Abstract
The superfamily of light-harvesting chlorophyll a/b-binding (Lhc) proteins in higher plants and green algae is composed of more than 20 different antenna proteins associated either with photosystem I (PSI) or photosystem II (PSII). Several distant relatives of this family with conserved chlorophyll-binding residues and proposed photoprotective functions are induced transiently under various stress conditions. Whereas “classical” Lhc proteins contain three-transmembrane α-helices, their distant relatives span the membrane with between one and four transmembrane segments. Here, we report the identification and isolation of a novel member of the Lhc family from Arabidopsis with one predicted transmembrane α-helix closely related to helix I of Lhc protein from PSI (Lhca4) that we named Ohp2 (for a second one-helix protein of Lhc family described from higher plants). We showed that the Ohp2 gene expression is triggered by light stress and that the Ohp2 transcript and protein accumulated in a light intensity-dependent manner. Other stress conditions did not up-regulate the expression of the Ohp2 gene. Localization studies revealed that Ohp2 is associated with PSI under low- or high-light conditions. Because all stress-induced Lhc relatives reported so far were found in PSII, we propose that the accumulation of Ohp2 might represent a novel photoprotective strategy induced within PSI in response to light stress.
The superfamily of light-harvesting chlorophyll a/b-binding (Lhc) proteins in higher plants and green algae is composed of more than 20 different members associated with photosystem I (PSI) or photosystem II (PSII). The primary function of these proteins is the absorption of light through chlorophyll excitation and the transfer of the absorbed energy to photo-chemical reaction centers (Green and Durnford, 1996). On the basis of the three-dimensional structure determined at 3.4-Å resolution for one member of the Lhc family from higher plants (Kühlbrandt et al., 1994), it was proposed that all Lhc proteins in higher plants and green algae have three transmembrane α-helices, where helices I and III are evolutionarily related to each other and held together by ion pairs formed by charged residues.
In the past few years, several distant relatives of Lhc protein family with conserved chlorophyll-binding residues and a transient expression pattern related to various stress conditions have been described from higher plants, algae, or cyanobacteria (Adamska, 2001). These distant relatives include the four-helix PsbS protein of PSII (Funk, 2001) and a subfamily of proteins called early light-induced proteins (Elips; Adamska, 1997, 2001; Montané and Kloppstech, 2000). The Elip subfamily consists of three-helix Elips, two-helix stress-enhanced proteins (Seps), and one-helix proteins (Ohps) also called high-light-induced proteins (Hlips) or small chlorophyll a/b-binding-like proteins (Scps) in prokaryotic organisms (Adamska, 2001). All Elip subfamily members are short-lived proteins (Meyer and Kloppstech, 1984; Grimm and Kloppstech, 1987; Adamska et al., 1992) that accumulate in thylakoid membranes under photoinhibitory conditions (Adamska et al., 1992; Pötter and Kloppstech, 1993), when the expression of other Lhc proteins is down-regulated (Montané et al., 1997; Heddad and Adamska, 2000). Therefore, it was proposed that members of the Elip family might play a protective role within the thylakoids during light stress, either by transient binding of free chlorophyll molecules and preventing the formation of free radicals and/or by acting as sinks for excitation energy (Montané and Kloppstech, 2000; Adamska, 2001).
At ambient temperatures, photoinhibition occurs primarily at the level of PSII and involves reversible inactivation of PSII due to arrest of electron transport within this complex followed by irreversible damage to subunits of the PSII reaction center (Barber and Andersson, 1992; Prasil et al., 1992; Andersson and Aro, 2001). Photoinhibition of PSI in higher plants has been observed mainly in combination with chilling stress (Hihara and Sonoike, 2001). Thus, it is not surprising that all light stress-induced Lhc relatives investigated so far were found to be associated with PSII (Adamska and Kloppstech, 1991; Levy et al., 1992; Funk et al., 1995), which is supposed to be more sensitive to light-induced damage than PSI and needs protection.
In this work, we have isolated the first distant relative of the Lhc family in Arabidopsis that accumulates in response to light stress in PSI. This protein contained a single predicted transmembrane α-helix related to helix I of the Lhc protein from PSI (Lhca4), and we named it Ohp2 (for a second one-helix Lhc protein described from higher plants). The first described one-helix Lhc protein (called here Ohp1) is a 69-amino acid-long protein present in the thylakoid membrane in substoichiometric amounts as compared with PSI and PSII (Jansson et al., 2000).
We demonstrate here that the Ohp2 gene is expressed under low-light conditions but that the amount of Ohp2 transcripts and protein increased in a light intensity-dependent manner. Furthermore, the accumulation of Ohp2 in PSI was specifically triggered by light stress and not by other stress conditions, such as cold stress, heat shock, mechanical wounding, desiccation, high salt, and oxidative stress. Furthermore, some of these stress conditions reduced the amount of Ohp2 transcripts but did not influence the amount of protein in thylakoid membranes.
RESULTS
Ohp2 Is a Distant Relative of Lhc Protein Family with a Single Predicted Transmembrane α-Helix
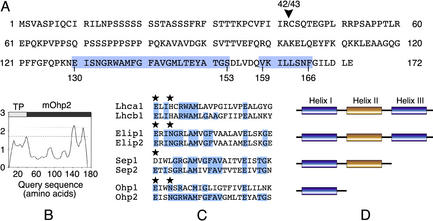
A BLAST search of the Arabidopsis expressed sequence tag (EST) database with an Elip consensus sequence (Adamska, 2001) resulted in the identification of a gene encoding a novel Elip-like protein that we named Ohp2. We designed primers based on the EST sequence (TC87219) and used PCR to amplify the coding region of the Ohp2 gene from an Arabidopsis cDNA library. The amplified 519-bp cDNA sequence encoded a protein predicted to be composed of 172 amino acids that contained a putative N-terminal transit peptide (amino acids 1–42) typical of proteins imported into chloroplasts (Fig. 1A). The nucleotide sequence for the Ohp2 gene has been deposited in the GenBank database under accession number AY057393. The genomic sequence of Ohp2 was found on chromosome I (GenBank accession no. AC015446, protein identity no. AAG12539).
Figure 1.
Predicted primary and secondary structure of Ohp2 from Arabidopsis. A, Deduced amino acid sequence of Ohp2. The arrowhead indicates the position of a predicted processing site (between amino acids 42 and 43), and the positions of potential hydrophobic regions (amino acids 130–153 and 159–166) are shown on a blue background. B, Hydropathy plot of the translated cDNA sequence of Ohp2. TP, Transit peptide; mOhp2, mature Ohp2. C, Sequence alignment of the conserved helix I of Lhca1 (GenBank accession no. M85150), Lhcb1.1 (GenBank accession no. X03907), Elip1 (GenBank accession no. U89014), Elip2 (GenBank accession no. Z97336), Sep1 (GenBank accession no. AF133716), Sep2 (GenBank accession no. AF133717), Ohp1 (GenBank accession no. AF054617), and a novel Ohp2 protein (GenBank accession no. AY057393) from Arabidopsis. The sequences were aligned manually with the assistance of the multiple alignment program ClustalW. Identical amino acids are shown on a blue background, and conserved amino acid residues involved in chlorophyll ligation in Lhcb2 (Kühlbrandt et al., 1994) are marked by asterisks. D, Location of the conserved region in various Lhc family members is marked in blue.
