Abstract
The plant growth hormone gibberellin (GA) is important for many aspects of plant growth and development. Although most genes encoding enzymes at each step of the GA biosynthetic pathway have been cloned, their regulation is less well understood. To assess how up-regulation of early steps affects the biosynthetic pathway overall, we have examined transgenic Arabidopsis plants that overexpress either AtCPS or AtKS or both. These genes encode the enzymes ent-copalyl diphosphate synthase (CPS) and ent-kaurene synthase, which catalyze the first two committed steps in GA biosynthesis. We find that both CPS and CPS/ent-kaurene synthase overexpressors have greatly increased levels of the early intermediates ent-kaurene and ent-kaurenoic acid, but a lesser increase of later metabolites. These overexpression lines do not exhibit any GA overdose morphology and have wild-type levels of bioactive GAs. Our data show that CPS is limiting for ent-kaurene production and suggest that conversion of ent-kaurenoic acid to GA12 by ent-kaurenoic acid oxidase may be an important rate-limiting step for production of bioactive GA. These results demonstrate the ability of plants to maintain GA homeostasis despite large changes in accumulation of early intermediates in the biosynthetic pathway.
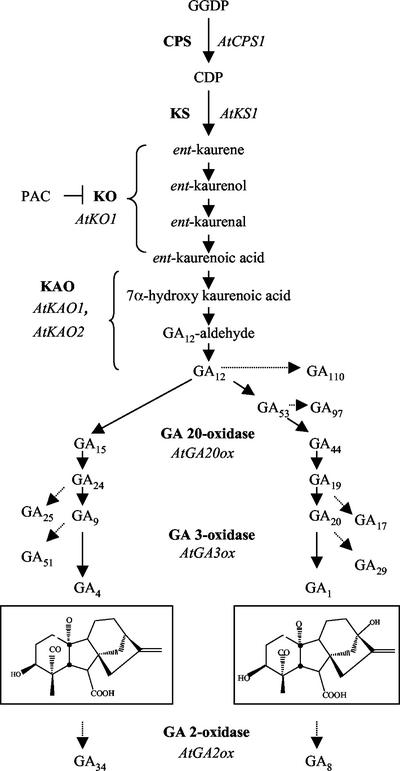
The plant growth hormone GA is important for processes including seed germination, stem growth, and flower and fruit development. The GA biosynthetic pathway has been characterized, and genes encoding most GA biosynthetic enzymes have been cloned in Arabidopsis and other species (Hedden and Phillips, 2000). In the first portion of the pathway, the precursor geranylgeranyl diphosphate (GGDP) is converted to ent-kaurene in a two-step process catalyzed by ent-copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS; Fig. 1). These enzymes are the products of the Arabidopsis genes AtCPS and AtKS, respectively (Sun et al., 1992; Yamaguchi et al., 1998b). Subsequent reactions catalyzed by the cytochrome P450 enzymes ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO) produce ent-kaurenoic acid (KA) and GA12 from ent-kaurene (Helliwell et al., 1999, 2001a). The final portion of the pathway branches from GA12 to form non-13-hydroxylated metabolites and bioactive GA4 or early 13-hydroxylation metabolites and bioactive GA1. These steps are catalyzed by GA 20-oxidase and GA 3-oxidase enzymes (encoded by multiple AtGA20ox and AtGA3ox genes, respectively). GA 13-oxidase, which converts GA12 to GA53 to form the 13-hydroxylated metabolites, has not been identified in any plant species.
Figure 1.
GA biosynthesis in Arabidopsis. Solid arrows, Metabolic steps in GA biosynthesis. Dashed-line arrows, Inactivation steps. Bold text, Enzymes catalyzing these reactions. Italics, Arabidopsis gene names. T bar, Inhibition of the indicated enzyme. Boxed structures, Bioactive GAs. Brackets, Multiple steps catalyzed by one enzyme. The conversion of GA12 to GA53 is catalyzed by GA 13-oxidase, which has not been cloned in Arabidopsis. CDP, ent-Copalyl diphosphate. GA 20-oxidase catalyzes multiple steps, from GA12 to GA9 and GA53 to GA20. PAC, paclobutrazol.
Plants that have been treated with an excess of GA hormone have a characteristic GA overdose morphology. This includes increased hypocotyl length, paler leaves, elongated petioles, taller stem height, and earlier flowering time (Jacobsen and Olszewski, 1993). Plants engineered to overexpress GA biosynthetic genes have also been generated to investigate effects on morphology and GA biosynthesis. Transgenic plants that overexpress GA 20-oxidase, in either Arabidopsis (Huang et al., 1998; Coles et al., 1999) or Solanum tuberosum (Carrera et al., 2000), show various aspects of this GA overdose morphology. Overexpression of Cucurbita maxima GA 20-oxidase, whose major product in Arabidopsis is inactive GA17 and GA25, results in a decrease in bioactive GA4 in Arabidopsis and a slight decrease in stem elongation (Xu et al., 1999). These results indicate that modification of expression of downstream enzymes in GA biosynthesis can affect both GA levels and plant morphology.
Overexpression experiments have also been attempted with the AtCPS gene of Arabidopsis. Because AtCPS encodes the enzyme catalyzing the first committed step in GA biosynthesis and because it shows a highly regulated, tissue-specific expression pattern (Silverstone et al., 1997a), it is a potential regulatory point for control of overall levels of bioactive GAs. However, Arabidopsis plants that overexpress the AtCPS cDNA under the control of a cauliflower mosaic virus (CaMV) 35S promoter have only subtle morphological variations from wild type (WT; Sun and Kamiya, 1994). In vitro studies of ent-kaurene produced from labeled precursor GGDP or intermediate ent-copalyl diphosphate suggest that CPS and KS may interact (Duncan and West, 1981). Thus, it might be necessary to overexpress both AtCPS and AtKS to produce an increase in ent-kaurene, which could lead to an increase in bioactive GAs and a GA overdose morphology.
Rate-limiting steps have not been clearly defined for the GA biosynthetic pathway. Therefore, overproduction of enzymes at the beginning of the pathway may be informative because increased flux through the pathway could identify steps at which metabolite accumulates. To determine the effect of increased levels of enzymes catalyzing early steps in GA biosynthesis, we examined transgenic Arabidopsis plants that overexpress either AtCPS or AtKS or both AtCPS and AtKS. We investigated their morphology, ent-kaurene, KA and GA content, response to a GA biosynthesis inhibitor, and expression of downstream GA biosynthetic genes. We report here that CPS and CPS/KS overexpressors (OEs) produce increased levels of ent-kaurene while maintaining normal morphology and WT levels of bioactive GA.
