Abstract
Previously we reported (R.T. Mullen, C.S. Lisenbee, J.A. Miernyk, R.N. Trelease [1999] Plant Cell 11: 2167–2185) that overexpressed ascorbate peroxidase (APX), a peroxisomal membrane protein, sorted indirectly to Bright Yellow-2 cell peroxisomes via a subdomain of the endoplasmic reticulum (ER; peroxisomal endoplasmic reticulum [pER]). More recently, a pER-like compartment also was identified in pumpkin (Cucurbita pepo) and transformed Arabidopsis cells (K. Nito, K. Yamaguchi, M. Kondo, M. Hayashi, M. Nishimura [2001] Plant Cell Physiol 42: 20–27). Here, we characterize more extensively the localization of endogenous Arabidopsis peroxisomal APX (AtAPX) in cultured wild-type Arabidopsis cells (Arabidopsis var. Landsberg erecta). AtAPX was detected in peroxisomes, but not in an ER subcompartment, using immunofluorescence microscopy. However, AtAPX was detected readily with immunoblots in both peroxisomal and ER fractions recovered from sucrose (Suc) density gradients. Most AtAPX in microsomes (200,000g, 1 h pellet) applied to gradients exhibited a Mg2+-induced shift from a distribution throughout gradients (approximately 18%–40% [w/w] Suc) to ≥42% (w/w) Suc regions of gradients, including pellets, indicative of localization in rough ER vesicles. Immunogold electron microscopy of the latter fractions verified these findings. Further analyses of peroxisomal and rough ER vesicle fractions revealed that AtAPX in both fractions was similarly associated with and located mostly on the cytosolic face of the membranes. Thus, at the steady state, endogenous peroxisomal AtAPX resides at different levels in rough ER and peroxisomes. Collectively, these findings show that rather than being a transiently induced sorting compartment formed in response to overexpressed peroxisomal APX, portions of rough ER (pER) in wild-type cells serve as a constitutive sorting compartment likely involved in posttranslational routing of constitutively synthesized peroxisomal APX.
Peroxisomes are single-membrane-bound organelles that in plants participate in major tissue-specific functions such as fatty acid β-oxidation (oilseed cotyledons), photorespiration (mature leaves), and ureide metabolism (root nodules; Olsen, 1998; Reumann, 2000). Reactive oxygen byproducts of these processes are removed in part by a membrane-associated ascorbate peroxidase (APX) that cycles electrons through ascorbate to effectively remove hydrogen peroxide and regenerate NAD+ (del Río et al., 1998; Corpas et al., 2001; Donaldson, 2002). A peroxisomal isoform of APX has been identified and characterized partially in several different plant species including Arabidopsis (Zhang et al., 1997; Wang et al., 1999), barley (Hordeum vulgare; Shi et al., 2001), cotton (Gossypium hirsutum; Bunkelmann and Trelease, 1996; Mullen et al., 2001b), cucumber (Cucumis sativus; Corpas et al., 1994; Corpas and Trelease, 1998), pea (Pisum sativum; Jiménez et al., 1997; López-Huertas et al., 1999), pumpkin (Cucurbita pepo; Yamaguchi et al., 1995a, 1995b; Nito et al., 2001), and spinach (Spinacia oleracea; Ishikawa et al., 1998). Collectively, these studies indicate that peroxisomal APX functions as a singlepass peroxisomal membrane protein (PMP) with its N-terminal active site domain facing the cytosol, where it is thought to scavenge hydrogen peroxide that escapes the peroxisome or that is produced in the cytosol.
Arabidopsis possesses at least five different isoforms of APX, with two located in the cytosol, two in the chloroplast, and one in the peroxisomal membrane (Jespersen et al., 1997; Shigeoka et al., 2002). Zhang et al. (1997) identified the latter in a yeast two-hybrid system as a candidate 14-3-3-interacting protein and called it APX3; hereafter in this paper, it is referred to as Arabidopsis peroxisomal APX (AtAPX). The corresponding single-copy gene coded for a predicted protein with 85% of its amino acid sequence identical to cotton peroxisomal APX, including the unique C-terminal region containing a putative transmembrane domain (TMD; Bunkelmann and Trelease, 1996). Zhang et al. (1997) determined that peroxisomal AtAPX transcript levels increased slightly in response to various chemical and environmental stresses, and Wang et al. (1999) found that tobacco (Nicotiana tabacum) plants overexpressing peroxisomal AtAPX were protected specifically against peroxisome-derived oxidative stress. Although these and another study (Shi et al., 2001) provide important functional information for peroxisomal AtAPX in planta, cell biological aspects related to its posttranslational intracellular sorting and association with organelle membranes have not been elucidated.
A growing body of evidence from recent studies mostly of yeast and some on human and plant PMPs have implicated the endoplasmic reticulum (ER) as a normal sorting compartment in an indirect sorting pathway to peroxisomes (Titorenko and Rachubinski, 2001a, 2001b; Trelease, 2002). For example, the N-terminal 16 amino acids of the Hansenula polymorpha peroxisomal biogenesis protein, or peroxin (Pex), Pex3p, were sufficient for sorting a reporter protein to ER (Baerends et al., 1996). Similarly, proliferation of ER membranes was observed in mammalian cells overexpressing the human ortholog of Pex3p (Kammerer et al., 1998), as well as in Brewer's yeast (Saccharomyces cerevisiae) overexpressing Pex15p (Elgersma et al., 1997). Salomons et al. (1997) also showed that peroxisomal proteins accumulated within the ER in H. polymorpha cells treated with brefeldin A (BFA). That BFA interferes with the formation of coated vesicles at ER and Golgi membranes (Nebenführ et al., 2002) suggests the involvement of vesicle transport in peroxisomal sorting and/or biogenesis. In support of this hypothesis, the AAA-ATPases Pex1p and Pex6p from Pichia pastoris were localized in cell fractionation experiments to vesicles that were distinct from peroxisomes (Faber et al., 1998), and Just and coworkers demonstrated that rat liver Pex11p may promote vesiculation of peroxisomes by recruiting ADP-ribosylation factor and coatomer (Passreiter et al., 1998; Anton et al., 2000). Although similar experimental data are not yet available for plant proteins, a set of 15 Arabidopsis Pex orthologs, including Pex1p, Pex3p, Pex6p, and Pex11p, have been identified through recent comparisons of human, mouse, and yeast Pex sequences with Arabidopsis sequences (Baker et al., 2000; Mullen et al., 2001a; Charlton and López-Huertas, 2002). The majority of these orthologs are predicted PMPs that function as receptor, docking, and/or proliferation factors for matrix and membrane protein import and organelle biogenesis (Charlton and López-Huertas, 2002).
Cottonseed peroxisomal APX, a non-Pex PMP, is the only plant PMP that has been shown to sort indirectly to peroxisomes (Mullen et al., 1999). In that study, in vitro translated peroxisomal APX inserted posttranslationally into maize (Zea mays) rough ER membranes in an ATP-dependent but signal recognition particle-independent manner. Furthermore, transiently expressed, epitope-tagged peroxisomal APX sorted in vivo to a reticular/circular compartment that colocalized with a portion of ER that was stained with the general ER stain 3,3′-dihexyloxacarbocyanine iodide. In the presence of BFA, the transiently expressed peroxisomal APX accumulated within the reticular/circular compartment, and after removal of BFA, the APX appeared within peroxisomes. Collectively, these results were interpreted to indicate that peroxisomal APX was synthesized in the cytosol on unbound polysomes and then sorted indirectly to peroxisomes by first traveling to a subdomain of the ER, which was named peroxisomal ER (pER). Later, Mullen and Trelease (2000) showed that targeting of transiently expressed peroxisomal APX to pER and peroxisomes was dependent upon its C-terminal TMD and adjacent basic amino acid residues, cluster of which together constituted a membrane peroxisomal targeting signal.
In a study aimed at analyzing the subcellular localization of pumpkin peroxisomal APX, Nito et al. (2001) found that the pumpkin protein did not comigrate in Suc gradients with markers of pumpkin cotyledon rough ER. In Arabidopsis plants stably transformed with the pumpkin peroxisomal APX, they localized with immunogold microscopy the expressed protein in the boundary membranes of cotyledon peroxisomes and in unidentified smooth-surfaced membranous structures, but not in rough ER or any other common organelle in the cells. In a separate immunofluorescence study with cultured Bright Yellow-2 (BY-2) cells, Mullen et al. (2001b) found that stably expressed cottonseed peroxisomal APX was localized to a diffuse membranous compartment that resembled reticular/circular pER. Authors of both studies suggested that the non-peroxisomal structures were smooth-surfaced vesicles either that were derived from the main ER or that were actually subdomain(s) of the main ER, i.e. pER.
Mullen et al. (2001b) also reported temporal, expression-induced morphological changes of pER in cultured BY-2 cells, i.e. a shift from reticular to more circular pER, and proposed that this phenomenon represented an artifact of protein overexpression. Lisenbee et al. (2003) concurred with this suggestion. They found that the circular portion of the fluorescence image was a consequence of oligomerization (zippering) of overexpressed green fluorescent protein-APX protruding from aggregated organelles, and they concluded that authentic pER is composed only of a reticular compartment. Gong et al. (1996) and Wright et al. (1988) described aberrant proliferations of ER membranes (karmellae) under similar overexpression conditions. Persons such as South and Gould (1999) and South et al. (2000, 2001), who were unable to provide experimental evidence for the involvement of ER in peroxisomal biogenesis, cautioned against interpretations made from studies based on the sorting of overexpressed proteins. Thus, consideration of rough ER or portions thereof (e.g. pER) as an authentic sorting compartment for PMPs must confront potential artifacts in transformed cells. In the current study, we apply a combined microscopic and cell fractionation approach to examine in wild-type (non-transformed) Arabidopsis cells the biochemical association, topology, and subcellular localizations of endogenous AtAPX.