Hydropathy plots revealed (Fig. 1B) that the mature Ohp2 is composed of 130 amino acids and differs from Elips and Seps by the presence of a single transmembrane α-helix located at the C terminus of this protein (amino acids 130–153). A second predicted hydrophobic segment present between amino acids 159 and 166 is too short to span the membrane, however it is not excluded that this region might be embedded into the lipid bilayer. Furthermore, the transmembrane domain of Ohp2 is strongly conserved in all Lhc proteins and their distant relatives as shown in Figure 1C. This conserved domain is located in transmembrane α-helices I and III of Lhcb proteins and Elips (Adamska, 2001), and in the helix I of Seps (Heddad and Adamska, 2000) or Ohps/Hlips/Scps (Dolganov et al., 1995; Funk and Vermaas, 1999; Adamska, 2001) as shown schematically in Figure 1D. Comparison of helices I and III of Lhc and Elip family members revealed that helix I of Ohp2 is the most closely related to helix I of one-helix Elip-like proteins from cyanobacteria and algae and Lhc protein of PSI (Lhca4) from Arabidopsis, barley (Hordeum vulgare), Scot's pine (Pinus sylvestris), and tomato (Lycopersicon esculentum; 64%–70% identity and 76% similarity). It contains several universally conserved amino acid residues present in nearly all known Lhc proteins and their relatives (Fig. 1C), such as two Glu (E), one Arg (R), and one Met (M). Two conserved amino acids, either a Gln (E) or an Asp (D) and an Asn (N) or a His (H), reported to participate in a chlorophyll binding in Lhc proteins and their relatives (Kühlbrandt et al., 1994; Green and Kühlbrandt, 1995) are present in Ohp2 sequence at highly conserved positions (Fig. 1C). However, to bind chlorophylls, the Ohp2 protein would need to form homo- or heterodimeric structures. Experimental evidence was provided that at least two different heterodimeric complexes are formed between one-helix proteins HliA/HliB and HliC/HliD in the cyanobacterium Synechocystis sp. PCC6803 under light stress conditions (He et al., 2001).
Ohp2 Is an Integral Thylakoid Membrane Protein Located within PSI
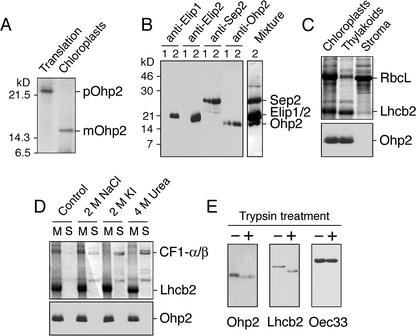
The presence of an N-terminal transit peptide and a predicted transmembrane domain in the Ohp2 sequence suggested that this protein is imported into the plastid and inserted into thylakoid membranes. To confirm the chloroplast location of Ohp2 experimentally, we performed in vitro import of radioactively labeled Ohp2 precursor (pOhp2) into isolated, intact pea (Pisum sativum) chloroplasts. The results demonstrated that labeled Ohp2 was present within trypsin-treated chloroplasts as proven by autoradiography (Fig. 2A). During the import assay, the 22-kD Ohp2 precursor (pOhp2) was processed to a mature protein (mOhp2) with an apparent molecular mass of 17 kD (Fig. 2A).
Figure 2.
Location of Ohp2 in thylakoid membranes. A, In vitro import of radioactively labeled Ohp2 into isolated intact pea chloroplasts. pOhp2, Precursor of Ohp2; mOhp2, mature protein. B, Testing of the specificity of polyclonal antibodies raised against overexpressed Ohp2. Leaves were exposed to low-light (lines 1, 100 μmol m–2 s–1) or to light stress (lines 2, 2,500 μmol m–2 s–1) conditions for 3 h, and thylakoid membranes were isolated and used for western blotting with antibodies raised against Elip1, Elip2, Sep2, or Ohp2 from Arabidopsis. For comparison, the western-blot membrane was incubated with a mixture of all four antibodies (Mixture). C, Location of Ohp2 in the thylakoid membrane assayed by immunoblotting. As references, proteins of chloroplast, thylakoid, and stromal fractions were separated by SDS-PAGE, and the distribution of the ribulose-1,5-bisphosphate carboxylase large subunit (RbcL) and the chlorophyll a/b-binding protein of PSII (Lhcb2) was analyzed by Coomassie Blue staining. D, The integral membrane location of Ohp2 was verified by incubation of membranes in the absence (control) or presence of 2 m NaCl, 2 m KI, or 4 m urea. The membrane pellet (lanes M) and supernatant (lanes S) containing extracted peripheral membrane proteins were analyzed by SDS-PAGE followed by Coomassie Blue staining, and the presence of Ohp2 was assayed by immunoblotting. As references, the distribution of the α- and β-subunits of the CF1 ATP synthase complex (CF1-α/β) and the Lhcb2 is shown. E, A protease protection assay was carried out after addition of trypsin (50 μg mL–1) to the isolated thylakoid membranes (1 mg chlorophyll mL–1) and incubation of samples at 4°C for 30 min. The protection of Ohp2 against degradation by trypsin was tested by immunoblotting. As references, the sensitivity of the Lhcb2 toward trypsin treatment and the protection of the lumen located 33-kD protein from the oxygen-evolving complex (Oec33) are shown.
We produced a polyclonal antibody against the recombinant protein expressed in Escherichia coli and tested the specificity of this antibody using thylakoid membranes isolated from control and light stress-treated leaves (Fig. 2B). For comparison, immunoblot analysis using antibodies raised against Elip1, Elip2, and Sep2 were performed. The results revealed (Fig. 2B) that the Ohp2 antibody specifically cross-reacted with a protein with an apparent molecular mass of 17-kD present in thylakoid membranes under ambient light conditions that accumulated in response to light stress. No other proteins from Elip or Lhc families were recognized by this antibody, confirming the specificity of the cross-reaction. Furthermore, apparent molecular masses of Elips, Sep2, and Ohp2 and their expression patterns under low- and highlight conditions differed for all these proteins. Although Elip1 and Elip2 were detected in thylakoid membranes only after light stress treatment, Sep2 and Ohp2 were present also under low-light conditions, but their level increased significantly after exposure of leaves to light stress (Fig. 2B).
