Abstract
l-Fucose (l-Fuc) is a monosaccharide constituent of plant cell wall polysaccharides and glycoproteins. The committing step in the de novo synthesis of l-Fuc is catalyzed by GDP-d-mannose 4,6-dehydratase, which, in Arabidopsis, is encoded by the GMD1 and GMD2 (MUR1) genes. To determine the functional significance of this genetic redundancy, the expression patterns of both genes were investigated via promoter-β-glucuronidase fusions and immunolocalization of a Fuc-containing epitope. GMD2 is expressed in most cell types of the root, with the notable exception of the root tip where strong expression of GMD1 is observed. Within shoot organs, GMD1::GUS expression is confined to stipules and pollen grains leading to fucosylation of the walls of these cell types in the mur1 mutant. These results suggest that GMD2 represents the major housekeeping gene for the de novo synthesis of GDP-l-Fuc, whereas GMD1 expression is limited to a number of specialized cell types. We conclude that the synthesis of GDP-l-Fuc is controlled in a cell-autonomous manner by differential expression of two isoforms of the same enzyme.
l-Fuc is a monosaccharide constituent of various glycoproteins and polysaccharides synthesized by plant cells. It is found predominantly in xyloglucan, a hemicellulosic polysaccharide that is believed to cross-link cellulose microfibrils (Bacic et al., 1988; Carpita and Gibeaut, 1993). l-Fuc is also present in the pectic polysaccharides rhamnogalacturonan I and II and in root mucilage, which is believed to lubricate the root as it travels through the soil matrix in addition to providing protection during periods of drought (Greenland, 1979; Rougier, 1981; Baldo et al., 1983).
The localization of l-fucosylated xyloglucan polymers within root cell walls has been accomplished with the use of an antibody directed against the terminal l-Fuc epitope of this hemicellulose (Puhlmann et al., 1994). These studies have shown that l-Fuc is found in almost all cells walls of the developing Arabidopsis root tip, although in different amounts (Freshour et al., 1996). More intense labeling was found in the epidermal and lateral root cap cells, which may be due to the presence of a thicker cell wall. Immunogold labeling and electron microscopy established that outer lateral root cap cell walls were heavily labeled, whereas interior-facing walls of these cells were not (Freshour et al., 1996). Terminal l-Fuc-containing epitopes were also found in most cells from mature portions of the root but were absent from the radial cross walls of endodermal cells (Freshour et al., 1996). These studies suggest that the synthesis of fucosylated polysaccharides is differentially regulated at the cellular and whole-root level in Arabidopsis. If this is the case, the de novo synthesis of l-Fuc may be tightly regulated to provide necessary precursors when and where they are needed during the development of the Arabidopsis root.
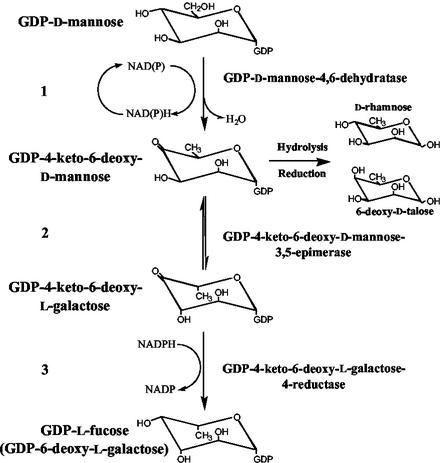
The biosynthesis of l-Fuc occurs through the conversion of GDP-d-Man to GDP-l-Fuc in three catalytic steps: 4,6-dehydration, 3,5-epimerization, and 4-reduction (for review, see Feingold and Avigad, 1980; Reiter and Vanzin, 2001). These activities are carried out by two enzymes, a GDP-d-Man 4,6-dehydratase and a GDP-4-keto-6-deoxy-d-Man (GDP-KDM) 3,5-epimerase-4-reductase (synonymous with GDP-l-Fuc synthase) in bacteria, animals, and plants (Tonetti et al., 1996; Bonin et al., 1997; Sturla et al., 1997; Ohyama et al., 1998; Sullivan et al., 1998; Bonin and Reiter, 2000; see Fig. 1 for details). Characterization of the l-Fuc-deficient mur1 mutant of Arabidopsis led to the cloning of a gene, GMD2 (MUR1), encoding a plant GDP-d-Man 4,6-dehydratase (Bonin et al., 1997). This mutant was isolated in a screen for deficiencies in monosaccharide composition of leaf tissue (Reiter et al., 1993, 1997). Analyses of other tissues of mur1 plants revealed that l-Fuc is virtually absent from stems, flowers, and siliques but is only reduced by about 40% in roots (Reiter et al., 1993). These results suggested that a root-specific 4,6-dehydratase might be present in Arabidopsis. Another coding region, designated GMD1, with high sequence similarity to GMD2 both on the nucleotide and amino acid levels, was isolated from a cDNA library (Bonin et al., 1997). The present work describes the biochemical characterization of the GMD1 protein and the determination of expression patterns of the two isoforms of GDP-d-Man 4,6-dehydratase in Arabidopsis.
Figure 1.
Schematic representation of the de novo pathway for the synthesis of GDP-l-Fuc. The procedure for the quantitation of GDP-KDM is indicated at the right.