RESULTS
Immunofluorescence Localization of Peroxisomal APX in Arabidopsis Cells
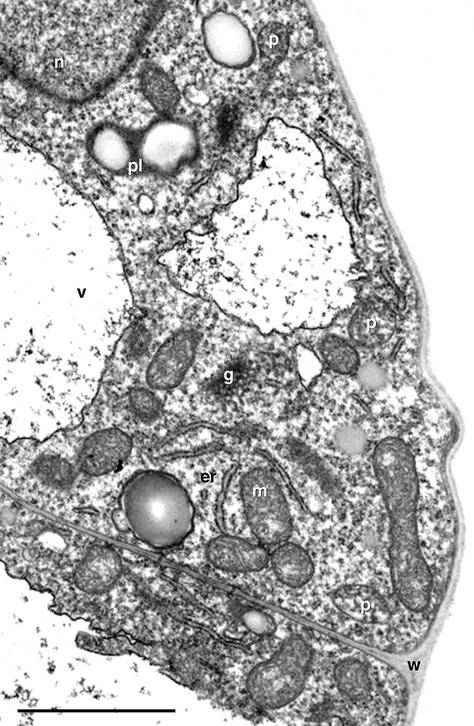
Figure 1 shows the ultrastructure of organelles that are prevalent in wild-type Arabidopsis suspension cultured cells. These cells house all typical plant organelles including abundant peroxisomes and rough ER sheets (shown in profile view), and unbound polysomes occupy much of the cytoplasmic area. Peroxisomes appear in section view as circular (approximately 0.5 μm in diameter) or elongate organelles with a granular, noncrystalline matrix that is delineated by a single boundary membrane.
Figure 1.
Electron micrograph of a portion of an Arabidopsis suspension cell illustrating profiles of rough ER (er), peroxisomes (p), mitochondria (m), plastids with starch grains (pl), and Golgi bodies (g) dispersed throughout the cytoplasm. n, Nucleus; v, vacuole; w, cell wall. Bar = 2 μm.
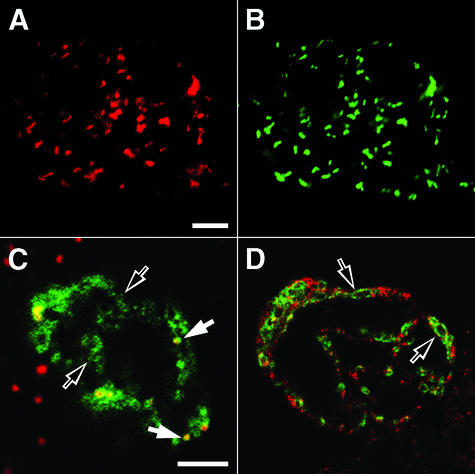
Figure 2 is a grouping of immunofluorescence images showing the localization of endogenous and overexpressed peroxisomal APX within representative wild-type and transiently transformed cells, respectively. The clumped group of wild-type cells in Figure 2A displays numerous rod-shaped fluorescence images within their cytoplasm attributable to application of anti-cucumber peroxisomal APX and rhodamine-conjugated secondary antibodies. The same pattern of rod-shaped organelles was observed in the same cells dual-labeled with anti-tobacco catalase and cyanine (Cy) 2-conjugated secondary antibodies (Fig. 2B). Comparison of the two images convincingly shows that the anti-cucumber peroxisomal APX antibodies recognize endogenous AtAPX within pre-existing peroxisomes. This selective labeling of peroxisomes was consistent regardless of cell culture age, primary antibody preparation (crude serum or affinity-purified IgGs), secondary antibody conjugates, or concentration and length of antibody applications. Control experiments included applying preimmune serum in place of the primary IgGs, omitting primary IgGs, and adding irrelevant IgGs. In all cases, the rod-shaped structures did not exhibit an immunofluorescence signal (representative images not shown). We conclude that at steady state, endogenous peroxisomal AtAPX is detectable by immunofluorescence microscopy within pre-existing peroxisomes, but not within any sort of preperoxisomal compartment. Implicit in this conclusion is the important observation that representative images such as that shown in Figure 2A do not include labeling of membranous reticular structures indicative of the pER observed in hemagglutinin epitope-tagged APX (HA-APX) transformed BY-2 cells (Mullen et al., 1999). The confocal image presented in Figure 2C shows a HA-APX-transformed Arabidopsis cell dual labeled with anti-HA (green) and anti-catalase (red) antibodies. Overexpressed HA-APX is localized to punctate and reticular structures. Only the punctate portion of HA-APX is colocalized with endogenous catalase in peroxisomes (yellow/orange color). In a different transformed cell (Fig. 2D), neither the punctate nor the reticular HA-APX fluorescence (green) is colocalized with the endogenous ER marker protein calnexin (red). These results are consistent with the localizations of overexpressed peroxisomal APX reported for BY-2 cells (Mullen et al., 1999). This is particularly significant because the comparisons reveal that transformed Arabidopsis cells also possess the reticular compartment identified as pER.
Figure 2.
Cucumber peroxisomal APX IgGs recognize endogenous peroxisomal AtAPX in Arabidopsis peroxisomes but not ER, whereas transiently expressed HA-APX sorts to peroxisomes and reticular pER. Wild-type Arabidopsis cells were formaldehyde-fixed, perforated with pectolyase and cellulase, and permeabilized with Triton X-100 before immunolabeling for endogenous peroxisomal and ER marker proteins. In separate experiments, wild-type cells were bombarded with a hemagglutinin epitope-tagged HA-APX and allowed to express the transgene for 20 h before formaldehyde fixation. A and B, Fluorescence images of four clumped wild-type cells dual labeled for endogenous peroxisomal APX (rabbit anti-peroxisomal APX IgGs; A) and catalase (mouse anti-catalase hybridoma medium; B). C, Merged confocal optical sections of a cell transiently expressing HA-APX (green) and exhibiting both punctate (white arrows) and reticular (black arrows) fluorescence patterns attributable to binding of mouse anti-HA epitope antibodies. Expressed HA-APX colocalizes (yellow) with endogenous catalase (rabbit anti-catalase IgGs) in peroxisomes (red). D, In a separate HA-APX-transformed cell, the green reticular fluorescence (black arrows) attributable to expressed HA-APX does not colocalize with the red reticular fluorescence of endogenous ER calnexin (rabbit anti-calnexin serum). Bars = 5 μm.
Separation of Peroxisomal AtAPX in Suc Density Gradients
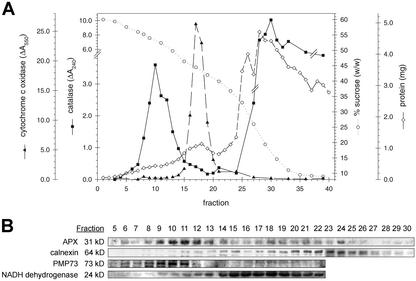
Cell fractionation experiments were performed to test the hypothesis derived from our immunofluorescence studies that peroxisomal AtAPX resides within peroxisomes and possibly in another membrane compartment in wild-type cells. Figure 3A shows representative results of organelles and vesicles separated in a linear Suc density-gradient experiment. The applied sample was a low-speed (1,500g, 15 min) supernatant that contained intact organelles (except nuclei and plastids) and organelle-derived membrane vesicles (e.g. from Golgi, ER, etc.). Peroxisomes equilibrated at approximately 1.25 g mL–1 (fraction 10) as evidenced by peak catalase activity, whereas mitochondria equilibrated at approximately 1.20 g mL–1 (fraction 17) as shown by peak cytochrome c oxidase activity. The greatest amount of protein accumulated in fractions at the top of the gradient (fractions 25 and higher).
Figure 3.
Peak levels of endogenous peroxisomal AtAPX protein segregate in Suc density gradient (30%–59% [w/w]) fractions with peak peroxisomal catalase activity and PMP73 protein, whereas other AtAPX protein partially co-exists with calnexin-marked ER in and on top of the gradient. A, The representative gradient profile shows protein content, Suc concentration (percentage w/w), and activities per fraction of peroxisomal catalase and mitochondrial cytochrome c oxidase. To calculate micromoles per minute per fraction, divide ΔA per minute values (ordinates) by 0.036 or by 21 for catalase or cytochrome c oxidase, respectively. B, Immunoblot analyses (chemiluminescence) of trichloro-acetate (TCA)-precipitated gradient fractions (expanded scale; 25 μg of protein) comparing the distributions of endogenous peroxisomal AtAPX protein with ER calnexin, peroxisomal PMP73, and mitochondrial NADH dehydrogenase proteins.