To further explore the chloroplast location of Ohp2, we performed immunoblot analysis with purified, intact chloroplasts and their subfractions. The results revealed (Fig. 2C, bottom) that Ohp2 is located in the thylakoid membrane fraction. As a reference, the distribution of the RbcL and one of the major antenna proteins of PSII (Lhcb2) is shown by Coomassie Blue staining (Fig. 2C, top). Whereas the RbcL is known to be a soluble stromal enzyme, the Lhcb2 is a polytopic protein located within the thylakoid membrane.
To prove an integral membrane location of Ohp2, isolated thylakoid membranes were washed with salt or the chaotropic agent urea to release extrinsic membrane proteins (Boudreau et al., 1997). The presence of Ohp2 in the thylakoid pellet fraction confirmed an integral membrane location of this protein (Fig. 2D, bottom). Similarly, the Lhcb2 protein was found in the thylakoid membrane, whereas the peripherally located α- and β-subunits of the CF1 ATP synthase were partially or completely removed from the membrane by salt or urea washes (Fig. 2D, top).
Interestingly, in some of the immunoblots, the Ohp2 antibody recognized two distinct bands migrating close to each other in SDS gels (Fig. 2D, bottom). This might be the result of a posttranslational modification or partial proteolysis of Ohp2. Several potential phosphorylation sites and a conserved ATP/GTP-binding motif located between amino acids 84 and 91 were predicted in the Ohp2 sequence (not shown).
The topology of Ohp2 in the thylakoid membrane was investigated by its susceptibility to added trypsin (Fig. 2E, left). The primary specificity of trypsin is directed against -P1-P′1-peptide bonds, where P1 represents either Lys (K) or Arg (R) and P′1 is a nonspecific amino acid residue (Bond, 1996). On the basis of the deduced amino acid sequence of Ohp2 (Fig. 1A, top) and the topology reported for Lhc from higher plants (Kühlbrandt et al., 1994), we expected that the 12-amino acid thylakoid lumen-exposed C-terminal part of Ohp2 would be protected but that the 87-amino acid N-terminal stroma-exposed part will be accessible to trypsin digestion. Incubation of the isolated thylakoid membranes with trypsin resulted in a slightly higher mobility of Ohp2 protein in SDS gels (Fig. 2E, left), indicating that approximately 1-kD fragment of this protein was removed by trypsin. Similarly, a 2-kD N-terminal stroma-exposed fragment of Lhcb2 was removed by trypsin (Fig. 2E, middle). In contrast, the 33-kD oxygen-evolving complex protein (Oec33), known to be located at the lumenal side of the membrane, was protected against trypsin digestion (Fig. 2E, right). This suggests that the N terminus of Ohp2 is not completely free in the stroma but might be partially protected by binding to other proteins to form a complex.
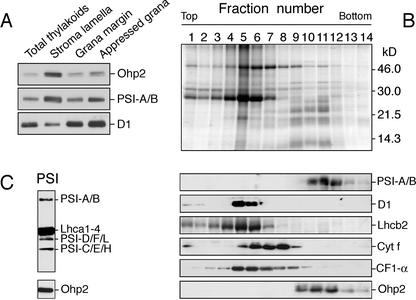
Further fractionation of the thylakoid membranes into appressed and nonappressed membrane regions, using differential centrifugation and aqueous polymer two-phase partition (Andersson and Anderson, 1980), demonstrated that the vast majority of Ohp2 was present in the stroma lamellae. Lower levels of this protein were detected in fractions enriched in the appressed and nonappressed regions of grana stacks (Fig. 3A, top). The distribution of the subunits A/B from the PSI reaction center and the D1 protein from PSII reaction center was in accordance with the previously published data (Morrissey et al., 1986). While the vast majority of the D1 protein was detected in the grana region, the majority of the PSI-A/B was found in the stroma lamellae (Fig. 3A).
Figure 3.
Association of Ohp2 with PSI. A, Localization of Ohp2 in appressed and nonappressed regions of thylakoid membranes assayed by immunoblotting. As references, the distribution of subunits-A/B from the PSI reaction center (PSI-A/B) and the D1 protein from PSII (D1) reaction center were assayed. B, Location of Ohp2 in PSI. Thylakoid membranes isolated from control leaves grown under ambient light conditions (100 μmol m–2 s–1), were solubilized with n-dodecyl β-d-maltoside, and the released protein complexes were separated by a Suc density gradient centrifugation. The protein composition in collected fractions was analyzed on SDS-gel stained by Coomassie Blue (top). The distribution of PSI, PSII, and its antenna, cytochrome b/f, and ATP synthase complexes were analyzed by immunoblotting using polyclonal antibodies directed against the A and B subunits of PSI reaction center (PSI-A/B), the D1 protein of PSII reaction center, the chlorophyll a/b-binding protein of PSII (Lhcb2), the subunit f of the cytochrome b6/f complex (cyt f), and the α-subunit of the CF1 ATP synthase complex (CF1-α; middle panels). Location of Ohp2 was assayed by immunoblotting (bottom). C, Composition of the fraction 11 tested by antibodies directed against subunits A, B, C, D, E, F, H, and L of PSI (PSI-A/B, PSI-D/F/L, or PSI-C/E/H, respectively), light-harvesting antenna proteins of PSI (Lhca1–4), and Ohp2.
The multiprotein complexes, such as PSI and PSII, which are both connected with the antenna systems, cytochrome b/f, and ATP synthase, mediate photo-synthetic reactions within the thylakoid membrane. To explore the association of Ohp2 with one of these complexes, we disrupted the thylakoid membrane organization with Triton X-100 detergent and separated the protein complexes released by Suc density gradient centrifugation (Steinback et al., 1982). Immunoblot analysis using an anti-Ohp2 antibody revealed that this protein was enriched in fractions containing PSI (data not shown). The association with PSI was further confirmed by the isolation of PSI complex after solubilization of thylakoid membranes with n-dodecyl β-d-maltoside (Takahashi et al., 1991; Fischer et al., 1997; Fig. 3B, top). The identity of isolated complexes was confirmed by immunoblotting using antibodies raised against prominent subunits of PSI, PSII and its antenna, cytochrome b/f, and the ATP synthase (Fig. 3B, middle). Immunoblot analysis using the anti-Ohp2 antibody revealed that this protein is located mainly in fractions 10 to 12 containing reaction center proteins of PSI (Fig. 3B, bottom). Neither PSII nor the cytochrome b/f or ATP synthase complex contained any traces of Ohp2. In contrast, three other members from the Elip subfamily, Elip1, Elip2, and Sep2, were found in fractions enriched in antenna proteins of PSII (not shown).