RESULTS
Generation of Overexpression Lines
The reaction catalyzed by CPS is a possible step limiting the flow of metabolite through the GA biosynthetic pathway because the substrate, GGDP, is a common precursor for other products, including carotenoids, the phytol chain of chlorophyll, and the growth hormone abscisic acid (Takahashi et al., 1991). We have previously generated transgenic Arabidopsis plants that accumulate CPS protein at a much higher level than WT plants (Sun and Kamiya, 1994). The CPS overexpression construct used was sufficient to rescue the GA-deficient ga1-3 mutant (null for AtCPS; Sun and Kamiya, 1994). However, these transgenic plants did not exhibit a GA overdose phenotype in the ga1-3 background. We have now generated additional overexpression lines in the Landsberg erecta (Ler) WT background to better examine the effects of increased CPS expression. To further increase metabolite flux in the early GA biosynthetic pathway, we also attempted to overproduce both CPS and KS proteins in double transgenic plants. As an additional comparison, we made transgenic plants overexpressing the AtKS gene alone to examine the effects of its overexpression relative to those of AtCPS overexpression.
To generate lines overexpressing AtCPS, Ler (WT) plants were transformed with the pGA1-49 CPS overexpression construct (Sun and Kamiya, 1994). This construct contains a CaMV 35S promoter with dual enhancer plus a 5′ non-translated region (NTR) from tobacco etch virus (TEV) fused translationally to the AtCPS cDNA at its ATG start site. TEV-NTR is known to increase translation (Carrington and Freed, 1990) and was effective to overproduce the CPS protein (Sun and Kamiya, 1994). T2 seeds were screened for 3:1 (resistant:sensitive) kanamycin resistance, indicating a single insertion site of the transgene. Seven independent transgenic lines were rescreened at the T3 generation to identify homozygotes.
To overproduce KS protein in Arabidopsis, a cassette containing the CaMV 35S promoter-TEV-NTR-AtKS cDNA was cloned into the binary vector pDHB321. This vector has the bar gene conferring resistance to BASTA in plants. We introduced this plasmid construct into WT and one CPS OE via Agrobacterium tumefaciens-mediated transformation. In the T2 generation, we selected four and six transgenic lines in WT and CPS OE background, respectively, that segregated approximately 3:1 for BASTA resistance versus sensitivity. Homozygotes were selected by examining segregation at the T3 generation.
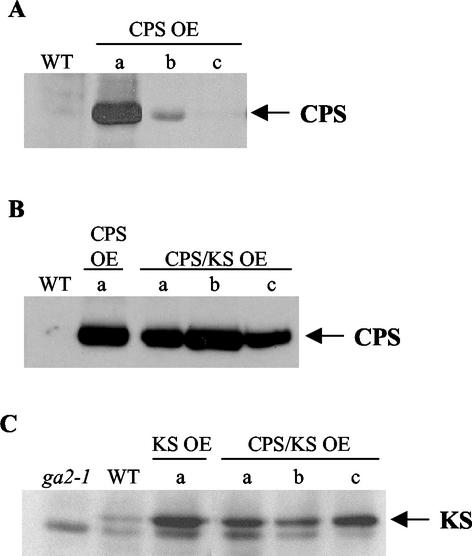
Protein levels of all lines were determined by immunoblot analysis, using anti-CPS antisera (Sun and Kamiya, 1994) and/or anti-KS antisera generated against full-length Arabidopsis KS protein. CPS and KS protein levels of selected lines are shown (Fig. 2). Lines overexpressing high, moderate, or low levels of CPS are shown (CPS OE-a, -b, or -c, respectively; Fig. 2A). WT CPS protein is not detectable under our conditions. However, CPS OE-a and -b clearly show higher protein levels than WT. The double OEs were generated from CPS OE-a, and all show similar levels of CPS expression as in that line (Fig. 2B). Unlike CPS, KS protein is detectable in WT, and all overexpression lines shown have higher KS levels than WT. The mutation in ga2-1 causes a truncation in KS, resulting in a 74-kD protein, approximately the same size as the background band seen below KS in all lanes (Fig. 2C). Expression of the KS OE construct is sufficient to complement the ga2-1 mutant, suggesting that the transgene does produce active KS protein (data not shown).
Figure 2.
CPS and KS protein levels in overexpression lines. A, CPS levels in WT and CPS OE plants. B, CPS levels in CPS OE and CPS/KS OE plants. C, KS levels in WT, KS OE, and CPS/KS OE plants. Immunoblots show 30 (A and C) or 35 (B) μg of total protein extract per lane. Blots were probed with anti-CPS (A and B) or anti-KS (C) antisera. The 76-kD CPS protein is indicated by the arrow in A and B and is not detectable in WT. The upper band in C (indicated by the arrow) is the 88-kD KS protein, not visible in the ga2-1 mutant lane. ga2-1 protein is approximately 74 kD, similar in size to the lower band in C that represents a nonspecific background protein.
Although it is not possible to determine the absolute degree of CPS overexpression in our conditions, we used recombinant CPS and KS protein to determine amounts of each enzyme present in the overexpression lines. Dilutions of protein from WT and CPS or KS OEs were compared with known amounts of recombinant protein for quantification. From this method, we estimate that the amount of CPS in line CPS OE-a is approximately 1 ng μg–1 total protein. The amount of KS is estimated to be 0.1 ng μg–1 total protein in WT and 0.5 ng KS μg–1 total protein in line KS OE-a (data not shown).
CPS, KS, and CPS/KS OEs Have WT Morphology
WT plants treated with exogenous GA and transgenic plants overexpressing the downstream GA biosynthetic gene AtGA20ox1 showed aspects of GA overdose morphology (Huang et al., 1998; Coles et al., 1999; Carrera et al., 2000). To determine whether our overexpression constructs would affect plant morphology, we characterized plants with regard to traits affected by GA, including hypocotyl length, final height, rosette diameter, flowering time, silique length, and number of seeds per silique. Seven CPS OE lines (CPS OE-a, -b, and five others), three KS OE lines (KS OE-a, -b, and one other), and three CPS/KS OE lines (CPS/KS OE-a, -b, and -c) were examined for this analysis. None of the characteristics examined was significantly different from WT for any of the overexpression lines (data not shown). This could be due to any of several possibilities, including inability of the overexpressed proteins to produce more ent-kaurene, limitation on metabolite flux through the biosynthesis pathway by downstream enzymes, or increased catabolism of GA hormone.
ent-Kaurene and GA Measurements
The lack of GA overdose morphology in the CPS and/or KS OEs suggests that bioactive GA levels may not be increased in these plants. Plants from selected transgenic lines were grown on soil to rosette stage (d 21 or 22) for combined gas chromatography (GC)-selected ion monitoring (SIM) analysis to determine their levels of kaurenoids and GAs. For initial ent-kaurene measurements, plants were treated with the GA biosynthesis inhibitor paclobutrazol (PAC) 4 d before harvest to promote the accumulation of sufficient levels of ent-kaurene for detection. Because high levels of ent-kaurene were found in the PAC-treated tissue, especially in overexpression lines, measurements were repeated once using untreated rosette tissue to determine normal ent-kaurene accumulation. A total of five lines was used for this analysis: transgenic lines expressing medium or high levels of CPS; CPS OE-b and -a, respectively; KS overexpression line KS OE-a; double overexpression line CPS/KS OE-a; and control WT.