Figure 3B presents immunoblot analyses of the same gradient fractions. As predicted, anti-cucumber peroxisomal APX antibodies recognized peak levels of 31-kD polypeptides within the same fractions that exhibited peak peroxisomal catalase enzyme activities and peroxisomal PMP73 protein (Corpas et al., 1994). These were well separated from the fractions possessing mitochondrial NADH dehydrogenase protein (Fig. 3B). Pooled portions of these fractions were prepared for electron microscopy. Fractions 9 through 12 were highly enriched with peroxisomes, and fractions 15 through 19 were purified mitochondria (micrographs not shown). Although some AtAPX protein was detected in most gradient fractions, higher levels consistently were detected at the top of the gradients in the region around fraction 24 (approximately 1.16 g mL–1). The relative amount of AtAPX protein in these fractions at the top of the gradient actually is higher than that shown because the protein content is substantially higher in these fractions and the immunoblots were prepared from a constant amount of protein (25 μg protein) per lane. The ER marker calnexin also migrated into the gradient, but was at relatively low levels in the peroxisomal regions. As expected, high levels, especially when considering the high protein content, were in the fractions just into and at the top of the gradient where microsomes typically occur in these gradients. In this region, there is a partial co-existence of AtAPX and calnexin, which suggests that the two proteins exist in some different and some similar microsomal vesicles. The near absence of calnexin protein in the peroxisomal region indicates that the AtAPX observed in this region was in the peroxisomes, not in contaminating ER vesicles. In summary, the results of these cell fractionation experiments demonstrate that at steady state, endogenous peroxisomal AtAPX polypeptides exist in both peroxisomes and ER (vesicles).
Mg2+-Induced Shift Separation and Immunogold Localization of Peroxisomal AtAPX within Rough ER Vesicles
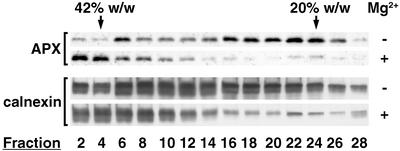
The results described above prompted experiments to examine the apparent association of peroxisomal AtAPX specifically with rough ER vesicles. The Mg2+-dependent association of ribosomes with ER membranes results in the migration of rough vesicles to greater densities in Suc gradients containing Mg2+ (Lord, 1983). Figure 4 shows immunoblot data for gradient fractions (with/without Mg2+) probed with antibodies against peroxisomal APX and calnexin. When microsomal membranes were stripped of bound ribosomes with 2 mm EDTA, both peroxisomal AtAPX and calnexin were found in most of the gradient fractions, although AtAPX was more abundant in fraction six (39% [w/w] Suc) and in fractions 16 to 24 (28%–20% [w/w] Suc). When ribosomemembrane associations were maintained with 5 mm MgCl2, most of the AtAPX shifted with calnexin to heavier densities in fractions two to four (≥42% [w/w] Suc). A pellet collected from the bottom of the tubes also possessed considerable AtAPX and calnexin immunoreactivity (data not shown). Interestingly, in all replicate experiments with added Mg2+, a small portion of AtAPX consistently remained in the 20% (w/w) region of the gradients (shown in Fig. 4). Separate experiments employing longer centrifugation times (up to 4 h; data not shown) showed that AtAPX was at equilibrium in both regions of the gradients, i.e. not all of the AtAPX in the bottom of the gradient pelleted out of the 42% region, nor did the AtAPX in the 20% region migrate further into the gradient. Collectively, these data indicate that after cellular disruption, the non-peroxisomal portion of peroxisomal AtAPX resides mostly within rough ER vesicles (≥42% [w/w]) and partly within unidentified structures in the approximately 20% fractions (electron microscopy shown below).
Figure 4.
The majority of microsomal AtAPX exhibits a Mg2+-induced shift, as does calnexin in rough ER vesicles. Arabidopsis suspension cells were ruptured in homogenization medium containing either 5 mm MgCl2 (+Mg2+) or 2 mm EDTA (–Mg2+). Microsomes (200,000g, 60 min) were applied to 15% to 45% (w/w) linear Suc gradients containing 5 mm MgCl2 or 2 mm EDTA. Gradient fractions (0.5 mL) were analyzed for protein content and then were TCA-precipitated for application to SDS gels (25 μg protein lane–1). Immunoblot analyses (chemiluminescence) for AtAPX and calnexin were performed as in Figure 3B.
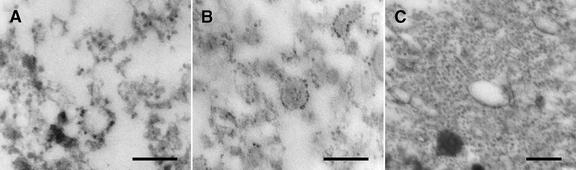
Three different samples with peroxisomal AtAPX immunoblot reactivity that were derived from MgCl2 gradient experiments were prepared for electron microscopy (Fig. 5). The materials in these fractions were collected by centrifugation onto filters, and these samples were sectioned perpendicular to the filter so that we could assess any obvious stratification of materials within the samples. Both the pellet collected from the bottom of the gradient tubes (Fig. 5A) and the 42% region (Fig. 5B) were composed almost entirely of ribosome-studded membranes that varied only in the relative abundance of ribosomes bound to the vesicles. The vesicles in the pellet sample exhibited a greater abundance of ribosomes than the vesicles at the top of the 42% sample (comparative data not calculated). In contrast, the 20% region of the gradient (Fig. 5C) was heterogeneous, consisting mostly of unbound ribosomes (polysomes?), smooth membrane vesicles, and unidentified osmiophilic bodies.
Figure 5.
Electron microscopy reveals that the fractions containing Mg2+-shifted AtAPX consist almost entirely of rough ER vesicles. A, Rough vesicles collected from the pellet fraction at the bottom of the tubes (below the formed gradients). B, Rough vesicles collected from the 42% region of the gradients. C, Unbound ribosomes (polysomes?), smooth vesicles, and osmiophilic bodies collected from the 20% region of the gradients. Bars = 0.2 μm.
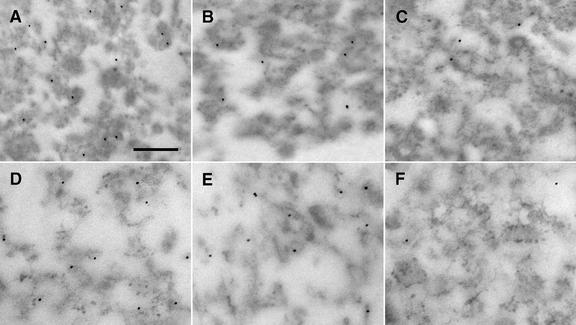
Immunogold microscopic analyses were made of nonosmicated portions of the three samples represented in Figure 5. Figure 6 shows representative immunogold electron micrographs that were obtained with the pellet (top row) and 42% (w/w) Suc (bottom row) samples. Table I presents the results of gold particle counts normalized and averaged per square micrometer sectional area for each of the immunogold analyses represented in Figure 6. Two controls were employed. In one case, preimmune serum was used instead of the primary antibodies (Fig. 6, C and F), and in the other case, the primary antibodies were omitted (images not shown). The micrographs shown in Figure 6, C and F, actually are not representative; rather, they were selected to show at least some of the few bound gold particles to confirm addition of the gold-conjugated antibody to the sections. As apparent in Figure 6, A, B, D, and E, virtually all of the gold particles were bound to the membranes/ribosomes of the rough vesicles, not within the vesicles or not to non-vesicle areas of the sections. Accordingly, only those Protein A-gold particles observed over vesicular material were scored and included in the values for AtAPX and calnexin binding presented in Table I. Both peroxisomal AtAPX (e.g. Fig. 6, A and D) and calnexin (e.g. Fig. 6, B and E) were detected in the sections at levels significantly greater than the nonspecific binding observed in control samples. Gold particle binding to AtAPX was higher than binding to calnexin. Attempts were made in dual labeling experiments (10 and 20 nm Protein A-gold particles) to ascertain whether calnexin and AtAPX were localized in the same rough microsomes. Unfortunately, the results were ambiguous, and this question remains unanswered. Nonosmicated samples from the 20% region of the gradient (e.g. Fig. 5C) also were probed with anti-APX IgGs and Protein A-gold, but the frequency of gold particle binding was not significantly greater in any portion of the heterogeneous sample than in control samples. Thus, the site of peroxisomal AtAPX localization in the 20% sample is not specifically known.
Figure 6.
Peroxisomal AtAPX and calnexin are detected via immunogold microscopy in rough ER vesicles collected from Mg2+-shifted gradient fractions. A through C, Nonosmicated pellet sample. D through F, Nonosmicated 42% region. Rough vesicles were decorated by 10-nm gold particles marking sites of section-exposed AtAPX (A and D) and calnexin (B and E). C and F, Negative controls in which sections were probed with preimmune serum and gold-conjugated Protein A. Bar = 0.2 μm.
Table I.
Frequency of Protein A-gold particle binding to antigens in thin sections of rough ER vesicles in gradient fractions similar to those shown in Figure 6
| Antigen | Particles/μm2a |
|---|---|
| Pellet | |
| AtAPX | 9.8 ± 3.2b |
| Calnexin | 4.3 ± 1.4 |
| Preimmune serum | 1.5 ± 0.8 |
| Omit primary IgGs | 0.4 ± 0.2 |
| 42% Region | |
| AtAPX | 14.9 ± 3.2 |
| Calnexin | 8.5 ± 3.7 |
| Preimmune serum | 0.9 ± 0.4 |
Section surface area. b Mean number of gold particles ± SD.