The composition of fraction 11 was verified further by immunoblotting with antibodies raised against purified PSI complex from spinach (Spinacia oleracea; Tjus and Andersson, 1991). Several structural subunits of PSI reaction center and antenna system were recognized by these antibodies, confirming the presence of PSI complex in this fraction (Fig. 3C). No changes in the localization of Ohp2 were observed under light stress conditions as proven by the isolation of protein complexes from leaves exposed to light stress (2,000 μmol m–2 s–1) for 4 h; under these conditions, Ohp2 was enriched in PSI complexes (not shown).
Ohp2 Transcript and Protein Accumulate in Response to Light Stress
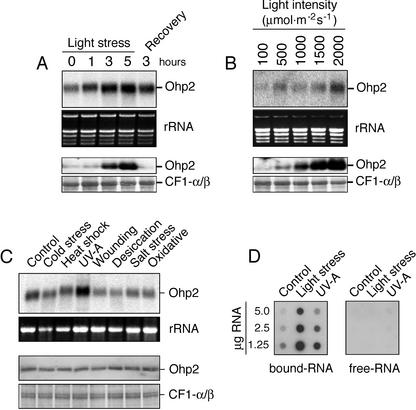
It was shown recently that the expression of Elip subfamily members in eukaryota (Adamska et al., 1992; Pötter and Kloppstech, 1993; Heddad and Adamska, 2000) and in prokaryota (Dolganov et al., 1995; He et al., 2001) was induced or strongly enhanced in response to light stress. In contrast, the “typical” Lhc proteins responded to light stress by reduction of their transcript level (Montané et al., 1997; Heddad and Adamska, 2000). To analyze the effect of light stress on Ohp2 gene expression, the Ohp2 transcript (Fig. 4A, top) and protein (Fig. 4A, bottom) levels were assayed in leaves of Arabidopsis exposed to high light or recovery conditions for different periods of time. Northern blots revealed (Fig. 4A, top) that Ohp2 transcripts were present under low-light conditions, but the amount of these transcripts increased significantly after exposure of leaves to light stress. The accumulation of Ohp2 transcripts occurred almost linearly with an illumination time up to 5 h, and the transfer of leaves to lowintensity light (recovery conditions) resulted in a degradation of these transcripts (Fig. 4A, top). The ethidium bromide-stained pattern of ribosomal RNAs confirmed that equal amounts of RNA were loaded in each lane (Fig. 4A, middle).
Figure 4.
Expression of Ohp2 under various stress conditions. A, Kinetics of accumulation of Ohp2 transcripts (top panels) and proteins (bottom panels) in response to light stress (2,500 μmol m–2 s–1) and recovery at low-intensity light (100 μmol m–2 s–1). B, Light intensity-dependent gene and protein expression of Ohp2 assayed by northern (top panels) and western (bottom panels) blotting, respectively, after 3 h of light stress exposure. C, Expression pattern of Ohp2 under different stress conditions assayed by northern (upper panels) and western (lower panels) blotting, respectively, after 3 h of stress exposure. As references, the rRNA pattern in the gel visualized by staining with ethidium bromide and the level of α- and β-subunits of the CF1 ATP synthase stained by Coomassie Blue are shown. D, Distribution of Ohp2 mRNAs between free (free-RNA) and polysome-bound (bound-RNA) fractions assayed by dot-blot hybridization after exposure of leaves to low light (Control, 100 μmol m–2 s–1), high light (Light stress, 2,500 μmol m–2 s–1), or UV-A irradiation (UV-A, 20 μmol m–2 s–1) for 3 h.
To prove whether an increase in Ohp2 transcript level is accompanied by the accumulation of the corresponding protein, we performed immunoblot analysis. The results revealed (Fig. 4A, bottom) that the Ohp2 level in thylakoid membranes was enhanced by the light stress treatment and a lowering of the light intensity during recovery resulted in a rapid degradation of this protein. Only traces of Ohp2 were detected in control leaves collected before light stress treatment and in leaves transferred to recovery conditions for 3 h. The amounts of CF1-α/β assayed as a control did not change significantly during the light stress treatment (Fig. 4A, bottom).
To test the dependence of Ohp2 expression on photon fluency rates, leaves of Arabidopsis were exposed to increasing light intensities, and the transcript and protein levels were assayed by northern (Fig. 4B, top) or western (Fig. 4B, bottom) blotting. The results demonstrated that both Ohp2 transcripts and protein accumulated in a light intensity-dependent manner up to 2,000 μmol m–2 s–1.
In addition to light stress, several other stress conditions, such as desiccation, osmotic stress, cold stress, or salt stress, have been shown to induce the expression of Elip subfamily members in various plant species (Adamska, 2001). However, the expression of Elip1, Elip2, Sep1, and Sep2 genes in Arabidopsis was shown to be specific for light stress (Heddad and Adamska, 2000). To test the effect of various stresses on expression of the Ohp2 gene, the amount of Ohp2 transcript and protein was assayed in leaves of Arabidopsis exposed to cold stress, heat shock, UV-A irradiation, mechanical wounding, desiccation, high salt, or oxidative stress. As a control, leaves were exposed to ambient light and temperature, and the amount of Ohp2 transcript and protein expressed under this condition was used as a reference. The results revealed (Fig. 4C, top) that only exposure of leaves to UV-A irradiation resulted in the accumulation of Ohp2 transcripts as compared with the control. Exposure of leaves to cold stress, wounding, desiccation, salinity, or oxidative stress resulted in the reduction of the Ohp2 transcript level (Fig. 4C, top). No significant changes in the Ohp2 transcript level were assayed during heat shock.
Because UV-A light is absorbed by a blue-light receptor (Christie and Briggs, 2001) that was reported to promote the accumulation of Elip in mature green pea leaves exposed to light stress (Adamska et al., 1992), we expected that a similar regulation of Ohp2 gene expression might exist in Arabidopsis. Surprisingly, immunoblot analysis (Fig. 4C, bottom) showed that no significant changes in the Ohp2 level were assayed during illumination with UV-A light, although the level of corresponding transcripts was strongly enhanced (compare with Fig. 4C, top). Furthermore, the other stress conditions tested also did not influence Ohp2 levels in thylakoid membranes.
To determine whether Ohp2 transcripts induced by UV-A light are actively translated, we analyzed the distribution of Ohp mRNAs between polysome-bound and -free fractions (Fig. 4D). For comparison, polysomal fractions from control leaves or leaves exposed to high-intensity white light were isolated and analyzed. During the first step of protein synthesis in the cytoplasm, mRNAs are integrated into the ribosomal complex, and polysomes are formed. Analysis of the mRNA content of polysomes is assumed to be indicative of active protein synthesis. Our results revealed (Fig. 4D) that the vast majority of Ohp2 mRNAs was detected in the polysomal fraction under all light conditions tested, confirming their simultaneous translation into corresponding proteins. Only traces of Ohp2 transcripts were detected as free mRNAs for all light treatments. Notably, a much higher amount of Ohp2 transcripts was induced in response to illumination with high-intensity white light than with UV-A (Fig. 4D).