Because ent-kaurene is the immediate product of the reactions catalyzed by CPS and KS, its level of accumulation should suggest whether the overexpressed proteins are functional. The medium (CPS OE-b) and high (CPS OE-a) OEs show a 30- and 1,008-fold increase in ent-kaurene levels, respectively, compared with untreated tissue (Table I). The CPS/KS double overexpression line shows the greatest increase, with 1,888-fold more ent-kaurene than WT. However, ent-kaurene levels in the KS OE line are comparable with WT. This suggests that it is CPS rather than KS that is limiting for ent-kaurene production, which is consistent with reports that AtCPS mRNA is expressed at much lower levels than AtKS mRNA in WT plants (Silverstone et al. 1997; Yamaguchi et al. 1998b). ent-Kaurene is converted to KA by the cytochrome P450 enzyme KO. We measured KA in WT and transgenic lines with high ent-kaurene levels. Our results show that, when compared with WT, the OEs have a similar degree of KA accumulation as ent-kaurene (Table I).
Table I.
ent-Kaurene and KA levels in rosette plants with or without PAC treatment
| Line | ent-Kaurenea + PAC | ent-Kaureneb - PAC | ent-KAc - PAC | |||
|---|---|---|---|---|---|---|
| WT | 200 | (1)d | 4.1 | (1) | 13 | (1) |
| CPS OE-a | 36,777 | (184) | 4,034 | (1,008) | 14,462 | (1,112) |
| CPS OE-b | 4,047 | (20) | 119 | (30) | 109 | (8) |
| KS OE-a | 203 | (1) | 2.7 | (0.7) | nde | |
| CPS/KS OE-a | 69,479 | (347) | 7,550 | (1,888) | 12,521 | (963) |
Data for ent-kaurene + PAC represent the average of two measurements in nanograms per gram fresh wt. bNos. represent single measurements in nanograms per gram fresh wt. cKA values represent measurements from one experiment in nanograms per gram dry wt. This has been repeated once with similar results. dParentheses denote fold change relative to WT. end, Not determined.
The later portion of the GA biosynthesis pathway branches downstream of GA12. The non-13-hydroxylation branch and the early 13-hydroxylation branch are parallel portions of the pathway that produce bioactive GA4 and GA1, respectively (Fig. 1). We measured GA levels in both branches (Table II). Relative amounts of GA12 in WT and transgenic lines correspond with the trend observed for earlier metabolites. The GA12 level was near WT in KS OE-a, 5- to 9-fold of WT level in the CPS overexpression lines, and 10 times greater than WT in the CPS/KS double OE. Similarly, the non-13-hydroxylated metabolites GA15 and GA24 were somewhat elevated in the CPS lines and in the double OE. Notably, however, levels of GA9, bioactive GA4, and inactivated GA34 and GA51 were similar to WT for all transgenic lines measured.
Table II.
GA levels in WT and CPS and/or KS OE lines
Values represent one measurement per compound per line, as determined by GC-SIM. Measurements have been repeated at least once with similar results. Italicized nos. represent those compounds with at least 3-fold increased levels relative to WT.
| Line | Non-13-Hydroxylated GAs
|
13-Hydroxylated GAs
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GA12 | GA15 | GA24 | GA9 | GA4a | GA34b | GA51b | GA53c | GA44 | GA19 | GA20 | GA1a | GA8b | GA29b | |
| ng g-1 dry wt | ||||||||||||||
| WT | 30.5 | 4.2 | 38.5 | 1.4 | 3.7 | 13.3 | 7.6 | 6.7 | 0.8 | 7.9 | 0.3 | 0.4 | 1.2 | 0.2 |
| CPS OE-a | 387.3 | 11.9 | 172.6 | 1.4 | 3.3 | 10.9 | 6.2 | 31.0 | 1.3 | 13.2 | 0.3 | 0.2 | 0.6 | 0.2 |
| CPS OE-b | 179.9 | 7.3 | 90.9 | 1.1 | 3.6 | 11.1 | 16.6 | 23.1 | 1.6 | 13.0 | 0.3 | 0.3 | 0.9 | 0.2 |
| KS OE-a | 29.8 | 3.8 | 34.5 | 1.3 | 4.1 | 8.6 | ndd | nd | 0.8 | 9.1 | 0.4 | 0.3 | 1.0 | 0.2 |
| CPS/KS OE-a | 328.3 | 12.1 | 160.0 | 1.0 | 2.7 | 9.7 | 17.1 | 27.9 | 1.3 | 9.6 | 0.2 | 0.2 | 0.6 | 0.1 |
Indicates bioactive forms of GA. b Indicates inactivated forms of GA. c GA53 data represent a single measurement. d nd, Not determined.
Overall metabolite levels were higher in the non-13-hydroxylated pathway than in the parallel 13-hydroxylation pathway. This may be expected because GA4 is the primary active GA in Arabidopsis (Talón et al., 1990). In the 13-hydroxylated branch, the early intermediate GA53 was present at higher levels in the CPS and CPS/KS lines relative to WT. However, amounts of GA44, GA19 and GA20, bioactive GA1, and inactive GA8 and GA29 were not significantly different between OEs and WT plants (Table II). Thus, although CPS and CPS/KS OEs do produce more ent-kaurene and early GA intermediates, they have WT levels of bioactive GAs, consistent with their morphology.
Increased Resistance of CPS OEs to GA Biosynthesis Inhibitor
PAC impairs GA biosynthesis as a competitive inhibitor of KO (Rademacher, 1991). Treatment with PAC blocks oxidation of ent-kaurene, the product of the reactions catalyzed by CPS and KS (Fig. 1). WT plants treated with appropriate concentrations of PAC show aspects of a GA-deficient phenotype, including decreased germination efficiency, reduced hypocotyl length, and delayed flowering time because of a decrease in synthesis of bioactive GA. However, plants with elevated levels of ent-kaurene may be more resistant to PAC because the higher amount of ent-kaurene could compete with PAC to allow the synthesis of some bioactive GA.