Membrane Association and Topology of Peroxisomal AtAPX
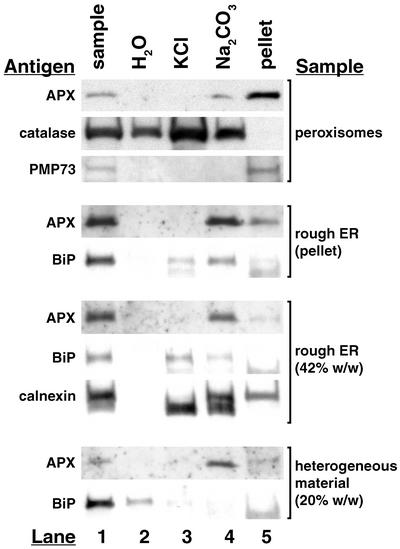
The various vesicle fractions (shown in Fig. 5), including peroxisomes, were compared in examinations of the association of peroxisomal AtAPX with the vesicle/organelle membranes. The results are shown in Figure 7. Lane 1 documents the presence of the various antigens in each of the four untreated samples. Interestingly, binding protein (BiP) was present in the samples taken from the 20% region of the gradient, suggesting that ER or ER-derived transport vesicles were in this sample. Lane 2 shows that as expected, peroxisomal AtAPX was not solubilized in water (buffer) in any of the samples subjected to aqueous hypotonic treatment. As controls, it was expected that most of the matrix catalase would have been released into the water upon bursting of the peroxisomes, and that BiP would have been released from the lumen of the rough ER vesicles. The results show, however, that these were not the cases. Only some of the catalase was released upon bursting in water (lane 2) and more was released upon subsequent treatments of the peroxisomes with KCl (lane 3) and alkaline carbonate (lane 4). No catalase remained in the final pellet as expected (lane 5). The reason for the sequential solubilization is not clear, although catalase is known to aggregate and therefore may not have been readily solubilized. The aqueous solubilization of BiP from the 20% sample but not from rough-surfaced vesicles in the pellet and in the 42% samples may reflect greater stability of the rough-surfaced vesicles compared with the BiP-containing structures in the 20% sample (smooth vesicles?). BiP was not found in any of the final pellets as expected (Lane 5).
Figure 7.
Peroxisomal AtAPX is similarly associated with membranes of isolated peroxisomes, rough ER vesicles, and membranes within the 20% sample. Untreated samples (lane 1) were subjected to hypotonic shock in water (lane 2) and then treated sequentially with 0.2 m KCl (lane 3) and alkaline (pH 11.5) sodium carbonate (lane 4), leaving a final pellet solubilized in dodecyl-β-d-maltoside detergent (lane 5). The extracted proteins were TCA-precipitated and then subjected to SDS-PAGE (15 μg of protein) and analyzed on blots (chemiluminescence) using antibodies specific for peroxisomal APX and positive control marker proteins.
Data shown in lane 3 of Figure 7 indicate that peroxisomal AtAPX was not bound ionically to membranes of any of the samples (not solubilized in KCl). The anti-calnexin antibody that was used in this study also recognizes lumenal calreticulin (50 kD, lane 3) and the membrane-bound calnexin (64 kD, lane 1; Coughlan et al., 1997). Thus, lumenal calreticulin and BiP were released from KCl-treated rough ER vesicles in the 42% sample as was BiP from the rough ER vesicles in the pellet. Anti-calnexin antibodies were not used for the pellet fraction. Lane 4 shows that peroxisomal AtAPX was solubilized to greater or lesser degrees in all samples treated with alkaline carbonate. Lane 5 shows that AtAPX remained associated with the final membrane pellets derived from peroxisomes (like peroxisomal membrane PMP73) and the rough ER vesicle samples (like calnexin in the 42% sample). The similar behaviors of membrane-associated peroxisomal AtAPX in all four samples provide evidence that some biogenetic relationship exists among the various membranes.
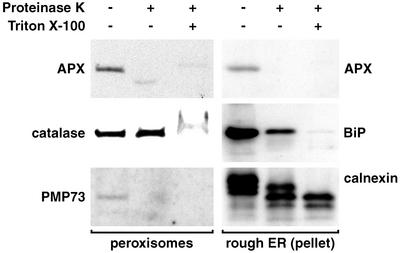
Figure 8 shows immunoblot results of protease digestion experiments designed to elucidate the topological orientation of peroxisomal AtAPX in peroxisomal and rough ER vesicle membranes. In the absence of Triton X-100, exposed peroxisomal AtAPX in replicate experiments was digested in both peroxisomal and rough ER membrane samples, as was membrane-bound PMP73 (positive PMP control) in peroxisomes. As soluble protein controls, catalase and lumenal BiP in peroxisomes and ER vesicles, respectively, were protease-protected until membranes were detergent-solubilized. Membrane-bound calnexin (top band, lane 1, 64 kD) was digested only partially without added detergent (top band, lane 2, 59 kD) and then was digested completely after addition of detergent (lane 3). This behavior is in accordance with that of Arabidopsis calnexin described by Huang et al. (1993). The data indicate that a majority of membrane-bound peroxisomal AtAPX is on the outside (i.e. cytosolic face) of peroxisomes and rough ER vesicles, whereas calnexin faces the ER lumen as was expected.
Figure 8.
Peroxisomal AtAPX resides on the cytosolic face of peroxisomes and rough ER vesicles. Intact peroxisomes and rough ER vesicles from the pellet fraction (Fig. 5A) were treated with proteinase K (4:1 [w/w] sample protein:proteinase K) with (+) and without (–) presolubilization in Triton X-100. Treated samples then were TCA-precipitated and subjected to SDS-PAGE (15 μg of protein) and immunoblot (chemiluminescence) analyses. Proteinase K and Triton X-100 were omitted from control reactions.
DISCUSSION
An important intent of this study was to determine whether in Arabidopsis wild-type cells rough ER and/or a portion (subdomain) thereof served as a sorting compartment for endogenous peroxisomal AtAPX. Suspension-cultured Arabidopsis cells allowed direct comparisons with our previous work on sorting of overexpressed cottonseed peroxisomal APX in BY-2 cells (Mullen et al., 1999; Mullen and Trelease, 2000; Mullen et al., 2001b). Our antibodies, raised against cucumber peroxisomal APX, cross-reacted specifically with peroxisomal AtAPX in Arabidopsis cells. The data presented in Figures 2, A and B, 3, 7, and 8 collectively provide the first demonstration that an endogenous peroxisomal AtAPX is associated with the boundary membrane of peroxisomes in Arabidopsis cells. Nito et al. (2001) localized overexpressed pumpkin APX in Arabidopsis peroxisomes, not the endogenous AtAPX. We conclude that at steady state, endogenous peroxisomal AtAPX is a membrane-associated, mostly cytosolic-facing protein that is present in both peroxisomes and a portion of rough ER. Whether a pER subdomain of rough ER is the actual sorting compartment is discussed next.
Peroxisomal AtAPX Is Localized to Peroxisomes and ER-Derived Vesicle Membranes in Wild-Type Cells
Nito et al. (2001) presented data similar to our Figure 3 showing that pumpkin peroxisomal APX and BiP occur together in fractions collected from the top of Suc gradients. From time-course results, they concluded that pumpkin peroxisomal APX was transported to glyoxysomes via ER during postgerminative seedling growth. Their conclusion, however, is couched with other experimental results that differ from ours. Pumpkin peroxisomal APX did not exhibit a Mg2+-induced shift as did BiP; instead it shifted to less-dense (approximately 29% [w/w]) Suc, indicating that the APX was localized in membranous structures other than rough ER vesicles. In situ immunogold microscopy of wild-type cotyledons did not reveal the identity of the structures, whereas examination of pumpkin-APX-transformed Arabidopsis plants revealed overexpressed protein in glyoxysomal membranes and an unidentified smooth-surfaced membrane structure, but not in rough ER. The unidentified structure was considered to be smooth ER or specialized subdomains of ER similar to pER previously described in BY-2 cells by us (Mullen et al., 1999). In cultured Arabidopsis cells (Fig. 2, C and D), reticular pER subdomains also were observed only when peroxisomal APX was overexpressed. Thus, the results from Nishimura's group with pumpkin and Arabidopsis plants and ours with cultured BY-2 and Arabidopsis cells generally are in agreement in terms of ER (subdomain) involvement in the sorting of peroxisomal membrane APX to pre-existing peroxisomes. This agreement persists even with the apparent discrepancy in subcellular localization of peroxisomal APX in rough/smooth ER vesicles.
The Mg2+-induced shift data presented in Figure 4 are representative of results obtained from five separate experiments to ensure the reliability of the results. The Mg2+-induced shift of calnexin is similar to those described by Matsuoka et al. (1997) with cultured tobacco cells and by Lu and Hrabak (2002) with Arabidopsis plants, indicating that the rough ER vesicle shift is not an anomaly related to the cultured Arabidopsis cells. Representative electron micrographs of the two regions where AtAPX shifted (pellet and 42%, Fig. 5) show that the material in both are composed of similar, highly purified rough ER vesicles. These observations are in line with the observed Mg2+-induced shift of vesicles with bound ribosomes (polysomes).
Just as consistently, a small portion of the AtAPX remained in the 20% (w/w) Suc region. We wondered whether this AtAPX was in the same structure(s) as was pumpkin APX that was recovered in the 29% region of the Mg2+ gradients (Nito et al., 2001). However, the AtAPX did not exhibit a shift to the 20% region, whereas the pumpkin APX shifted from 35% to 29% (w/w) Suc. The vesicles observed in the pooled 20% region (Fig. 5C) appeared to be smooth surfaced, and the ribosomes appeared to be clustered into polysomes not bound to membrane vesicles. These were distinctly different from the material in the more homogeneous pellet and 42% region (Fig. 5, A and B). Unbound polysomes are prevalent in the cytosol of Arabidopsis cells (Fig. 1). Thus, the vesicles in the 20% sample would not be expected to undergo a Mg2+-induced shift. It is apparent that insufficient comparative microscopic/biochemical data are available to decide whether the non-rough ER, APX-containing structures are the same in Arabidopsis and pumpkin.