DISCUSSION
Several light stress-induced one-helix proteins related to Lhc family were found in cyanobacteria Synechocystis sp., Synechoccocus sp., and Anabaena sp., Glaucocystophyta (Cyanophora paradoxa), Cryptophyta (Guillardia theta), red algae (Cyanidium caldarium), and Porphyra purpurea and in higher plants rice (Oryza sativa) and Arabidopsis (Adamska, 2001; Heddad and Adamska, 2002). This suggests that a one-helix distant relative of Lhc proteins present in ancient cyanobacteria might be a progenitor of higher plant and algal antenna proteins (Montané and Kloppstech, 2000; Heddad and Adamska, 2002). According to this scenario the ancient antenna systems seemed to be composed of light stress-induced proteins, the function of which was not light harvesting but the dissipation of absorbed energy in the form of heat or fluorescence (Montané and Kloppstech, 2000). Thus, the Ohp2 in Arabidopsis would represent a relict of such an ancient energy-dispersing antenna system, which is still present and functional in plants in addition to more recently evolved Lhc proteins with light-harvesting functions.
We demonstrated that Ohp2 was present in the thylakoid membranes under low-light conditions and that its level increased in response to light stress in a light intensity-dependent manner. The transfer of plants to low-intensity light for recovery resulted in a drastic decrease in the amount of Ohp2 in thylakoid membranes, despite the presence of significant transcript levels. This indicates that the regulation of Ohp2 gene expression occurs at the level of both transcript and protein accumulation. The protein level could be controlled by different Ohp2 translation rates under light stress and recovery conditions or by changes in the stability of the Ohp2 protein. Analyses of Ohp2 transcripts in UV-A favor the second possibility. UV-A light was sufficient to promote accumulation of Ohp2 transcripts, but not the corresponding protein, even though the transcripts appeared to be actively translated based on their association with polysomes (Fig. 4, C and D).
We demonstrated that the accumulation of Ohp2 transcript and protein was triggered specifically by high-intensity light and that other stress conditions did not up-regulate the expression of the Ohp2 gene. However, exposure of leaves to high-salinity stress (0.9–2.0 m NaCl), a longer desiccation period (12 h), or cold stress treatment (8 h), resulted in the reduction of Ohp2 transcript levels without significant changes in the amount of corresponding protein (not shown). A similar down-regulation of the transcript but not the protein level was reported for Sep2 from Arabidopsis in response to cold stress (Heddad et al., 2001). In this respect, Ohp2 and Sep2 differ from their homologs present in wheat (Triticum aestivum; Wcr12, for wheat cold-regulated 12-kD protein), barley (HV60, for low molecular mass Elip of H. vulgare), in the green algae Dunaliella bardawil (Cbr, for carotene biosynthesis-related) or in the cyanobacterium Synechocystis sp. PCC6803 (HliA, HliB, and HliC) that were reported to be induced by cold stress in the absence of high light (Adamska, 2001; He et al., 2001). Furthermore, conversely to Ohp2 from Arabidopsis, some Elip family members in different plant species were also reported to be induced by salinity or desiccation stress (Adamska, 2001). The up-regulation of the transcript level by high-intensity light was reported for Ohp1 from Arabidopsis (Jansson et al., 2000) and for four Hlips from the cyanobacterium Synechocystis sp. PCC6803 (He et al., 2001). However, the protein level of Hlips was also enhanced during nitrogen limitation, sulfur deprivation, and low temperature (He et al., 2001). The protein level of Ohp1 was not investigated in Arabidopsis.
We demonstrated that Ohp2 is located in PSI (Fig. 3B). There is an earlier report (Cronshagen and Herzfeld, 1990) showing that in etiolated pea seedlings exposed to light for 48 h, the majority of Elip was found in the PSI fraction. However, progressing localization studies by cross-linking and immunoprecipitation revealed that Elip in pea is present in the nonappressed region of thylakoid membranes and associated with PSII complex (Adamska and Kloppstech, 1991). Other members of Elip family from Arabidopsis, such as Elip1, Elip2, and Sep2 were also found to be located in PSII (M. Heddad and I. Adamska, unpublished data). Nothing is known about the location of Ohp1 in Arabidopsis or its homolog proteins in algae or cyanobacteria.
It was reported that under light stress conditions, a mobile pool of Lhcb1 and Lhcb2 moves from PSII to PSI due to the reversible phosphorylation of Lhc proteins by a thylakoid-bound kinase (Allen and Forsberg, 2001; Haldrup et al., 2001). The protein subunit H of the PSI reaction center was shown to be involved in the docking of phospho-Lhc proteins when they act as a part of the light-harvesting antenna of PSI (Lunde et al., 2000). Our data revealed that there was no light-dependent (low versus high light) difference in the localization of Ohp2, indicating that the state transition process did not influence the location of Ohp2.
Although the three-dimensional structure of the PSI reaction center has been determined by x-ray crystallography to 2.5-Å resolution for the cyanobacterium Synechococcus elongatus (Jordan et al., 2001), very little information exists about the dynamic nature of PSI as compared with PSII. The PSI has been long believed to be resistant to photoinhibition, however, it was recently found that PSI is quite sensitive to light especially at chilling temperatures (Hihara and Sonoike, 2001). Also the composition and size of the antenna system of PSI have been assumed to be slightly affected by changes in the environment. The accumulation of Ohp2 in response to light stress in PSI indicates that this photosystem undergoes a dynamic rearrangement of its organization, which possibly takes place in the antenna system. Recently, it was demonstrated that iron deficiency induces the formation of an antenna ring around trimeric PSI in cyanobacteria (Bibby et al., 2001; Boekema et al., 2001). This antenna is formed transiently from 18 CP43′ molecules (products of the iron stress-induced IsiA gene) with significant homology to one of the chlorophyll a-binding proteins of PSII.
Many protective mechanisms against photoinhibition of PSII have been reported (Barber and Andersson, 1992; Prasil et al., 1992; Andersson and Aro, 2001) where the degradation and replacement of the photodamaged D1 protein of PSII reaction center is the most efficient one. The photoinhibition of PSI is far more dangerous for the chloroplast because the recovery from photoinhibition is not complete even after 1 week (Hihara and Sonoike, 2001). Thus, accumulation of Ohp2 in PSI exposed to photoinhibitory light might represent one of the strategies to prevent or lower light stress-induced damage. It was proposed that proteins of Elip subfamily might have a protective function within PSII under light stress conditions either by binding free chlorophyll molecules and preventing the formation of free radicals and/or by acting as sinks for excitation energy (Montané and Kloppstech, 2000; Adamska, 2001). An alternative function involving the regulation of tetrapyrrole biosynthesis in connection with chlorophyll availability was proposed for ScpB and ScpE from Synechocystis sp. PCC6803 (Xu et al., 2002).