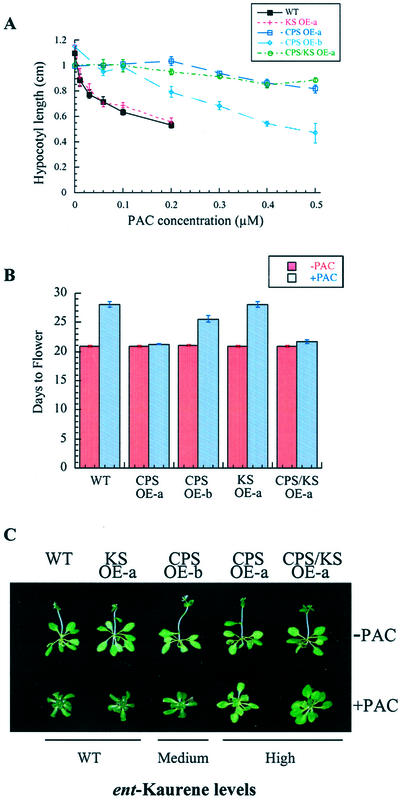
We compared the effect of PAC on WT and CPS/KS OEs during seedling growth and initiation of flowering. Hypocotyl elongation was assayed with plants grown on plates containing varying concentrations of PAC that still allow germination of all lines tested. Plants were incubated in the dark to promote longer hypocotyl growth for easier measurement. CPS OE-a and CPS/KS OE-a showed high levels of resistance to PAC and maintained long hypocotyls even at 0.5 μm PAC (Fig. 3A). KS OE-a responded similarly to WT, with hypocotyl length decreasing at increasing PAC, whereas CPS OE-b showed an intermediate response between that of WT and high-CPS OEs. CPS overexpression also conferred resistance to PAC during rosette development and transition to flowering. WT plants grown on 1 μm PAC developed a GA-deficient phenotype, with smaller rosettes, dark-green leaves, and delayed flowering (Fig. 3, B and C). KS OEs responded similarly to WT for flowering time and rosette development, whereas CPS OE-b showed only a slight increase in PAC resistance (Fig. 3, B and C). PAC-treated CPS OE-a and CPS/KS OE-a flowered within 1 d of untreated plants and had more resistant rosette development (Fig. 3, B and C). Despite the increased resistance of CPS OE-a and CPS/KS OE-a to PAC, these plants showed delayed inflorescence elongation (Fig. 3C). CPS and CPS/KS OE lines also showed increased PAC resistance at seed germination (data not shown).
Figure 3.
Increased resistance of CPS/KS OE lines to PAC. A, Hypocotyl length of 6-d-old etiolated seedlings as an average of 10 seedlings ± se. WT and KS OE-a plants have reduced germination at PAC concentrations higher than 0.2 μm. B, Flowering time was determined by the appearance of the first flower bud. Values are indicated as ± se for the average of 20 plants per line. Plants were grown in long days (LDs) on Murashige and Skoog plates (–PAC, red bars) or Murashige and Skoog plus 1 μm PAC (+PAC, blue bars). Experiments in A and B have been repeated once each with similar results. C, Phenotypes of 26-d-old rosette plants representative of each line from the experiment described in B. Relative ent-kaurene levels in these lines are indicated below the photographs. Although the influorescence on PAC-treated CPS OE-a and CPS/KS OE-a is not clearly visible in this photograph, these plants have flower buds.
Expression of Downstream GA Biosynthetic Genes in CPS/KS Overexpression Lines
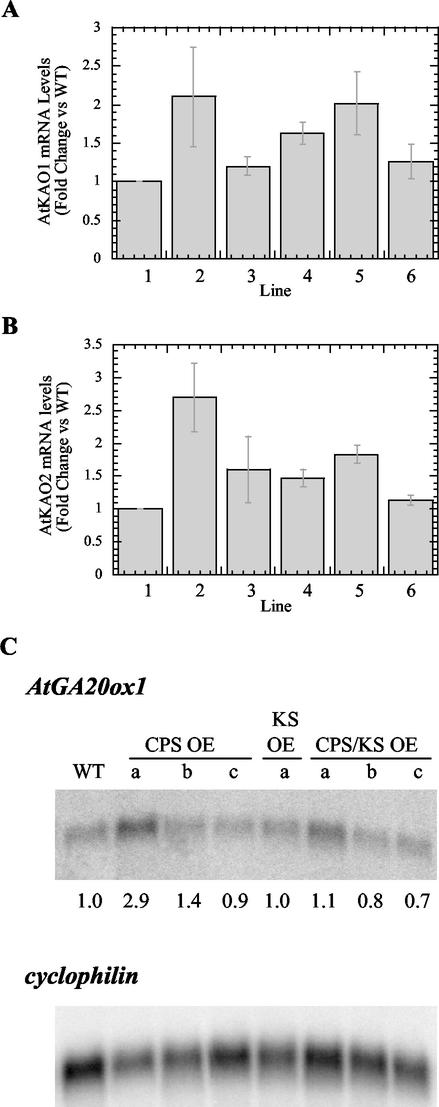
Because the greatest difference in metabolite accumulation between the CPS and CPS/KS OEs versus WT occurs between KA and GA12, the reaction catalyzed by KAO may be rate limiting. This could be because endogenous KAO levels are insufficient to handle excess metabolite or because AtKAO mRNA levels are down-regulated in the overexpression lines. Two genes encoding KAO have been identified in Arabidopsis, AtKAO1 and AtKAO2 (Helliwell et al., 2001a), but it is not known whether they are under negative feedback regulation in response to GA. To determine whether AtKAO transcript levels are altered in the OEs, we measured the expression of these genes in rosette tissue using quantitative reverse transcription (RT)-PCR (qPCR; Fig. 4, A and B). The data show that the transcript level of neither gene is decreased significantly in the OEs as compared with WT.
Figure 4.
Expression of downstream GA biosynthetic genes in CPS and/or KS overexpression lines. A, AtKAO1 mRNA levels in OE lines, relative to WT. Lines are as follows: 1, WT; 2, CPS OE-a; 3, CPS OE-b; 4, KS OE-a; 5, CPS/KS OE-a; and 6, CPS/KS OE-b. B, AtKAO2 mRNA levels in OE lines, relative to WT. Lines are designated as in A. For both A and B, numbers for WT, CPS OEa, and CPS OEb represent the average of duplicate qPCR reactions for each of three separate batches of tissue. Numbers for KS OE-a and CPS/KS OE lines represent the average of three qPCR reactions from a single batch of tissue. Bars = se. Total RNA was extracted from rosette tissue, and the amount of RNA in each reaction was determined by normalization to the housekeeping gene Act11. C, Autoradiogram of RNA blot containing 1 μg of poly(A+) RNA per lane. The blot was probed with a radiolabeled antisense AtGA20ox1 RNA probe (top) and then re probed with a random prime-labeled cyclophilin cDNA probe (bottom). Numbers below the top blot represent relative amounts of message based on loading control. WT was arbitrarily set to 1.0.