Immunogold microscopy of peroxisomal AtAPX and calnexin showed that these proteins were located, albeit separately, in the membranes of the rough ER vesicles found in the pellets and 42% regions (Fig. 6). Protein A-gold binding per square micrometer section area was higher for APX than calnexin in both fractions (Table I), but this comparison likely is not meaningful. The frequency of gold binding was lower than was expected but convincing for localization of the two antigens in these isolated rough microsomes. Unfortunately, however, the low frequency precluded analyses of the data to decide whether calnexin and AtAPX were in the same vesicles, and/or whether peroxisomal AtAPX was in a subdomain of the main ER (pER?). The AtAPX apparently is in relatively low abundance in the cell cytoplasm, which confirms the stated conclusion of Nito et al. (2001) for pumpkin and explains our inability to detect at the steady-state endogenous peroxisomal AtAPX in ER with immunofluorescence microscopy (Fig. 2, A and B) and with electron immunogold microscopy of sectioned cells (data not shown). The cell fractionation procedure and Mg2+ shift served to concentrate the membranous structures with peroxisomal AtAPX.
Peroxisomal AtAPX Is Situated Similarly in Peroxisomal and ER Vesicle Membranes
The data presented in Figure 7 demonstrate that peroxisomal AtAPX was stably associated with the KCl-washed membranes isolated from peroxisomes, rough ER vesicles, and the heterogeneous material. Washing of each of these fractions with alkaline carbonate released at least a portion (peroxisomes and rough ER vesicles), and sometimes all (heterogeneous 20% [w/w] Suc material), of the bound AtAPX. Alkaline carbonate treatment also released portions of APX from peroxisomal membranes of several other plant species (Corpas et al., 1994; Yamaguchi et al., 1995b; Bunkelmann and Trelease, 1996). This behavior is surprising because the results suggest that the protein is not integrally associated with membranes (Fujiki et al., 1982), yet peroxisomal APX proteins are believed to be tail-anchored proteins (Zhang et al., 1997; Mullen and Trelease, 2000, and refs. given below). Thus, we hesitate to conclude from these observations that peroxisomal AtAPX is a peripheral, rather than integral, component of peroxisomes and ER vesicles. Instead, we suggest that at least for this membrane protein, solubilization in alkaline carbonate does not indicate a peripheral membrane association.
Support for our supposition comes from studies with yeast PMPs. For example, Brewer's yeast Pex13p, a recognized integral membrane component of the docking complex, was solubilized partially in carbonate (Elgersma et al., 1996). In other examples, P. pastoris Pex14p (Johnson et al., 2001) and H. polymorpha Pex3p (Haan et al., 2002), both membrane proteins which do not possess TMDs, were resistant to carbonate extraction. The explanation for the latter two unexpected results was that PpPex14p and HpPex3p associate tightly with other integral membrane PMPs. The authors concluded that results from alkaline carbonate treatments do not always reveal unequivocally the correct membrane association of PMPs. In our case, we were most interested in comparing results with ER and peroxisomal membranes. It was of interest to find that the treatment removed a greater proportion of peroxisomal AtAPX from rough ER membranes than from peroxisomal membranes (Fig. 7). This observation suggests that AtAPX forms a unique and perhaps more tentative association with these non-peroxisomal membranes. Experiments designed to elucidate the topological orientation of peroxisomal AtAPX showed that the majority of the polypeptide was anchored on the cytosolic face of peroxisomal and rough ER membranes (Fig. 8). We were unable to determine the topology of AtAPX that resided in the 20% region of the gradient because proper control reactions could not be established. Perhaps AtAPX is not associated with membranes in this region. The cytosolic orientation of peroxisomal APX has been demonstrated within glyoxysomes of various oilseed species (Yamaguchi et al., 1995a; Jiménez et al., 1997; Ishikawa et al., 1998; Mullen et al., 2001b). This tail-anchored topology places the active site in close proximity to the boundary membrane, where it is thought to scavenge H2O2 that escapes the peroxisomal matrix by passive diffusion (Yamaguchi et al., 1995a; Corpas et al., 2001; Donaldson, 2002). One might speculate that the cytosolic orientation of peroxisomal AtAPX in rough ER predicts a similar function in this and other (smooth membrane) non-peroxisomal compartments. Although electron transfer reactions among ER membrane proteins are required for disulfide bond formation in the ER lumen (Frand et al., 2000), the ER likely does not provide the H2O2 substrates that are required for APX activity, and none of the seven types of APX that exist in Arabidopsis are predicted to function in/at the ER (Jespersen et al., 1997). Nonetheless, the trend that emerges from the results in Figures 7 and 8 is that the association and orientation of peroxisomal AtAPX is similar among all membranes in which the protein has been detected. These similarities strongly suggest a biogenetic relationship between ER and peroxisomes in wild-type cells.
MATERIALS AND METHODS
Plasmid Description
pRTL2/HA-APX codes for a single copy of the hemagglutinin epitope appended to the N terminus of the entire cottonseed (Gossypium hirsutum) peroxisomal APX polypeptide; its construction was described by Mullen et al. (1999).
Suspension Cell Cultures and Microprojectile Bombardment
Arabidopsis var. Landsberg erecta suspension-cultured cells were a gift from Steven Neill (University of West England, Bristol, UK) and were grown in a Murashige and Skoog medium composed of Murashige and Skoog salt and vitamin mixture (4.4 g L–1; Invitrogen, Carlsbad, CA), Suc (30 g L–1), α-naphthalene acetic acid (4.2 mg L–1), and kinetin (0.02 mg L–1) adjusted to pH 5.5 (Desikan et al., 1996). Cell cultures were maintained on a platform shaker (150 rpm) in the dark at 25°C as 50-mL cultures in 125-mL Erlenmeyer flasks that were subcultured weekly. All chemicals, except the Murashige and Skoog components, were purchased from Sigma-Aldrich (St. Louis).
For transient transformation via microprojectile bombardment, pelleted 4-d-old cells were resuspended in an equal (pellet) volume of transformation medium, which consisted of growth medium without α-naphthalene acetic acid or kinetin, plus 250 mm sorbitol and 250 mm mannitol. In petri dishes, resuspended cells were spread on sterile filter papers prewetted with transformation medium and equilibrated in the dark for 1 h before biolistic bombardment. Column-purified (Qiagen, Valencia, CA) plasmid DNA (10 μg) was precipitated onto M-17 tungsten particles according to the deagglomeration and isopropanol modifications of Sawant et al. (2000). DNA-coated particles were accelerated with a PDS-1000/He Particle Delivery System (Bio-Rad Laboratories, Hercules, CA) as described for BY-2 cells (Banjoko and Trelease, 1995). Bombarded cells were held in covered petri dishes for approximately 20 h to allow transient expression of the introduced gene (Fig. 2, C and D).
Immunofluorescence Microscopy
Wild-type cells were fixed 4 d post-subculture by adding an equal volume of 8% (w/v) formaldehyde (prepared fresh from paraformaldehyde; Ted Pella, Redding, CA) directly to a portion (7–10 mL) of the swirling cell culture, which was then inverted continuously for 1 h. After pelleted cells were washed in four changes of phosphate-buffered saline (PBS), cell walls were perforated and partially digested in 0.1% (w/v) Pectolyase Y-23 (Seishin Pharmaceutical, Tokyo) and 0.1% (w/v) Cellulase RS (Karlan Research Products, Santa Rosa, CA) in PBS for 2 h at 30°C with inversion rocking, followed by three washes in PBS. Bombarded cells (see above paragraph) were scraped from filter papers into 15-mL conical tubes and were fixed and perforated/digested similarly except fixation was in 4% (w/v) formaldehyde diluted in 0.5× transformation medium (final concentrations).
Bombarded cells (Fig. 2, C and D) were processed further using our standard 1-mL volume method (Lee et al., 1997), whereas wild-type cells (Fig. 2, A and B) were processed further using an “on-slide” method adapted from Kandasamy et al. (1999). In our standard 1-mL volume method, 0.1 mL (sedimented volume) of fixed, perforated/digested cells (scraped from plates) in 1.5-mL microfuge tubes were permeabilized in 0.3% (v/v) Triton X-100 (Sigma-Aldrich) in PBS for 15 min, washed in four changes of PBS, and then incubated for 1 h each in primary and fluorophore-conjugated secondary antibodies diluted in PBS. For the on-slide method, fixed, perforated/digested wild-type cells suspended in PBS were applied to poly-l-Lys-coated (Sigma-Aldrich) glass microscope slides for 5 min, and then the slides were submerged in two changes of fresh PBS. The adhered cells were permeabilized in 0.5% (v/v) Triton X-100 in PBS for 30 min and then washed in three changes of PBS. Primary (2 h application) and secondary (1 h application) antibodies diluted in Tris-buffered saline (20 mm Tris-HCl, pH 7.4, and 180 mm NaCl) containing 0.05% (v/v) Tween 20 (TBST) and 5% (w/v) protease-free bovine serum albumin (BSA; #A-3059, Sigma-Aldrich) were applied to slides arranged horizontally. The on-slide method typically required twice the concentration of primary and secondary antibodies that were used in the standard 1-mL method.