Recently, it was shown that the PsbS protein of PSII contributes to photoprotective energy dissipation rather than photosynthetic light harvesting (Li et al., 2000). Also, a cyanobacterial mutant lacking all four Hli genes gradually lost its photosynthetic capacity and died in high light, confirming the proposal that the Elip family members are required for survival and acclimation of cells to the absorption of excess light energy (He et al., 2001). All of these data support the concept that whereas the “classical” Lhc proteins are involved in light harvesting, their distant relatives induced in response to light stress participate in the dissipation of excess energy. This might represent a short-term strategy to cope with changing light intensities. Our data indicate that this strategy is not restricted to PSII but might also operate in the PSI antenna system.
MATERIALS AND METHODS
Growth of Plants and Stress Conditions
Arabidopsis cv Columbia plants were grown in a growth chamber on soil at 25°C at a light intensity of 100 μmol m–2 s–1 under short-day conditions. Light stress treatment was performed on mature leaves detached from 4- to 5-week-old plants, floated on water, and exposed to a light intensity of 2,500 μmol m–2 s–1 for different times as mentioned in figures. For cold or heat stress, detached leaves floated on water were transferred for 3 h to incubators set at 4°C or 42°C, respectively. Wounding stress was obtained by cutting the leaves into 5 mm2 segments, which were then incubated for 3 h on water at room temperature at a light intensity of 10 μmol m–2 s–1. Desiccation stress was performed on leaves dehydrated on 3MM paper (Whatmann, Clifton, NJ) at room temperature at a light intensity of 10 μmol m–2 s–1. After 3 h of incubation, the relative water content of leaves was reduced by 50% (w/w). Leaves subjected to a high salt or an oxidative stress were submerged for 3 h in 500 mm NaCl or in 2% (v/v) H2O2 solutions, respectively. UV-A treatment was performed by illumination of detached leaves with a UV lamp (366 nm) at a light intensity of 20 μmol m–2 s–1 for 3 h. Plant material was collected and either immediately used for extractions or frozen in liquid nitrogen and stored at –70°C for further preparations.
Gene Cloning, Sequencing, and Data Analysis
The Ohp2 gene from Arabidopsis was amplified by PCR using the CD4-13 λ-ZipLox cDNA library obtained from the Arabidopsis Biological Resource Center at Ohio State University (Columbus). Specific PCR primers were designed on the basis of a sequence present in the EST cDNA database (TC87219). The amplified 519-bp fragment (including stop codon) corresponding to the coding region of Ohp2 cDNA was ligated into a pCR2.1 vector using a TA cloning kit (Invitrogen AB, Groningen, the Netherlands), and the insert sequence was verified by sequencing of both cDNA strands (CyberGene, Stockholm). Similarity searches were done using the ADVANCED BLAST program. Alignment of amino acid sequences was performed using ClustalW with a manual correction of gaps. The transmembrane regions were predicted using the dense alignment surface method, and the hydropathy plot was raised according to Kyle and Doolittle (1982). Prediction of the subcellular location and determination of the processing site were analyzed by the TARGETP v.1.01 program. Protein pattern and motif predictions were performed by using sequence motif search and protein motif fingerprint databases. The software programs used are accessible on the Internet (http://ca.expasy.org/tools/, http://www.genome.ad.jp/, and http://www.cbs.dtu.dk/services/TargetP).
Expression of Ohp2 in Escherichia coli and Production of Polyclonal Antibodies
The Ohp2 cDNA was amplified using a pair of primers (5′-TCAGTAGCTTCACCGATTCAAT-3′ and 5′-TTCCAAGTCTAGAATGCCGAAA-3′) and the pCR2.1 plasmid as template. The Ohp2 protein was expressed in E. coli as a fusion protein with the N-terminal attached thioredoxin and C-terminal attached His-tag (His6) using a kit (pBAD/Topo ThioFusion Expression Systems, Invitrogen AB) according to the manufacturer's protocol. Recombinant Ohp2 protein was extracted with 6 m guanidinium hydrochloride, purified under denaturing conditions by affinity chromatography on Ni-NTA-agarose column (Qiaexpress, Qiagen GmbH, Hilden, Germany), and eluted from the column with 8 m urea, 0.1 m NaH2PO4, and 0.01 m Tris-HCl, pH 4.5. The Ohp2 fractions were separated by SDS-PAGE and transferred to a nitrocellulose membrane (Protran, Schleicher & Schuell GmbH, Dassel, Germany) before raising polyclonal antibodies in rabbits (BioGenes, Berlin).
In Vitro Transcription, Translation, and Import
Plasmid pCR2.1 containing the Ohp2 insert was linearized with the restriction enzyme BamHI and was used for in vitro transcription with T7-RNA polymerase according to Sambrook et al. (1989). In vitro translation was performed in a wheat (Triticum aestivum) germ lysate (Promega, Falkenberg, Sweden) in the presence of [35S]Met (1,220 Ci mmol–1, Amersham Biosciences, Uppsala). In vitro imports into isolated intact chloroplasts were performed according to Adamska and Kloppstech (1991).
Fractionation of Chloroplasts and Thylakoid Membranes
Intact chloroplasts were isolated on a Percoll gradient and separated into soluble stroma and thylakoid membrane fractions as described (Heddad and Adamska, 2000). For separation of peripheral and integral membrane proteins, the thylakoid membranes were incubated with 2 m NaCl, 2 m KI or 4 m urea for 30 min at 4°C and pH 11.0 in darkness under gentle stirring as described (Boudreau et al., 1997). After centrifugation for 15 min at 14,000 rpm, the thylakoid pellet (containing integral membrane proteins) and the supernatant (containing peripheral membrane proteins) were separated by SDS-PAGE, and the Ohp2 content was analyzed by immunoblotting.
The topology of Ohp2 in the thylakoid membrane was tested by protease protection assay. Isolated thylakoid membranes were resuspended in 50 mm HEPES, pH 8.0, and 330 mm sorbitol at a chlorophyll concentration of 1 mg mL–1 and incubated for 30 min at 4°C with 50 μg mL–1 trypsin. Protection of Ohp2 against degradation by trypsin was tested by immunoblotting.
For the isolation of appressed and nonappressed membrane regions, thylakoids were mechanically disrupted with a Yeda press, and membrane fragments were separated by differential centrifugation and aqueous polymer two-phase partitioning as described by Andersson and Anderson (1980).