Previous work has shown that the AtGA20ox1 and AtGA3ox1 genes, encoding enzymes that catalyze the final steps in GA biosynthesis, are under negative feedback regulation (Chiang et al., 1995; Phillips et al., 1995; Xu et al., 1995; Silverstone et al., 1998). In a GA-deficient background, such as the ga1-3 mutant, these genes are up-regulated; in the presence of exogenous GA, these genes are down-regulated. It is possible that feedback control also acts in the CPS and KS overexpression lines, such that down-regulation of GA 20-oxidase (AtGA20ox) and/or GA 3-oxidase (AtGA3ox) may compensate for increased ent-kaurene production. This would limit any change in overall bioactive GA levels; therefore, plants might not show GA overdose morphology. Our results indicate that although GA24 levels are higher in overexpression lines than in WT, GA9 levels are the same in all lines (Table II). AtGA20ox encodes the enzyme catalyzing the conversion of GA24 to GA9; therefore, decreased expression of this gene might account for the lack of accumulation of downstream GA intermediates. We examined transcript levels of AtGA20ox1 in the overexpression lines by RNA-blot analysis. Figure 4C shows that message levels of AtGA20ox1 are not significantly decreased in any line. The increased AtGA20ox1, AtKAO1, and AtKAO2 transcript levels observed in CPS OE-a are likely not significant because CPS/KS OE-a also has similar levels of GA intermediates to CPS OE-a but no significant change in AtGA20ox1 expression. Levels of AtKO and AtGA3ox1 mRNA are also not decreased in the overexpression lines (data not shown). This indicates that for the genes examined, negative regulation at the transcript level does not account for the WT levels of bioactive GA in CPS/KS OEs.
DISCUSSION
Although genes encoding the enzymes for most steps in GA biosynthesis have been cloned in Arabidopsis, the regulation of the GA biosynthetic pathway is less well understood. Overproduction of enzymes catalyzing the first committed steps in GA biosynthesis may be one tool to aid our understanding of the control of GA biosynthesis. Because CPS is present at lower levels than downstream biosynthetic enzymes, overexpression of this protein may increase flux through the pathway and show possible rate-limiting steps downstream.
We have generated lines that produce high levels of CPS or KS protein or both CPS and KS in Arabidopsis. CPS and CPS/KS OEs have high levels of the early biosynthetic intermediates ent-kaurene and KA but have WT levels of bioactive GAs and normal morphology. However, KS OEs have WT ent-kaurene levels. This shows that CPS is limiting for ent-kaurene biosynthesis, consistent with data from Arabidopsis and C. maxima, indicating that CPS expression is more limited than KS expression (Silverstone et al., 1997b; Smith et al., 1998; Yamaguchi et al., 1998b). Calculation of reaction velocities with recombinant C. maxima enzymes has shown a 2.6-fold higher rate for KS than for CPS, which also supports a limiting role for CPS (Kawaide et al., 2000). Work with the fungus Phaeosphaeria sp. L487 demonstrated that addition of purified C. maxima KS but not CPS enhanced ent-kaurene production by the fungal CPS/KS enzyme in vitro (Kawaide et al., 2000), suggesting that in that system, it is KS activity that is limiting for ent-kaurene production. The difference in regulation of ent-kaurene production between Phaeosphaeria sp. L487 and Arabidopsis may be due to the fact that Phaeosphaeria sp. L487 has a bifunctional KS (fungal CPS/KS) that converts GGDP to ent-kaurene, whereas Arabidopsis and other plants have two separate enzymes that may be present in differing stoichiometries. Immunoblots containing recombinant CPS or KS standards suggest that the absolute amount of KS protein in WT plants is much greater than that of CPS (data not shown). This may explain why KS overexpression without CPS overexpression is insufficient to increase ent-kaurene production.
The ability of plants to maintain homeostasis of GA hormone levels in the presence of a 1,000-fold increase in ent-kaurene and KA suggests that later steps in GA biosynthesis are tightly regulated. Overexpression of the penultimate enzyme in GA biosynthesis, GA 20-oxidase, does result in increased GA content (Huang et al., 1998; Carrera et al., 1999; Coles et al., 1999). Therefore, it is likely that some of the key regulation important for maintaining appropriate hormone levels in the CPS and CPS/KS OEs lies at or upstream of the reactions catalyzed by GA 20-oxidase.
Measurements of the GA biosynthetic intermediates from the overexpression lines give some indication as to which steps in the pathway may be rate limiting. The most significant change in accumulation of metabolite in the OEs as compared with WT occurs from KA to GA12. Lines with high levels of CPS (CPS OE-a and CPS/KS OE-a) accumulate at least 1,000 times more ent-kaurene and KA than WT but only approximately 10 times more GA12 than WT, a 100-fold decrease in accumulation. This difference identifies the portion of the pathway whose reactions are catalyzed by KAO as likely rate-limiting in the CPS and CPS/KS OE plants. RNA levels of AtKAO1 and AtKAO2 are not decreased in the overexpression lines (Fig. 4, A and B), and we cannot rule out the possibility of decreased KAO enzyme amount or activity. However, our data suggest that endogenous KAO levels may be insufficient to process the great increase in KA present in the CPS and CPS/KS OE lines, resulting in high levels of KA but less increase in GA12. Thus, although CPS appears limiting for ent-kaurene biosynthesis, KAO may be limiting for production of middle and later GA intermediates. Plants engineered to overproduce KAO, perhaps in combination with CPS and/or GA20ox, may have a more pronounced increase in bioactive GA than that observed with overexpression of AtGA20ox1 alone. However, it is still unclear whether KAO in WT plants would ever encounter such high substrate levels for its activity to be limiting.
Evidence from plants such as Alpidea amatysia, C. maxima, and Cucurbita pepo suggests that KA can be converted to kaurenolides and also be converted to GA12 (Fukui et al., 1977; Hedden and Graebe, 1981; Rustaiyan and Sadjadi, 1987). If this reaction also occurs in Arabidopsis, high levels of ent-kaurene and KA in the CPS overexpression lines may result in some synthesis of non-GA products, which could partially account for the decrease in GA12 accumulation.
Recently, a novel class of GA 2-oxidases has been identified in Arabidopsis (Schomberg et al., 2003). These enzymes, encoded by AtGA2ox7 and AtGA2ox8 genes, catalyze 2β-hydroxylation of C20-GAs, such as GA12 and GA53, to form inactive GAs, GA110 and GA97, respectively (Fig. 1). Further studies are necessary to examine if AtGA2ox7 and AtGA2ox8 play any role in creating a large difference in accumulation of KA and GA12 observed in the CPS OE lines. However, our data suggest that increased 2β-hydroxylation of C19-GAs in the overexpressors is unlikely, given that the inactivated forms of GA, including GA34, GA51, GA8, and GA29, are near WT in all lines.
Another possible limitation on metabolite flux through the GA biosynthetic pathway is that of transport of intermediates, subcellularly or across cell types. Although CPS and KS are localized to plastids (Sun and Kamiya, 1994; Yamaguchi et al., 1998b), later reactions in GA biosynthesis take place on the endoplasmic reticulum and in the cytosol (for review, see Hedden and Kamiya, 1997), requiring movement of metabolite to different subcellular compartments. Additional transport may be necessary between different cell types. Data from GUS staining and in situ hybridizations show that AtCPS is localized to the pro-vasculature, whereas AtKO and AtGA3ox1 are in cortical cells of Arabidopsis embryos during germination (Yamaguchi et al., 2001). These differences in localization of biosynthetic enzymes, along with the hydrophobic nature of ent-kaurene, have led to the suggestion that ent-kaurene may require a transporter (Yamaguchi et al., 2001), and this transport step could limit GA biosynthesis. However, because KA, the next intermediate after ent-kaurene, also accumulates to high levels in the CPS OEs, ent-kaurene transport may not be limiting under our conditions.