Antibody sources and concentrations were as follows (affinity-purified refers to IgGs eluted from Protein A-Sepharose columns; Kunce et al., 1988): rabbit anti-cucumber peroxisomal APX affinity-purified IgGs (1:500; Corpas et al., 1994); rabbit anti-cottonseed catalase affinity-purified IgGs (1:500; Kunce et al., 1988); rabbit anti-castor calnexin antiserum (1:500; provided by Sean Coughlan; Coughlan et al., 1997); mouse anti-HA epitope monoclonal antibody (1:300; Roche Diagnostics, Basel); mouse anti-salicylic acid-binding protein (catalase) monoclonal antibody (1:10; Chen et al., 1993); goat anti-rabbit rhodamine (1:1,000); goat anti-rabbit Cy5 (1:500); and goat anti-mouse Cy2 (1:500 or 1:1,000). All fluorophore-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (Westgrove, PA). Cells were coverslipped in 90% (v/v) glycerol with n-propyl gallate to prevent photobleaching of fluorescence. The samples used for non-confocal microscopy were flattened by placing lead weight on the coverslips before coating the coverslip with fingernail polish. The samples used for confocal microscopy were not flattened; coverslips were suspended over clear adhesive tape and coated with fingernail polish. Non-confocal fluorescence images (Fig. 2, A and B) were recorded to TMAX 400 film (Eastman Kodak, Rochester, NY) with an Axiovert 100 microscope (Carl Zeiss, Thornwood, NY) and digitized directly with a Super Coolscan LS-4000 slide scanner (Nikon Instruments, Melville, NY) before pseudocoloring green or red and assembling into figures using Photoshop 6.0 (Adobe Systems, Mountain View, CA). Confocal images (Fig. 2, C and D) were obtained with a TCS NT microscope (Leica, Heidelberg) and were analyzed as described previously (Mullen et al., 1999).
Sample Analyses, SDS-PAGE, and Immunoblots
Buoyant densities of Suc gradient fractions were measured at room temperature (RT) with an Abbe 2C refractometer (Bausch and Lomb, Rochester, NY). Amounts of protein in Suc gradient fractions were determined by the Coomassie Blue dye-binding method (standard assay) using bovine plasma gamma globulin as the standard (Bio-Rad). Catalase and cytochrome c oxidase activities were assayed as described by Ni et al. (1990) and Tolbert et al. (1968), respectively. Proteins were precipitated for SDS-PAGE in a final concentration of 10% (w/v) TCA for 15 min at 4°C. TCA pellets were resuspended in SDS sample buffer (62.5 mm Tris-HCl, pH 6.8, 10% [v/v] glycerol, 4% [w/v] SDS, 0.05% [v/v] β-mercaptoethanol, and 0.00125% [w/v] bromphenol blue), neutralized with solid Tris, and then boiled for 5 min before separation in 4% to 20% (w/v) gradient precast Mini-Protean II polyacrylamide gels (Bio-Rad). Separated proteins were transferred to polyvinylidene difluoride membranes (Immobilon P; Millipore, Bedford, MA) with a semidry transfer apparatus (Bio-Rad) according to Bunkelmann et al. (1995), except for the addition of 0.0375% (w/v) SDS to the transfer solutions and decreasing the transfer time to 90 min. Polyvinylidene difluoride membranes were incubated in 5% (w/v) nonfat dry milk (purchased from a local market) in TBST for either 1 h at RT or overnight at 4°C. Primary and secondary antibodies used were as follows: rabbit anti-cucumber peroxisomal APX IgGs (1:1,000), rabbit anti-cucumber PMP73 IgGs (1:1,000; Corpas et al., 1994), rabbit anti-wheat NADH dehydrogenase (NAD9) antiserum (1:10,000; provided by Jean-Michel Grienenberger; Lamattina et al., 1993), rabbit anti-castor calnexin antiserum (1:5,000), rabbit anti-cottonseed catalase IgGs (1:1,000 or 1:2,000), rabbit anti-maize BiP antiserum (1:1,000; provided by Eliot Herman), goat anti-rabbit alkaline phosphatase conjugate (1:10,000; Bio-Rad). All antibodies were diluted in 1% (w/v) nonfat dry milk in TBST and applied separately for 1 h at RT, with three exchanges of TBST after each antibody application. Immunoreactive proteins were visualized using an ImmunStar chemiluminescence kit according to the manufacturer's instructions (Bio-Rad). Luminescent blots were exposed to Kodak X-OMAT autoradiography film.
Suc Gradient Isolation of Organelles
Arabidopsis suspension cells (25-mL portions) harvested 7 d post-subculture by centrifugation were resuspended in 1.5 volumes of ice-cold homogenization medium (HM; 25 mm HEPES-KOH, pH 7.5, 0.4 m Suc, 3 mm dithiothreitol, and 0.5 mm phenylmethylsulfonyl fluoride) and each portion was separately ruptured immediately by passing once through a cold French pressure cell (1-inch piston diameter) at 1,000 p.s.i. Pooled homogenates were centrifuged in a JS-13 rotor (Beckman Coulter, Fullerton, CA) at 1,500g for 15 min at 4°C, and the resulting supernatants were loaded onto 25-mL linear gradients (30%–59% [w/w] Suc in 25 mm HEPES-KOH, pH 7.5) underlaid with a 5-mL cushion (59% [w/w] Suc). Each gradient received approximately 12.5 mL of supernatant material or the equivalent of approximately 25 mL of cell culture. The gradients were centrifuged in a Beckman vTi 50 rotor at 50,000g for 75 min at 4°C and then fractionated by hand into 1-mL fractions, which were treated as described above.
Mg2+ Shift Subfractionation of Microsomes
Arabidopsis cells were harvested as described above and then resuspended in HM supplemented with either 5 mm MgCl2 or 2 mm EDTA (Matsuoka et al., 1997). After one passage through the French pressure cell at 2,000 p.s.i., the pooled homogenates were centrifuged in the Beckman JS-13 rotor at 1,500g (15 min, 4°C), and the resulting supernatants at 20,000g (45 min, 4°C). Microsomes were pelleted from the 20,000g supernatant in a Beckman 50.2 Ti rotor at 200,000g for 60 min at 4°C. Microsomal pellets were resuspended in the appropriate HM and approximately 1 mL (the equivalent of approximately 50 mL cell culture) was layered onto each 13-mL linear gradient (15%–45% [w/w] Suc in 25 mm HEPES-KOH, pH 7.5, with either MgCl2 or EDTA). The gradients were centrifuged in a Beckman SW 28.1 rotor at 125,000g for 2.5 h at 4°C and then fractionated by hand into 0.5-mL fractions, which were treated as described above.
Membrane Association and Topological Orientation
The methods of Corpas et al. (1994) were modified slightly according to Nito et al. (2001) for the sequential treatments of Suc gradient-isolated peroxisomes, rough ER, and other microsomal subfractions. First, isolated organelles/vesicles were diluted in 3 volumes of 25 mm HEPES-KOH, pH 7.5, and incubated for 30 to 60 min at 4°C with inversion rocking. The suspensions were centrifuged in a Beckman 90 Ti rotor at 100,000g for 30 min (4°C) to pellet membranes and produce a supernatant fraction containing water-solubilized proteins. Membranes were completely resuspended in 200 mm KCl in 25 mm HEPES-KOH, pH 7.5, and were vortexed briefly every 10 min during a 30-min incubation at 4°C. The membranes were pelleted at 100,000g (30 min) to generate a KCl-soluble supernatant fraction. These membranes were incubated for 30 min at 4°C in 100 mm Na2CO3, pH 11.5, and then centrifuged as above to produce an alkaline carbonate-soluble fraction that was neutralized with 1 n HCl. The final pellet was incubated overnight at 4°C in 1.5% (w/v) dodecyl-β-d-maltoside detergent (Calbiochem, San Diego) in 50 mm Tris-HCl, pH 7.2, containing 750 mm aminocaproate (Corpas et al., 1994). The amount of protein in all supernatants and the final detergent-solubilized pellet was determined before TCA precipitation for SDS-PAGE and immunoblot analyses.
For topology experiments, 50 μg of peroxisomal or 1 mg of microsomal proteins in Suc was diluted (when necessary) with iso-osmotic 25 mm HEPES-KOH, pH 7.5, and then incubated for 60 min at 4°C with various amounts of proteinase K (5 mg mL–1 stock solution in deionized water; Invitrogen). Matrix/lumenal proteins were made protease-accessible in separate reactions with a 15-min (4°C) preincubation in 1% (v/v) Triton X-100 detergent. All reactions were carried out in the presence of 1 mm dithiothreitol (Roche Diagnostics), and separate control reactions omitted both detergent and protease. Protease activities were stopped with the addition of phenylmethylsulfonyl fluoride (Sigma-Aldrich) to a final concentration of 1 mm, after which the samples were TCA precipitated for SDS-PAGE and immunoblot analyses as described above.
Electron Microscopy
Wild-type Arabidopsis cells were prepared for electron microscopy according to the methods described by Sabba et al. (1999) for BY-2 cells. In brief, 7-d-post-subculture cells were fixed for 1 h at RT in a final concentration of 0.6% (v/v) glutaraldehyde (Electron Microscopy Sciences, Fort Washington, PA). A 10× fixative solution (6% [v/v] glutaraldehyde in 50 mm PIPES-NaOH, pH 7.2) was added directly to the swirling cell culture to assure rapid fixation in an aerobic environment that yields superior ultra-structural preservation of cellular structures. After two washes in 100 mm cacodylate, pH 7.2, cells were post-fixed for 2 h at RT in 2% (v/v) osmium tetroxide (Electron Microscopy Sciences) in 100 mm cacodylate, pH 7.2. After two washes in deionized water and en bloc staining overnight at 4°C in 2% (w/v) aqueous uranyl acetate (Ted Pella), cells were dehydrated in ethanol, infiltrated in Spurr's resin (Ted Pella), concentrated within the tips of BEEM capsules (spun in a microfuge at 13,000 rpm for 5 min), and then polymerized overnight at 60°C.