Fractionation of thylakoid membranes and isolation of multiprotein complexes was performed by a Suc density gradient centrifugation using Triton X-100 as the detergent (Steinback et al., 1982). Isolation of PSI complexes was performed by a Suc density gradient centrifugation using n-dodecyl β-d-maltoside as the detergent (Takahashi et al., 1991) with modifications described by Fischer et al. (1997). Thylakoid membranes were resuspended in water at a chlorophyll concentration of 0.8 mg mL–1, 10% (w/v) n-dodecyl β-d-maltoside was added to a final concentration of 0.9%, and the mixture was incubated for 20 min on ice. The solubilized thylakoids were centrifuged at 20,000g for 20 min, and the clear supernatant was loaded on a 0.1 to 1.0 m Suc density gradient containing 5 mm Tricine, pH 8.0, and 0.05% (w/v) n-dodecyl β-d-maltoside with a 2 m Suc cushion. The gradients were centrifuged at 170,000g for 17 h at 4°C and 0.7-mL fractions were collected using a peristaltic pump. Proteins in each fraction were precipitated with 5% (w/v, final concentration) trichloroacetic acid, washed with 80% (v/v) cold acetone, and resuspended in equal volumes of sample buffer.
RNA Isolation and Analysis
Total RNA was extracted from control or light stress-treated leaves using a RNeasy mini kit (Qiagen GmbH) according to the manufacturer's protocol. After separation of 5 μg RNA in a 1.2% (w/v) agarose gel, RNA was transferred to a Hybond-N+ membrane before the hybridization as described (Heddad and Adamska, 2000).
For the isolation of polysomes, frozen plant material (5 g) was ground in liquid nitrogen, and the resulting tissue powder was resuspended at 4°C in 50 mL of polysome buffer containing 400 mm KCl, 50 mm Tris-HCl, pH 8.3, 10 mm magnesium-acetate, 250 mm Suc, 2% (w/v) Triton X-100, and 0.005% (v/v) β-mercaptoethanol. The suspension was filtered through Miracloth (Calbiochem, Stockholm) and centrifuged at 15,000g for 10 min at 4°C. The supernatant was loaded onto a two-step gradient containing 5 mL of 0.7 m and 7 mL of 1.7 m Suc in polysome buffer, and gradients were centrifuged for 17 h at 200,000g and 4°C. The supernatant (containing free RNA) was collected and used for RNA isolation as described above. The pellet (containing polysomes) was resuspended in 2 mL of polysome buffer and centrifuged at 200,000g for 30 min at 4°C over a 0.5 mL 1.7 m Suc cushion. The pellet was used for isolation of polysome-bound RNA as described above.
For dot-blot hybridization, RNA was spotted on Hybond-N+ membrane at three different concentrations, 5.0, 2.5, and 1.25 μg, and the membrane was used for hybridization as described (Heddad and Adamska, 2000).
The cDNA probe was labeled with 32α-dCTP by using a megaprime DNA labeling kit (Amersham Biosciences). The signal on the filter was analyzed by using a Phosphorimager FLA3000 (Fujifilm, Fuji, Tokyo) or x-ray film (Cronex 5, Agfa, Mortsel, Belgium).
Protein Analysis
Isolated thylakoid membranes were separated by SDS-PAGE (Laemmli, 1970) generally using 14% (w/v) polyacrylamide mini gels (mini gel system, Hoeffer, San Francisco). The gels were loaded on an equal protein basis. Immunoblotting was carried out according to Towbin et al. (1979) using polyvinylidene difluoride membranes with 45-μm pores (Amersham Biosiences) and enhanced chemiluminescence (ECL, Amersham Biosciences) as the detection system.
Acknowledgments
We thank Patrick Dessi for critical reading of the manuscript, the Arabidopsis Biological Resource Center at Ohio State University for providing cDNA library, and Ralf Oelmüller for providing antibodies against α-subunit of CF1 ATP synthase.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.019281.
This work was supported by the Swedish Research Council, by the Carl Tryggers Foundation (grant to I.A.), and by the Kinander Donation from Stockholm University (grant to U.A.).
References
- Adamska I (1997) Elips: light-induced stress proteins. Physiol Plant 100: 794–805 [Google Scholar]
- Adamska I (2001) The Elip family of stress proteins in the thylakoid membranes of pro-and eukaryota. In EM Aro, B Andersson, eds, Advances in Photosynthesis and Respiration-Regulation of Photosynthesis, Vol 11. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 487–505 [Google Scholar]
- Adamska I, Kloppstech K (1991) Evidence for an association of the early light-inducible protein (Elip) of pea with photosystem II. Plant Mol Biol 16: 209–223 [DOI] [PubMed] [Google Scholar]
- Adamska I, Ohad I, Kloppstech K (1992) Synthesis of the early light-inducible protein is controlled by blue light and related to light stress. Proc Natl Acad Sci USA 89: 2610–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF, Forsberg J (2001) Molecular recognition in thylakoid structure and function. Trends Plant Sci 6: 317–326 [DOI] [PubMed] [Google Scholar]
- Andersson B, Anderson JM (1980) Lateral heterogeneity in the distribution of chlorophyll-protein complexes of the thylakoid membranes of spinach chloroplasts. Biochim Biophys Acta 593: 427–440 [DOI] [PubMed] [Google Scholar]
- Andersson B, Aro EM (2001) Photodamage and D1 protein turnover in photosystem II. In EM Aro, B Andersson, eds, Advances in Photosynthesis and Respiration-Regulation of Photosynthesis, Vol 11. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 377–393 [Google Scholar]
- Barber J, Andersson B (1992) Too much of a good thing: Light can be bad for photosynthesis. Trends Biochem Sci 17: 61–66 [DOI] [PubMed] [Google Scholar]
- Bibby TS, Nield J, Barber J (2001) Iron deficiency induces the formation of an antenna ring around trimeric photosystem I in cyanobacteria. Nature 412: 743–745 [DOI] [PubMed] [Google Scholar]
- Boekema EJ, Hifney A, Yakushevska AE, Piotrowski M, Keegstra W, Berry S, Michel KP, Pistorius EK, Kruip J (2001) Green plant photosystem I binds light-harvesting complex I on one side of the complex. Nature 412: 745–748 [DOI] [PubMed] [Google Scholar]
- Bond JS (1996) Commercially available proteases. In RJ Beynon, JS Bond, eds, Proteolytic Enzymes: A Practical Approach. IRL Press at Oxford University Press, Oxford, pp 232–240
- Boudreau E, Takahashi C, Lemieux C, Turmel M, Rochaix JD (1997) The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J 16: 6095–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Briggs WR (2001) Blue light sensing in higher plants. J Biol Chem 276: 11457–11460 [DOI] [PubMed] [Google Scholar]
- Cronshagen U, Herzfeld F (1990) Distribution of early light-inducible proteins in the thylakoids of developing pea chloroplasts. Eur J Biochem 193: 361–366 [DOI] [PubMed] [Google Scholar]