Work using green fluorescent protein fusion proteins has shown that KO, which converts ent-kaurene to KA, may be localized to the outer face of chloroplasts and the cytoplasm and/or endoplasmic reticulum. The authors suggest that this localization of KO may provide a way for biosynthetic intermediate to move from the plastid to the endoplasmic reticulum (Helliwell et al., 2001b). This might facilitate the transport of hydrophobic ent-kaurene to be metabolized further by endoplasmic reticulum-localized enzymes. In addition, because the cell-type localization of GA biosynthetic genes is known only for seeds, it is not clear whether this pattern remains at the rosette stage for which GA measurements have been performed here. It will be of interest to determine whether the difference in cell-type localization of GA biosynthetic genes persists at other developmental stages.
We found that the overexpression lines contain elevated GA12 and GA53 but normal GA9 and GA20 levels. As discussed for KAO, this could be because limited amounts of GA 20-oxidases cannot handle excess substrates, or because GA 20-oxidation activity is down-regulated in the transgenic lines. We showed that AtGA20ox1 and AtGA3ox1 mRNA levels in the OEs are not decreased (Fig. 4; data not shown), indicating that negative feedback regulation of these genes does not account for the normal active GA levels in the OEs. The lack of negative feedback regulation in the OEs may not be surprising because there is no significant increase in bioactive GAs in these lines. In addition, the unchanged expression of downstream biosynthetic genes indicates that levels of upstream biosynthetic intermediates such as ent-kaurene do not affect transcriptional regulation of AtGA20ox1 or AtGA3ox1. Thus, potential down-regulation of GA 20-oxidation activity would be attributed to changes in protein levels or protein activity of GA 20-oxidases and/or expression of other AtGA20ox genes (Phillips et al., 1995) that were not examined in this study.
In conclusion, the ability of plants to maintain GA homeostasis despite the very large increase in early metabolite in the CPS and CPS/KS OEs suggests that many levels of regulation exist to preserve appropriate levels of bioactive GA.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Experiments were conducted using Arabidopsis of the ecotype Ler. Seeds were stratified at 4°C for 3 to 4 d and sown on soil at 22°C under LDs (16 h of light, 8 h of dark). For plate growth assays, plants were grown on media containing Murashige and Skoog salts plus 2% (w/v) Suc and 0.8% (w/v) agar. Plants for GA analysis and kaurene measurements were grown on soil in LD conditions to d 21 or 22, within 1 to 2 d of the appearance of the first flower bud. Rosettes were then harvested and frozen in liquid nitrogen for storage at –80°C until lyophilization. Additional plants for ent-kaurene analysis were grown on soil to d 21, then drenched with a solution of PAC (Bonzi, Uniroyal, Middlebury, CT) at 8.75 mL Bonzi L–1 (=120 μm active ingredient PAC). Each flat, containing approximately 250 plants, was bottom watered with 1 L of PAC solution. Tissues were harvested at 4 d after treatment and frozen in liquid nitrogen.
Plasmid Construction and Plant Transformation
CPS overexpression lines were generated using pGA1-49, containing the 2.4-kb AtCPS cDNA under the control of the CaMV 35S promoter and TEV-NTR, in the pBIN19 vector conferring kanamycin resistance (Sun and Kamiya, 1994). This construct was introduced into WT plants using Agrobacterium tumefaciens-mediated vacuum infiltration (Bechtold et al., 1993).
For KS OEs, a translational fusion of the AtKS cDNA (2.4 kb) to the CaMV 35S-TEV-NTR was made. We first introduced an AflIII site at the starting ATG codon of the AtKS gene by PCR. A PCR-amplified partial AtKS coding region (0.3 kb), having AflIII and SacI sites at the 5′ and 3′ ends, respectively, was cloned into the NcoI site (compatible with an AflIII site) and SacI sites of pRTL2 vector (Restrepo et al., 1990; pGA2-5). The inserted DNA fragment was sequenced to confirm no PCR errors. The rest of the AtKS cDNA was obtained from pGA2-4 (Yamaguchi et al., 1998a). pGA2-4 was digested with SacI and KpnI, and the resulting 2.1-kb DNA fragment was inserted into the SacI-KpnI sites of pGA2-5. This plasmid, named pGA2-6, has the full-length AtKS cDNA fused at ATG to the TEV-NTR and the CaMV 35S promoter with duplicated enhancer. The whole translational fusion was transferred into the binary vector pDHB321 (gift from David Bouchez, Institut National de la Recherche Agronomique, Versailles, France) as follows. The plasmid pGA2-6 was digested with SphI, and the resulting 3.4-kb DNA fragment was blunt ended. A BamHI linker was ligated with the blunt-ended DNA fragment, which was then inserted into the BamHI site of pDHB321 to make pGA2-9. This construct was introduced into Ler WT and CPS OE-a plants via A. tumefaciens-mediated transformation using the floral dip method (Clough and Bent, 1998).
Selection of transformants was based on antibiotic or herbicide resistance. Transgenic plants containing pGA1-49 were screened on Murashige and Skoog with 50 μg mL–1 kanamycin. Plants containing pGA2-9 were screened on soil by spraying with a 1:10,000 (w/v) dilution of gluphosinate ammonium (BASTA; Hoechst Agro-Evo, Berlin) daily for 3 d, beginning after the appearance of the first true leaves (about 1 week). Rescreening at T3 was conducted on plates containing 50 μm gluphosinate ammonium (Crescent Chemicals, Happauge, NY).
Generation of Anti-KS Antibody
For production of KS protein, the pET/AtKS plasmid, containing the His-T7-tagged 2.5-kb AtKS cDNA (Yamaguchi et al., 1998a), was transformed into HMS174(DE3) Escherichia coli cells. Production of the recombinant AtKS protein was induced by isopropylthio-β-galactoside as described by Yamaguchi et al. (1998a). Induced E. coli were harvested and lysed by French press. Inclusion bodies containing insoluble KS protein were resuspended in 50 mm Tris and separated on a 10% (w/v) SDS-polyacrylamide gel. The 88-kD band corresponding to KS protein was excised from the gel for generation of antisera. Rat antibodies to the full-length KS protein were generated by Cocalico Biologicals (Reamstown, PA).