Isolated peroxisomes, mitochondria, and microsomal subfractions were prepared with slight modifications according to Turley and Trelease (1990). Pooled Suc gradient fractions were fixed (30 min, 4°C) in a final concentration of 0.75% (v/v) glutaraldehyde in iso-osmotic Suc in 25 mm HEPES-KOH, pH 7.5 (plus 5 mm MgCl2, microsomes only). The fixed organelles were diluted over a 30-min period at 4°C in the same fixative solution (without Suc) to approximately 15% (w/w) Suc and then were pelleted at 4°C onto a GA-7 filter (0.3-μm pore size; Metricel, Pall Corp., Ann Arbor, MI) in a Beckman JS-13 rotor at 15,000g for 30 min (peroxisomes and mitochondria) or in a Beckman SW 41 rotor at 107,000g for 45 min (microsomes). The filters were encased completely in warm 5% (w/v) aqueous agar and washed in two to three changes of 25 mm HEPES-KOH, pH 7.5, followed by post-fixation in buffered 1% (v/v) osmium tetroxide for 45 min at RT. The latter step was omitted from microsomal samples prepared for immunogold microscopy. After water washes, the samples were dehydrated in ethanol, infiltrated in LR White resin (Electron Microscopy Sciences), and then sliced into pie-shaped segments for embedment in gelatin capsules at 50°C under vacuum for at least 24 h.
All samples were ultrathin sectioned with a diamond knife (Diatome, Biel, Switzerland) on a Leica Ultracut R ultramicrotome. Cell fraction samples were oriented such that sections were made perpendicular to the filter so that stratification of centrifuged structures could be observed. Semithin (0.5-μm) sections were contrasted with toluidine blue to find high densities of packed cells or to locate isolated cell samples on filters. Ultrathin sections with silver-gold reflectance (70 nm) were collected on 200-mesh copper (wild-type Arabidopsis cells and isolated organelles) or on nickel (isolated microsomes for immunogold probing) grids. Collected sections were post-stained for 10 min with 2% (w/v) aqueous uranyl acetate and for 5 min with Reynold's lead citrate (Fisher Scientific, Loughborough, Leicestershire, UK) and then were observed and photographed to plate negatives (Kodak) with a Philips CM12S (FEI Company, Hillsboro, OR) transmission electron microscope operated at 60 or 80 kV.
Immunogold Electron Microscopy
Immunogold labeling of rough microsomes was performed as described (Bunkelmann et al., 1995), except Gly pretreatments were omitted and BSA was used instead of fetal calf serum as the blocking agent. All incubations were carried out at RT by floating each nickel grid on separate droplets of solutions. Nonspecific binding sites were blocked for 30 min with 1% (w/v) BSA dissolved in filter-sterilized (0.2 μm) PBS containing 0.01% (w/v) sodium azide. The grids were washed in three changes (3 min each) of PBS and then incubated for 60 min in primary antibodies diluted in blocking solution as follows: anti-cucumber peroxisomal APX preimmune or affinity-purified (Protein A) IgGs (1:50), and anti-castor calnexin antiserum (1:50). After five exchanges in PBS (2 × 3 min and 3 × 10 min), the grids were then incubated for 45 min in 10-nm gold-conjugated Protein A (Sigma-Aldrich) diluted 1:50 in blocking solution. Extensive washing in PBS and deionized water prepared the grids for post-staining with uranyl acetate (2–10 min) and often with lead citrate (1 min).
Photographic negative and digital images were captured at magnifications ranging from approximately 25,000× to 100,000× to reliably assess Protein A-gold particle distributions on sections over rough ER vesicular material in the sections. Gold particle counts were made by unaided eye observations of prints or negatives, and the data were normalized per square micrometer of section area.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
We are grateful to Steven Neill (University of West England, Bristol, UK) for providing the Arabidopsis suspension cells, and we thank all those who donated the antibodies used in this study. Daisy Savarirajan maintained the Arabidopsis cell cultures. Electron and confocal microscopies were performed in the Life Sciences Electron Microscopy Facility and the W.M. Keck BioImaging Laboratory at Arizona State University.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.019976.
This work was supported by the National Science Foundation (grant no. MCB–0091826 to R.N.T.) and in part by the William N. and Myriam Pennington Foundation. The Graduate Program in Molecular and Cellular Biology at Arizona State University funded a Research Assistantship for C.S.L.
References
- Anton M, Passreiter M, Lay D, Thai T-P, Gorgas K, Just WW (2000) ARF- and coatomer-mediated peroxisomal vesiculation. Cell Biochem Biophys 32: 27–36 [DOI] [PubMed] [Google Scholar]
- Baerends RJS, Rasmussen SW, Hilbrands RE, van der Heide M, Faber KN, Reuvekamp PTW, Kiel JAKW, Cregg JM, van der Klei IJ, Veenhuis M (1996) The Hansenula polymorpha PER9 gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J Biol Chem 271: 8887–8894 [DOI] [PubMed] [Google Scholar]
- Baker A, Charlton W, Johnson B, López-Huertas E, Oh J, Sparkes I, Thomas J (2000) Biochemical and molecular approaches to understanding protein import into peroxisomes. Biochem Soc Trans 28: 499–504 [PubMed] [Google Scholar]
- Banjoko A, Trelease RN (1995) Development and application of an in vivo plant peroxisome import system. Plant Physiol 107: 1201–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunkelmann JR, Corpas FJ, Trelease RN (1995) Four putative, glyoxysome membrane proteins are instead immunologically-related protein body membrane proteins. Plant Sci 106: 215–226 [Google Scholar]
- Bunkelmann JR, Trelease RN (1996) Ascorbate peroxidase: a prominent membrane protein in oilseed glyoxysomes. Plant Physiol 110: 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton W, López-Huertas E (2002) PEX genes in plants and other organisms. In A Baker, I Graham, eds, Plant Peroxisomes. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 385–426
- Chen Z, Ricigliano JW, Klessig DF (1993) Purification and characterization of a soluble salicylic acid-binding protein from tobacco. Proc Natl Acad Sci USA 90: 9533–9537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, del Río LA (2001) Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci 6: 145–150 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Bunkelmann J, Trelease RN (1994) Identification and immunochemical characterization of a family of peroxisome membrane proteins (PMPs) in oilseed glyoxysomes. Eur J Cell Biol 65: 280–290 [PubMed] [Google Scholar]
- Corpas FJ, Trelease RN (1998) Differential expression of ascorbate peroxidase and a putative molecular chaperone in the boundary membrane of differentiating cucumber seedling peroxisomes. J Plant Physiol 153: 332–338 [Google Scholar]
- Coughlan SJ, Hastings C, Winfrey R (1997) Cloning and characterization of the calreticulin gene from Ricinus communis L. Plant Mol Biol 34: 897–911 [DOI] [PubMed] [Google Scholar]
- del Río LA, Sandalio LM, Corpas FJ, López-Huertas E, Palma JM, Pastori GM (1998) Activated oxygen-mediated metabolic functions of leaf peroxisomes. Physiol Plant 104: 673–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Hancock JT, Coffey MJ, Neill SJ (1996) Generation of active oxygen in elicited cells of Arabidopsis thaliana is mediated by a NADPH oxidase-like enzyme. FEBS Lett 382: 213–217 [DOI] [PubMed] [Google Scholar]
- Donaldson RP (2002) Peroxisomal membrane enzymes. In A Baker, I Graham, eds, Plant Peroxisomes. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 259–278
- Elgersma Y, Kwast L, Klein A, Voorn-Brouwer T, van den Berg M, Metzig B, America T, Tabak HF, Distel B (1996) The SH3 domain of the Saccharomyces cerevisiae peroxisomal membrane protein Pex13p functions as a docking site for Pex5p, a mobile receptor for the import of PTS1-containing proteins. J Cell Biol 135: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, Kwast L, van den Berg M, Snyder WB, Distel B, Subramani S, Tabak HF (1997) Overexpression of Pex15p, a phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in S. cerevisiae, causes proliferation of the endoplasmic reticulum membrane. EMBO J 16: 7326–7341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber KN, Heyman JA, Subramani S (1998) Two AAA family peroxins, PpPex1p and PpPex6p, interact with each other in an ATP-dependent manner and are associated with different subcellular membranous structures distinct from peroxisomes. Mol Cell Biol 18: 936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frand AR, Cuozzo JW, Kaiser CA (2000) Pathways for protein disulphide bond formation. Trends Cell Biol 10: 203–210 [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Fowler S, Shio H, Hubbard AL, Lazarow PB (1982) Polypeptide and phospholipid composition of the membrane of rat liver peroxisomes: comparison with endoplasmic reticulum and mitochondrial membranes. J Cell Biol 93: 103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong FC, Giddings TH, Meehl JB, Staehelin LA, Galbraith DW (1996) Z-membranes: artificial organelles for overexpressing recombinant integral membrane proteins. Proc Natl Acad Sci USA 93: 2219–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haan GJ, Faber KN, Baerends RJS, Koek A, Krikken A, Kiel JAKW, van der Klei IJ, Veenhuis M (2002) Hansenula polymorpha Pex3p is a peripheral component of the peroxisomal membrane. J Biol Chem 277: 26609–26617 [DOI] [PubMed] [Google Scholar]