- Dolganov NA, Bhaya D, Grossman AR (1995) Cyanobacterial protein with similarity to the chlorophyll a/b-binding proteins of higher plants: evolution and regulation. Proc Natl Acad Sci USA 92: 636–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Sétif P, Rochaix JD (1997) Targeted mutations in the psaC gene of Chlamydomonas reinhardtii: Preferential reduction of FB at low temperature is not accompanied by altered electron flow from photosystem I to ferredoxin. Biochemistry 36: 93–102 [DOI] [PubMed] [Google Scholar]
- Funk C (2001) The PsbS protein: A Cab-protein with a function of its own. In EM Aro, B Andersson, eds, Advances in Photosynthesis and Respiration-Regulation of Photosynthesis, Vol 11. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 453–467 [Google Scholar]
- Funk C, Schröder W, Napiwotzki A, Thus S, Renger G, Andersson B (1995) PSII-S proteins of higher plants: a new type of pigment-binding protein. Biochemistry 34: 11133–11141 [DOI] [PubMed] [Google Scholar]
- Funk C, Vermaas W (1999) A cyanobacterial gene family coding for single helix proteins resembling part of the light-harvesting proteins from higher plants. Biochemistry 38: 9397–9404 [DOI] [PubMed] [Google Scholar]
- Green BR, Durnford DG (1996) The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 47: 685–714 [DOI] [PubMed] [Google Scholar]
- Green BR, Kühlbrandt W (1995) Sequence conservation of light-harvesting and stress-response proteins in relation to the three-dimensional molecular structure of LHCII. Photosynth Res 44: 139–148 [DOI] [PubMed] [Google Scholar]
- Grimm B, Kloppstech K (1987) The early light-inducible proteins of barley: characterization of two families of 2-h-specific nuclear-coded chloroplast proteins. Eur J Biochem 167: 493–499 [DOI] [PubMed] [Google Scholar]
- Haldrup A, Jensen PE, Lunde C, Scheller HV (2001) Balance of power: a view of the mechanism of photosynthetic state transitions. Trends Plant Sci 6: 301–305 [DOI] [PubMed] [Google Scholar]
- He Q, Dolganov N, Björkman O, Grossman AR (2001) The high light-inducible polypeptides in Synechocystis PCC6803: expression and function in high light. J Biol Chem 276: 306–314 [DOI] [PubMed] [Google Scholar]
- Heddad M, Adamska I (2000) Light stress-regulated two-helix proteins in Arabidopsis thaliana related to the chlorophyll a/b-binding gene family. Proc Natl Acad Sci USA 97: 3741–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddad M, Adamska I (2002) The evolution of light stress proteins in photosynthetic organisms. Comp Funct Genom 3: 504–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddad M, Dunaeva M, Adamska I (2001) Expression pattern of the stress-enhanced protein Sep2 during cold and salt stress. In Osmond B, Critchley C, eds, Proceedings of the 12th International Congress on Photosynthesis, S3–060. CSIRO Publishing, Collingwood, Australia
- Hihara Y, Sonoike K (2001) Regulation, inhibition and protection of photosystem I. In EM Aro, B Andersson, eds, Advances in Photosynthesis and Respiration-Regulation of Photosynthesis, Vol 11. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 507–531 [Google Scholar]
- Jansson S, Andersson J, Kim SJ, Jackowski G (2000) An Arabidopsis thaliana protein homologous to cyanobacterial high-light-inducible proteins. Plant Mol Biol 42: 345–351 [DOI] [PubMed] [Google Scholar]
- Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, Krauss N (2001) Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411: 909–917 [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W, Wang DN, Fujiyoshi Y (1994) Atomic model of plant light-harvesting complex by electron crystallography. Nature 367: 614–621 [DOI] [PubMed] [Google Scholar]
- Kyle J, Doolittle RF (1982) A simple method of displaying the hydropathic character of a protein. J Mol Biol 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Levy H, Gokhman I, Zamir A (1992) Regulation and light-harvesting complex II association of a Dunaliella protein homologous to early light-induced proteins in higher plants. J Biol Chem 266: 13698–13705 [PubMed] [Google Scholar]
- Li XP, Björkman O, Shih C, Grossman AR, Rosenquist ML, Jansson S, Niyogi KK (2000) A pigment-binding protein essential for regulation of photosynthetic light-harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Lunde C, Jensen PE, Haldrup A, Knoetzel J, Scheller HV (2000) The PSI-H subunit of photosystem I is essential for state transitions in plant photo-synthesis. Nature 408: 613–615 [DOI] [PubMed] [Google Scholar]
- Meyer G, Kloppstech K (1984) A rapidly light-induced chloroplast protein with a high turnover coded for by pea nuclear DNA. Eur J Biochem 138: 201–207 [DOI] [PubMed] [Google Scholar]
- Montané MH, Dreyer S, Triantaphylidès C, Kloppstech K (1997) Early light inducible proteins during long-term acclimation of barley to photooxidative stress caused by light and cold: high level of accumulation by posttranscriptional regulation. Planta 201: 293–301 [Google Scholar]
- Montané MH, Kloppstech K (2000) The family of light-harvesting-related proteins (LHCs, ELIPs, HLIPs): Was the harvesting of light their primary function? Gene 258: 1–8 [DOI] [PubMed] [Google Scholar]
- Morrissey PJ, McCauley SW, Melis A (1986) Differential detergent solubilization of integral thylakoid membrane complexes in spinach chloroplasts: localization of photosystem II, cytochrome b6-f complex and photosystem I. Eur J Biochem 160: 389–393 [DOI] [PubMed] [Google Scholar]
- Pötter E, Kloppstech K (1993) Effects of light stress on the expression of early light-inducible proteins in barley. Eur J Biochem 214: 779–786 [DOI] [PubMed] [Google Scholar]
- Prasil O, Adir N, Ohad I (1992) Dynamics of photosystem II: mechanism of photoinhibition and recovery process. In J Barber, ed, The Photosystems: Structure, Function and Molecular Biology, Vol 11. Elsevier, Amsterdam, pp 295–348 [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T (1989) Enzymes used in molecular cloning. In Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 1–5.78–5.79
- Steinback KE, Mullet JE, Arntzen CJ (1982) Fractionation of thylakoid membrane protein complexes by sucrose density gradient centrifugation. In RB Hallick, NH Chua, eds, Methods in Chloroplast Molecular Biology. Elsevier Biomedical Press, Amsterdam, pp 864–871
- Takahashi Y, Goldschmidt-Clermont M, Soen SY, Franzen LG, Rochaix JD (1991) Directed chloroplast transformation in Chlamydomonas reinhardtii: Insertional inactivation of the psaC gene encoding the iron sulfur protein destabilizes photosystem I. EMBO J 10: 2033–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjus SE, Andersson B (1991) Extrinsic polypeptides of spinach photosystem I. Photosynth Res 27: 209–219 [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Vavilin D, Funk C, Vermaas W (2002) Small Cab-like proteins regulation tetrapyrrole biosynthesis in the cyanobacterium Synechocystis sp. PCC6803. Plant Mol Biol 49: 149–160 [DOI] [PubMed] [Google Scholar]