeiss, Jena, Germany).
For imaging of leaf epidermis, fully expanded rosette leaves were cryoprepared, coated with gold, and viewed with a LEO982 FE scanning electron microscope (LEO, Thornwood, NY).
Cell Wall Analysis
Mature inflorescence stems of 10-week-old plants were used for cell wall preparation. Crystalline cellulose was assayed with the acetic-nitric anthrone reagent according to Updegraff (1969). Cell wall monosaccharides (as alditol acetates) were measured according to Hoebler et al. (1989). Cell wall composition was analyzed using a Finngan GCQ mass spectrometer equipped with a direct exposure probe (rhenium loop; Thermoquest, San Jose, CA) according to Morrison and Archibald (1998). Analysis conditions were as follows: ionization energy of 20 electron V, mass range of 50 to 500 m/z, scan time of 1 s, temperature increase of approximately 10°C s–1 to 700°C, and ion source temperature of 175°C. All samples were run in triplicate.
Map-Based Cloning
The fra5 and fra6 mutants were crossed with Arabidopsis ecotype Landsberg erecta to generate F2 plants for mapping of mutant loci according to Konieczny and Ausubel (1993). For fine mapping of the fra5 and fra6 loci, 746 and 685 F2 mapping plants were used, respectively. Cleaved CAPS markers were either obtained from the Arabidopsis database or developed based on information from the Cereon Arabidopsis polymorphic database.
For complementation analysis, the wild-type AtCesA7 and AtCesA8 genes were PCR amplified and confirmed by sequencing. They were cloned into the binary vector pBI101 (CLONTECH, Polo Alto, CA) and introduced into the fra5 and fra6 mutants, respectively, by the Agrobacterium tumefaciens-mediated transformation procedure (Bechtold and Bouchez, 1994). Transgenic plants were selected on kanamycin and grown to maturity for analysis of their ability to complement the mutant phenotypes.
Generation of fra5- or fra6-Overexpressing Plants
The full-length coding region of the fra5 or fra6 mutant cDNA was PCR amplified from cDNAs derived from stems of the fra5 or fra6 mutant, respectively. Amplified cDNAs were confirmed by sequencing and ligated downstream of the CaMV 35S promoter in the binary vector pBI121 (CLONTECH). The constructs were introduced into wild-type Arabidopsis plants (ecotype Columbia) by the A. tumefaciens-mediated transformation procedure. Transgenic plants were selected on kanamycin, and the T2 generation was used for further analyses. The control wild types used in all the analyses were transgenic plants transformed with the pBI121 vector.
Gene Expression Analysis
Five-day-old seedlings of transgenic plants were extracted for total RNA using an RNA isolation kit (Qiagen, Valencia, CA). One microgram of total RNA was treated with DNase I and reverse transcribed to synthesize first strand cDNA. One-twentieth of the cDNA synthesized was used for PCR amplification of genes of interest with gene-specific primers. The CesA7-int5 primer (5′-CACAGAGGATATTTTGACGGGATTC-3′) and the CesA7-int3 primer (5′-TCAGCAGTTGATGCCACACTTGGA-3′) were derived from the coding region of AtCesA7 cDNA, and they were used to determine the combined amount of the wild-type AtCesA7 and the fra5 mutant mRNAs in the fra5-overexpressing plants. The CesA7-int5 primer and the Nos-T primer (5′-ATCGCAAGACCGGCAACAGGATTC-3′), which was derived from the Nos terminator on the PBI121 vector, were used to analyze the level of the fra5 mutant mRNA. The CesA7-int5 primer and the CesA7-ext3 primer (5′-GTGAAAACACTCTCGACAAAGTACAG-3′), which was derived for the 3′-untranslated region of the AtCesA7 mRNA, were used to analyze the expression level of the endogenous AtCesA7 mRNA in the transgenic plants. The CesA8-int5 primer (5′-GCTATTCATGTCATTAGCTGTGGA-3′) and the CesA8-int3 primer (5′-TTAGCAATCGATCAAAAGACAGTT-3′) were derived from the coding region of the AtCesA8 cDNA, and they were used to determine the combined amount of the wild-type AtCesA8 and the fra6 mutant mRNAs in the fra6-overexpressing plants. The CesA8-int5 primer and the Nos-T primer were used to analyze the level of the fra6 mutant mRNA. The expression of a ubiquitin gene was used as an internal control to determine the efficiency of RT-PCR among different samples.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requestor.
Acknowledgments
We thank John Shields for his assistance in the scanning electron microscopy and Parastoo Azadi for pectin analysis. We thank the reviewers and the editor for their constructive comments and suggestions on the revision of the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.019331.
This work was supported by the U.S. Department of Agriculture (grant to Z.-H.Y.'s laboratory) and by the Department of Energy (grant no. DE–FG02–96ER20220 to the Complex Carbohydrate Research Center).