Immunoblots
Total protein from 3-week-old Arabidopsis rosette tissue was extracted as described by Silverstone et al. (2001). Protein was separated on 8% (w/v) SDS-PAGE gels and transferred to nitrocellulose membrane. Immunoblot analysis was performed as described previously (Sambrook et al., 1989). For detection of CPS protein, the membrane was incubated with 1,000-fold diluted 30-kD rabbit CPS antisera (primary antibody; Sun and Kamiya, 1994) and then with 5,000-fold diluted peroxidase-conjugated goat-anti-rabbit antisera (secondary antibody; Pierce Chemical Co., Rockford, IL). For detection of KS protein, the membrane was incubated with 500-fold diluted rat KS antisera (primary antibody), followed by 4,000-fold diluted peroxidase-conjugated goat-anti-rat antisera (secondary antibody; Pierce). Protein was detected by enhanced chemiluminescence using the Super-signal pico kit (Pierce) and was visualized by autoradiography.
ent-Kaurene Analysis
Frozen tissues were ground in a chilled mortar and pestle and transferred to a solution of 80% (v/v) methanol (MeOH). Tissue was filtered and partitioned three times against n-hexane. The n-hexane fraction was evaporated at 12°C to a small volume, applied to a silica gel column, and eluted with n-hexane. Clear fractions were pooled and evaporated for GC-SIM analysis (Auto Mass, JEOL, Tokyo) using a capillary column DB-1 mass spectrometer (J&W Scientific, Folsom, CA). Oven temperature was set to 80°C for the first minute, followed by an increase of 30°C min–1 to 200°C, and then 5°C min–1 to 300°C. ent-Kaurene levels were determined by comparison with peak area of di-deuterated internal standard (257/259).
Quantitative Analysis of ent-Kaurenoic Acid
Lyophilized tissues (0.1–0.5 g dry weight) were extracted with 80% (v/v) aqueous MeOH and homogenized with a blender. ent-[17,17-2H2]kaurenoic acid (KA) was added to the extract as an internal standard. Extracts were evaporated in vacuo, and the acidic ethyl acetate-soluble fraction was obtained after solvent partitioning. Samples were then dissolved in n-hexane and were fractionated by silica gel chromatography. The eluate with n-hexane containing 5% (v/v) acidic ethyl acetate was collected as a fraction containing KA. The KA fraction was evaporated to dryness, dissolved in MeOH, and then purified on a C18 cartridge (Varian, Palo Alto, CA), which was eluted with 90% (v/v) aqueous MeOH. The eluate was evaporated under vacuum and then dissolved in MeOH. The sample was applied to a DEA cartridge (Varian) and eluted with MeOH containing 2% (v/v) acetic acid. The eluate was then purified by HPLC (System Gold, Beckman Instruments, Fullerton, CA) using a C18 column (Capcell Pak C18, 6 ϕ × 250 mm; Shiseido, Tokyo). Samples were injected in 20% (v/v) MeOH and were eluted with 20% (v/v) MeOH for 5 min, a 20% to 100% (v/v) gradient of MeOH in water containing 0.1% (v/v) acetic acid for 10 min, and then 100% (v/v) MeOH at a flow rate of 1 mL min–1. The fraction containing KA (21–22 min) was trimethylsilylated with N-methyl-N-trimethylsilyltrifluoroacetamide and then analyzed by GC-SIM. Oven temperature was held at 80°C for the 1st min, then increased to 250°C at 30°C min–1, followed by further increase to 320°C at 5°C minute –1. The content of KA was calculated from peak areas of mass-to-charge ratio 374 (cold KA) and mass-to-charge ratio 376 ([17, 17-2H2]KA).
GA Measurements
Purification and analysis was as described (Gawronska et al., 1995; Silverstone et al., 2001), except as noted. Lyophilized tissue (3 g dry weight per line per analysis) was immersed in 80% (v/v) MeOH and allowed to sit for 4 d at room temperature before extraction. Di-deuterated standards were added to 16.7 ng g–1 dry weight except for GA19 and GA24, for which 33.3 ng g–1 dry weight was added during the first analysis, and 16.7 ng g–1 dry weight was added in the second analysis. Before anion-exchange chromatography, samples were purified by C18 columns, eluted with 80% (v/v) MeOH, and evaporated to dryness. Fractions expected to contain GA4, GA9, GA15, GA24, and GA53 were subjected to additional purification by DEA column after methylation. For GC-SIM, oven temperature was set as described for KA analysis.
PAC Assays
PAC for plate assays was supplied by Zeneca (Richmond, CA). All PAC solutions contained 0.01% (v/v) Tween 20 to promote penetration of PAC through the seed coat. Control solutions for these experiments also contained 0.01% (v/v) Tween 20. For the hypocotyl growth assay, seeds were surface sterilized and cold treated for 2 d. Ten to 20 seeds per line were sown at d 0 on Murashige and Skoog plates containing each concentration of PAC. After 3 h of light exposure to promote germination, plates were double wrapped in foil, placed in boxes, and incubated at 22°C in darkness. Etiolated hypocotyls were measured on d 6. Seeds for flowering time were sown on 100- × 20-mm petri plates containing either Murashige and Skoog or Murashige and Skoog plus 1 μm PAC. Plates were incubated in growth chambers (22°C in LD), and the position of the plates was rotated daily to account for possible variations in light intensity. Flowering time was determined by the first appearance of flower buds.
Quantitative qPCR and RNA-Blot Analysis
RNA extraction from 3-week-old rosettes was as described (Ausebel et al., 1990), using phenol/chloroform extraction and LiCl precipitation. Additional RNA extractions for qPCR used the RNeasy Plant Kit (Qiagen USA, Valencia, CA), with on-column DNase treatment. For quantitative qPCR, 500 ng of total RNA was used per reaction; RNA was amplified using a LightCycler, with the LightCycler-RNA Amplification Kit, SYBR Green I (Roche Molecular Biochemicals, Mannheim, Germany). The amplification was performed using the manufacturer's suggested protocol, with 6 mm MgCl2 and 5 μm forward and reverse primers in a 10-μL reaction. Primers for amplification were designed with the aid of Roche LightCycler Probe Design Software, and sequence specificity was determined by BLAST search against The Arabidopsis Information Resource database (http://www.arabidopsis.org). The 224-bp KAO1 product was amplified with primers KAO1f (5′-CTGACTCCTTCACTCGC-3′) and KAO1r (5′-CCTGAGACGCTGTGTT-3′); the 228-bp KAO2 product was amplified with primers KAO2f (5′-TCCATTTGGACCCTGAAATC-3′) and KAO2r (5′-TGTGAGGCAAGAACATCACC-3′). Amplification was normalized to Act11 (At3G12110) as a standard using primers ActF (5′-TACCTCAGCAGAGCGT-3′) and ActR (5′-GAACAGAACCTCCGGG-3′) to generate a 184-bp product.