- Huang L, Franklin AE, Hoffman NE (1993) Primary structure and characterization of an Arabidopsis thaliana calnexin-like protein. J Biol Chem 268: 6560–6566 [PubMed] [Google Scholar]
- Ishikawa T, Yoshimura K, Sakai K, Tamoi M, Takeda T, Shigeoka S (1998) Molecular characterization and physiological role of a glyoxysome-bound ascorbate peroxidase from spinach. Plant Cell Physiol 39: 23–34 [DOI] [PubMed] [Google Scholar]
- Jespersen HM, Kjærsgård IVH, Østergaard L, Welinder KG (1997) From sequence analysis of three novel ascorbate peroxidases from Arabidopsis thaliana to structure, function and evolution of seven types of ascorbate peroxidase. Biochem J 326: 305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez A, Hernández JA, del Río LA, Sevilla F (1997) Evidence for the presence of the ascorbate-glutathione cycle in mitochondria and peroxisomes of pea leaves. Plant Physiol 114: 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Snyder WB, Cereghino JL, Veenhuis M, Subramani S, Cregg JM (2001) Pichia pastoris Pex14p, a phosphorylated peroxisomal membrane protein, is part of a PTS-receptor docking complex and interacts with many peroxins. Yeast 18: 621–641 [DOI] [PubMed] [Google Scholar]
- Kammerer S, Holzinger A, Welsch U, Roscher AA (1998) Cloning and characterization of the gene encoding the human peroxisomal assembly protein Pex3p. FEBS Lett 429: 53–60 [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Meagher RB (1999) The late pollen-specific actins in angiosperms. Plant J 18: 681–691 [DOI] [PubMed] [Google Scholar]
- Kunce CM, Trelease RN, Turley RB (1988) Purification and biosynthesis of cottonseed (Gossypium hirsutum L.) catalase. Biochem J 251: 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamattina L, Gonzalez D, Gualberto J, Grienenberger JM (1993) Higher plant mitochondria encode an homologue of the nuclear-encoded 30-kDa subunit of bovine mitochondrial complex I. Eur J Biochem 217: 831–838 [DOI] [PubMed] [Google Scholar]
- Lee MS, Mullen RT, Trelease RN (1997) Oilseed isocitrate lyases lacking their essential type 1 peroxisomal targeting signal are piggybacked to glyoxysomes. Plant Cell 9: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisenbee CS, Karnik SK, Trelease RN (2003) Overexpression and mislocalization of a tail-anchored GFP redefines the identity of peroxisomal ER. Traffic (in press) [DOI] [PubMed]
- López-Huertas E, Corpas FJ, Sandalio LM, del Río LA (1999) Characterization of membrane polypeptides from pea leaf peroxisomes involved in superoxide radical generation. Biochem J 337: 531–536 [PMC free article] [PubMed] [Google Scholar]
- Lord JM (1983) Endoplasmic reticulum and ribosomes. In JL Hall, AL Moore, eds, Isolation of Membranes and Organelles from Plant Cells. Academic Press, London, pp 119–134
- Lu SX, Hrabak EM (2002) An Arabidopsis calcium-dependent protein kinase is associated with the endoplasmic reticulum. Plant Physiol 128: 1008–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Higuchi T, Maeshima M, Nakamura K (1997) A vacuolar-type H+-ATPase in a nonvacuolar organelle is required for the sorting of soluble vacuolar protein precursors in tobacco cells. Plant Cell 9: 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RT, Flynn CR, Trelease RN (2001a) How are peroxisomes formed? The role of the endoplasmic reticulum and peroxins. Trends Plant Sci 6: 256–261 [DOI] [PubMed] [Google Scholar]
- Mullen RT, Lisenbee CS, Flynn CR, Trelease RN (2001b) Stable and transient expression of chimeric peroxisomal membrane proteins induces an independent “zippering” of peroxisomes and an endoplasmic reticulum subdomain. Planta 213: 849–863 [DOI] [PubMed] [Google Scholar]
- Mullen RT, Lisenbee CS, Miernyk JA, Trelease RN (1999) Peroxisomal membrane ascorbate peroxidase is sorted to a membranous network that resembles a subdomain of the endoplasmic reticulum. Plant Cell 11: 2167–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RT, Trelease RN (2000) The sorting signals for peroxisomal membrane-bound ascorbate peroxidase are within its C-terminal tail. J Biol Chem 275: 16337–16344 [DOI] [PubMed] [Google Scholar]
- Nebenführ A, Ritzenthaler C, Robinson DG (2002) Brefeldin A: deciphering an enigmatic inhibitor of secretion. Plant Physiol 130: 1102–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Trelease RN, Eising R (1990) Two temporally synthesized charge subunits interact to form the five isoforms of cottonseed (Gossypium hirsutum) catalase. Biochem J 269: 233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nito K, Yamaguchi K, Kondo M, Hayashi M, Nishimura M (2001) Pumpkin peroxisomal ascorbate peroxidase is localized on peroxisomal membranes and unknown membranous structures. Plant Cell Physiol 42: 20–27 [DOI] [PubMed] [Google Scholar]
- Olsen LJ (1998) The surprising complexity of peroxisome biogenesis. Plant Mol Biol 38: 163–189 [PubMed] [Google Scholar]
- Passreiter M, Anton M, Lay D, Frank R, Harter C, Wieland FT, Gorgas K, Just WW (1998) Peroxisome biogenesis: involvement of ARF and coatomer. J Cell Biol 141: 373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S (2000) The structural properties of plant peroxisomes and their metabolic significance. Biol Chem 381: 639–648 [DOI] [PubMed] [Google Scholar]
- Sabba RP, Durso NA, Vaughn KC (1999) Structural and immunocytochemical characterization of the walls of dichlobenil-habituated BY-2 tobacco cells. Int J Plant Sci 160: 275–290 [Google Scholar]
- Salomons FA, van der Klei IJ, Kram AM, Harder W, Veenhuis M (1997) Brefeldin A interferes with peroxisomal protein sorting in the yeast Hansenula polymorpha. FEBS Lett 411: 133–139 [DOI] [PubMed] [Google Scholar]
- Sawant SV, Singh PK, Tuli R (2000) Pretreatment of microprojectiles to improve the delivery of DNA in plant transformation. BioTechniques 29: 246–248 [DOI] [PubMed] [Google Scholar]
- Shi WM, Muramoto Y, Ueda A, Takabe T (2001) Cloning of peroxisomal ascorbate peroxidase gene from barley and enhanced thermotolerance by overexpressing in Arabidopsis thaliana. Gene 273: 23–27 [DOI] [PubMed] [Google Scholar]
- Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y, Yoshimura K (2002) Regulation and function of ascorbate peroxidase isoenzymes. J Exp Bot 53: 1305–1319 [PubMed] [Google Scholar]
- South ST, Baumgart E, Gould SJ (2001) Inactivation of the endoplasmic reticulum protein translocation factor, Sec61p, or its homolog, Ssh1p, does not affect peroxisome biogenesis. Proc Natl Acad Sci USA 98: 12027–12031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- South ST, Gould SJ (1999) Peroxisome synthesis in the absence of preexisting peroxisomes. J Cell Biol 144: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- South ST, Sacksteder KA, Li X, Liu Y, Gould SJ (2000) Inhibitors of COPI and COPII do not block PEX3-mediated peroxisome synthesis. J Cell Biol 149: 1345–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA (2001a) Dynamics of peroxisome assembly and function. Trends Cell Biol 11: 22–29 [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA (2001b) The life cycle of the peroxisome. Nat Rev Mol Cell Biol 2: 357–368 [DOI] [PubMed] [Google Scholar]
- Tolbert NE, Oeser A, Kisaki T, Hageman RH, Yamazaki RK (1968) Peroxisomes from spinach leaves containing enzymes related to glycolate metabolism. J Biol Chem 243: 5179–5184 [PubMed] [Google Scholar]
- Trelease RN (2002) Peroxisomal biogenesis and acquisition of membrane proteins. In A Baker, I Graham, eds, Plant Peroxisomes. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 305–337
- Turley RB, Trelease RN (1990) Purification of plant peroxisomes in isoosmotic metrizamide. Physiol Plant 79: 570–578 [Google Scholar]
- Wang J, Zhang H, Allen RD (1999) Overexpression of an Arabidopsis peroxisomal ascorbate peroxidase gene in tobacco increases protection against oxidative stress. Plant Cell Physiol 40: 725–732 [DOI] [PubMed] [Google Scholar]
- Wright R, Basson M, D'Ari L, Rine J (1988) Increased amounts of HMG-CoA reductase induce “karmellae”: a proliferation of stacked membrane pairs surrounding the yeast nucleus. J Cell Biol 107: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Mori H, Nishimura M (1995a) A novel isoenzyme of ascorbate peroxidase localized on glyoxysomal and leaf peroxisomal membranes in pumpkin. Plant Cell Physiol 36: 1157–1162 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Takeuchi Y, Mori H, Nishimura M (1995b) Development of microbody membrane proteins during the transformation of glyoxysomes to leaf peroxisomes in pumpkin cotyledons. Plant Cell Physiol 36: 455–464 [Google Scholar]
- Zhang H, Wang J, Nickel U, Allen RD, Goodman HM (1997) Cloning and expression of an Arabidopsis gene encoding a putative peroxisomal ascorbate peroxidase. Plant Mol Biol 34: 967–971 [DOI] [PubMed] [Google Scholar]