References
- Amor Y, Haigler CH, Johnson S, Wainscott M, Delmer DP (1995) A membrane associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc Natl Acad Sci USA 92: 9353–9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R et al. (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Bouchez D (1994) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In I Potrykus, G Spangenberg, eds, Gene Transfer to Plants. Springer-Verlag, Berlin, pp 19–23 [DOI] [PubMed]
- Brown RM, Montezinos D (1976) Cellulose microfibrils: visualization of biosynthetic and orienting complexes in association with the plasma membrane. Proc Natl Acad Sci USA 73: 143–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn JE, Hocart CH, Birch RJ, Cork AC, Williamson RE (2002) Functional analysis of the cellulose synthase genes CesA1, CesA2, and CesA3 in Arabidopsis. Plant Physiol 129: 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Gibeaut DM, Bacic A, Findlay K, Roberts K, Hamilton A, Baulcombe DC, Fincher GB (2000) Virus-induced silencing of a plant cellulose synthase gene. Plant Cell 12: 691–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer DP (1999) Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol 50: 245–276 [DOI] [PubMed] [Google Scholar]
- Desprez T, Vernhettes S, Fagard M, Refrégier G, Desnos T, Aletti E, Py N, Pelletier S, Höfte H (2002) Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol 128: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M-A, Apperley DC, Evans BW, Huxham IM, Jardine WG, Viëtor RJ, Reis D, Vian B, Jarvis MC (1998) Fine structure in cellulose microfibrils: NMR evidence from onion and quince. Plant J 16: 183–190 [DOI] [PubMed] [Google Scholar]
- His I, Driouich A, Nicol F, Jauneau A, Höfte H (2001) Altered pectin composition in primary cell walls of korrigan, a dwarf mutant of Arabidopsis deficient in a membrane-bound endo-1,4-β-glucanase. Planta 212: 348–358 [DOI] [PubMed] [Google Scholar]
- Hoebler C, Barry JL, David A, Delort-Laval J (1989) Rapid acid-hydrolysis of plant cell wall polysaccharides and simplified quantitative determination of their neutral monosaccharides by gas-liquid chromatography. J Agric Food Chem 37: 360–367 [Google Scholar]
- Holland N, Holland D, Helentjaris T, Dhugga KS, Xoconostle-Cazares B, Delmer DP (2000) A comparative analysis of the plant cellulose synthase (CesA) gene family. Plant Physiol 123: 1313–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Laosinchai W, Itoh T, Cui X, Linder CR, Brown RM (1999) Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell 11: 2075–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Kurek I, Kawagoe Y, Jacob-Wilk D, Doblin M, Delmer D (2002) Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc Natl Acad Sci USA 99: 11109–11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison WH, Archibald DD (1998) Analysis of graded flax fiber and yarn by pyrolysis mass spectrometry and pyrolysis gas chromatography mass spectrometry. J Agric Food Chem 46: 1870–1876 [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H (1998) A plasma membrane-bound putative endo-1,4-β-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J 17: 5563–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann GJ, Pureveen JBM, Eijkel GB, Poorter H, Boon JJ (1992) Differences in relative growth rate in 11 grasses correlate with differences in chemical composition as determined by pyrolysis mass spectrometry. Oecologia 89: 567–573 [DOI] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA 93: 12637–12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Kawagoe Y, Hogan P, Delmer DP (2002) Sitosterol-β-glucoside as primer for cellulose synthesis in plants. Science 295: 147–150 [DOI] [PubMed] [Google Scholar]
- Read SM, Bacic T (2002) Prime time for cellulose. Science 295: 59–60 [DOI] [PubMed] [Google Scholar]
- Scheible W-R, Eshed R, Richmond T, Delmer D, Somerville C (2001) Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis ixr1 mutants. Proc Natl Acad Sci USA 98: 10079–10084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedletzky E, Shmuel M, Trainin T, Kalman S, Delmer D (1992) Cell-wall structure in cells adapted to growth on the cellulose-synthesis inhibitor 2,6-dichlorobenzonitrile: a comparison between 2 dicotyledonous plants and a gramineous monocot. Plant Physiol 100: 120–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12: 2529–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible W-R, Cutler S, Somerville CR, Turner SR (1999) The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11: 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SR, Somerville CR (1997) Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff DM (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Williamson RE, Burn JE, Birch R, Baskin TI, Arioli T, Betzner AS, Cork A (2001) Morphology of rsw1, a cellulose-deficient mutant of Arabidopsis thaliana. Protoplasma 215: 116–127 [DOI] [PubMed] [Google Scholar]
- Zhong R, Taylor JJ, Ye Z-H (1997) Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 9: 2159–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]