For RNA-blot analysis, poly(A+) RNA was purified from 0.5 to 1 mg of total RNA from the same samples used for qPCR. Purification was performed using the PolyATtract mRNA Isolation System (Promega, Madison, WI) according to the manufacturer's protocol. RNA gel-blot analysis was carried out using an antisense RNA probe (Yamaguchi et al., 1998a). A plasmid clone At2301 (Phillips et al., 1995) that contains a full-length cDNA for the AtGA20ox1 gene in pBluescript SK– was digested with EcoRI and HindIII. The excised 0.5-kb DNA fragment (corresponding to the first 0.5 kb of the AtGA20ox1 cDNA) was cloned into the EcoRI and HindIII sites of pBluescript SK– (pGA5-1). The AtGA20ox1 cDNA fragment was amplified by PCR from pGA5-1 using the forward and reverse M13 primers. This PCR product was used as a template to synthesize an antisense AtGA20ox1 RNA probe using T7 RNA polymerase. Loading of poly(A+) blots was normalized by hybridization with a random prime-labeled cyclophilin cDNA probe (Lippuner et al., 1994). All blots were exposed to a PhosphorImager screen for detection and quantification of signal (ImageQuant Software, Molecular Dynamics, Sunnyvale, CA).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank Andy Phillips (Long Ashton Research Station, University of Bristol, UK) for providing AtGA20ox1 cDNA clone. We are grateful to members of Tai-ping Sun's lab (Duke University, Durham, NC) and to Peter Hedden (IACR Long Ashton Research Station, Department of Agricultural Science, University of Bristol, Long Ashton, UK) for helpful discussions.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.021725.
This work was supported by the U.S. Department of Agriculture (grant no. 99–35304–8061) and by the National Science Foundation (grant no. INT–9603418).
References
- Ausebel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1990) Current Protocols in Molecular Biology. Green Publishing Associates/Wiley-Interscience, New York
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris 316: 1194–1199 [Google Scholar]
- Carrera E, Bou J, García-Martínez JL, Prat S (2000) Changes in GA 20-oxidase gene expression strongly affect stem length, tuber induction and tuber yield of potato plants. Plant J 22: 247–256 [DOI] [PubMed] [Google Scholar]
- Carrera E, Jackson SD, Prat S (1999) Feedback control and diurnal regulation of gibberellin 20-oxidase transcript levels in potato. Plant Physiol 119: 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington JC, Freed DD (1990) CAP-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J Virol 64: 1590–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang H-H, Hwang I, Goodman HM (1995) Isolation of the Arabidopsis GA4 locus. Plant Cell 7: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coles JP, Phillips AL, Crocker SJ, García-Lepe R, Lewis MJ, Hedden P (1999) Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J 17: 547–556 [DOI] [PubMed] [Google Scholar]
- Duncan JD, West CA (1981) Properties of kaurene synthetase from Marah macrocarpus endosperm: evidence for the participation of separate but interacting enzymes. Plant Physiol 68: 1128–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui H, Namori R, Koshimizu K, Yamazaki Y (1977) Plant growth regulators in seeds of Cucurbita pepo L.: II. Structures of gibberellins A39, A48, A49 and a new kaurenolide in Cucurbita pepo L. Agric Biol Chem 41: 181–187 [Google Scholar]
- Gawronska H, Yang Y-Y, Furukawa K, Kendrick RE, Takahashi N, Kamiya Y (1995) Effects of low irradiance stress on gibberellin levels in pea seedlings. Plant Cell Physiol 36: 1361–1367 [Google Scholar]
- Hedden P, Graebe JE (1981) Kaurenolide biosynthesis in a cell-free system from Cucurbita maxima seeds. Phytochemistry 20: 1011–1015 [Google Scholar]
- Hedden P, Kamiya Y (1997) Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol 48: 431–460 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Chandler PM, Poole A, Olive MR, Dennis ES, Peacock WJ (2001a) The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc Natl Acad Sci USA 98: 2065–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Poole A, Peacock WJ, Dennis ES (1999) Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol 119: 507–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Sullivan JA, Mould RM, Gray JC, Peacock J, Dennis ES (2001b) A plastid envelope location of Arabidopsis ent-kaurene oxidase links the plastid and endoplasmic reticulum steps of the gibberellin biosynthesis pathway. Plant J 28: 201–208 [DOI] [PubMed] [Google Scholar]
- Huang S, Raman AS, Ream JE, Fujiwara H, Cerny RE, Brown S (1998) Overexpression of 20-oxidase confers a gibberellin-overproducing phenotype in Arabidopsis. Plant Physiol 118: 773–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE (1993) Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaide H, Sassa T, Kamiya Y (2000) Functional analysis of the two interacting cyclase domains in ent-kaurene synthase for the fungus Phaeosphaeria sp. L487 and a comparison with cyclases from higher plants. J Biol Chem 275: 2276–2280 [DOI] [PubMed] [Google Scholar]
- Lippuner V, Chou IT, Scott SV, Ettinger WF, Theg SM, Gasser CS (1994) Cloning and characterization of chloroplast and cytosolic forms of cyclophilin from Arabidopsis thaliana. J Biol Chem 269: 7863–7868 [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Ukens S, Appleford NEJ, Lange T, Huttly A, Gaskin P, Graebe JE, Hedden P (1995) Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol 108: 1049–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher W (1991) Inhibitors of gibberellin biosynthesis: applications in agriculture and horticulture. In N Takahashi, BO Phinney, J MacMillan, eds, Gibberellins. Springer-Verlag, New York, pp 296–310
- Restrepo MA, Freed DD, Carrington JC (1990) Nuclear transport of plant potyviral proteins. Plant Cell 2: 987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustaiyan A, Sadjadi AS (1987) Kaurene derivatives from Alepidea amatynsia. Phytochemistry 28: 2106–2107 [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schomberg FM, Bizzell CM, Lee DJ, Zeevaart JAD, Amasino RA (2003) Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15: 151–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Chang C-w, Krol E, Sun T-p (1997a) Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J 12: 9–19 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T-p (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Jung H-S, Dill A, Kawaide H, Kamiya Y, Sun T-p (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Mak PYA, Casamitjana Martínez E, Sun T-p (1997b) The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146: 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MW, Yamaguchi S, Ait-Ali T, Kamiya Y (1998) The first step of gibberellin biosynthesis in pumpkin is catalyzed by at least two copalyl diphosphate synthases encoded by differentially regulated genes. Plant Physiol 118: 1411–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T-p, Goodman HM, Ausubel FM (1992) Cloning the Arabidopsis GA1 locus by genomic subtraction. Plant Cell 4: 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T-p, Kamiya Y (1994) The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6: 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Phinney BO, MacMillan J, editors (1991) Gibberellins. Springer-Verlag, New York
- Talón M, Koornneef M, Zeevaart JAD (1990) Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc Natl Acad Sci USA 87: 7983–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y-L, Li L, Gage DA, Zeevaart JAD (1999) Feedback regulation of GA5 expression and metabolic engineering of gibberellin levels in Arabidopsis. Plant Cell 11: 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y-L, Li L, Wu K, Peeters AJM, Gage DA, Zeevaart JAD (1995) The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc Natl Acad Sci USA 92: 6640–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Kamiya Y, Sun T-p (2001) Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J 28: 443–453 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RGS, Kamiya Y, Sun T-p (1998a) Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis Seeds. Plant Cell 10: 2115–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Sun T-p, Kawaide H, Kamiya Y (1998b) The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol 116: 1271–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]