RESULTS
Cloning of the GMD1 Gene
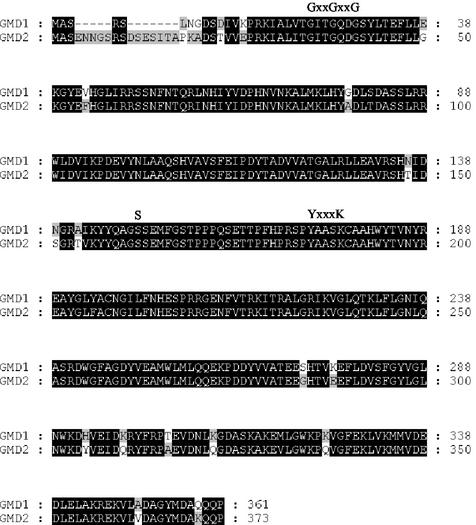
A cDNA copy of the GMD1 gene was cloned previously (Bonin et al., 1997). To isolate a full-length genomic clone corresponding to GMD1, an Arabidopsis λEMBL3 library was screened with the cDNA clone. From approximately 100,000 plaques used in the primary screen, five GMD1 genomic clones were identified. The insert from one of these clones was isolated, cloned into a plasmid vector, and sequenced. This sequence revealed an open reading frame of 361 amino acids showing 92% identity and 97% sequence similarity to GMD2 on the amino acid level (Fig. 2). On the nucleotide level, the GMD1 and GMD2 coding regions share 82% identity; however, the sequences upstream and downstream of the respective coding regions showed no significant sequence similarities, which is in line with the observation that GMD1 and GMD2 have markedly different expression patterns (see below). Recently, the GMD1 gene was sequenced by the Arabidopsis Genome Initiative (2000) and placed at the bottom of chromosome 5. The GMD1 sequences determined by us and by the Arabidopsis Genome Initiative were identical.
Figure 2.
Amino acid alignments between the Arabidopsis GMD1 and GMD2 proteins. Residues that are identical between the two sequences are highlighted by dark shading. The GxxGxxG motif involved in binding of the NAD+ cofactor and the catalytic triad consisting of a Ser residue and a YxxxK motif (Mulichak et al., 2002) are indicated.
GMD1 Is Expressed Mainly in Roots
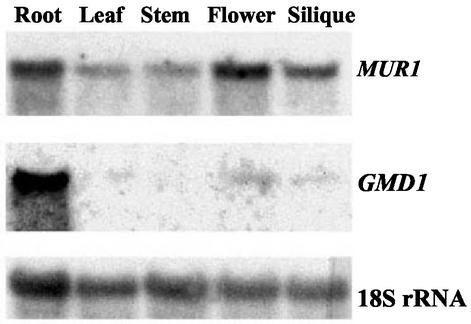
Based on our previous findings that the mur1 mutant contains substantial amounts of l-Fuc within its roots, we wished to determine if the GMD1 gene is transcribed within this tissue. Northern blots were performed to determine the transcript size and expression patterns of both GMD1 and GMD2. Total RNA was extracted from roots, leaves, stems, flowers, and siliques. The blot was probed with GMD1 and GMD2. GMD2 transcripts were highly abundant in roots and flowers and less abundant in leaves, stems, and siliques (Fig. 3). GMD1 appeared to be expressed mostly in roots, but after long exposures, weak hybridization was detectable in other tissues, particularly in flowers, even at very high stringency (Fig. 3). The approximate size of each transcript was 1.5 kb for both GMD1 and GMD2, which is in accordance with the longest cDNAs isolated.
Figure 3.
Northern-blot analysis of total RNA extracted from roots, leaves, stems, flowers, and siliques of wild-type Arabidopsis and probed with 32P-labeled GMD1 and GMD2 coding regions.
GDP-d-Man 4,6-Dehydratase Activity Is Present in mur1 Roots
To correlate GMD1 mRNA expression with GDP-d-Man 4,6-dehydratase activity in roots, crude protein extracts from mur1 and wild-type roots were assayed for this enzymatic activity. These assays revealed that mur1 root extracts produced less GDP-KDM than extracts from wild-type roots (44% conversion in the case of mur1 versus 57% conversion in the case of wild type). To assay the entire GDP-l-Fuc biosynthetic pathway in wild-type and mur1 roots, NADPH was added to the reaction mixture, permitting the conversion of GDP-KDM to GDP-l-Fuc (Bonin et al., 1997). These experiments indicated a reduction in the synthesis of l-Fuc in mur1 extracts compared with wild type (30% conversion in the case of mur1 versus 48% conversion in the case of wild type). Taken together, these results provide evidence that the partial Fuc-deficiency in mur1 roots is caused by a reduction in GDP-d-Man 4,6-dehydratase activity.
Recombinant GMD1 Protein Has GDP-d-Man 4,6-Dehydratase Activity in Vitro
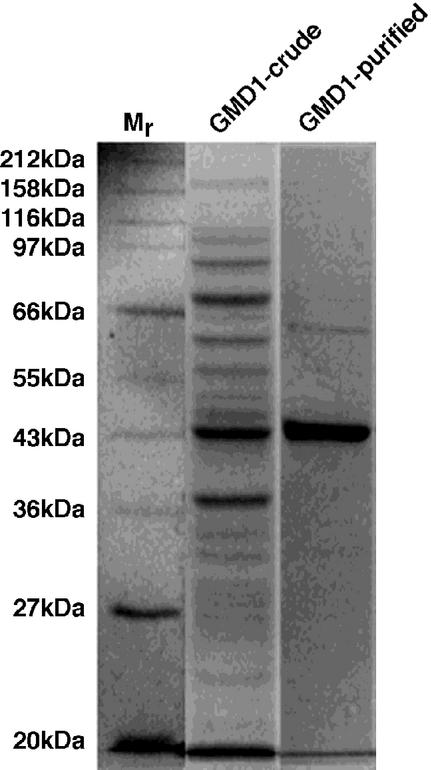
To verify the predicted function of the GMD1 protein, the open reading frame was cloned into the pET28b expression vector such that a carboxy-terminal His tag was incorporated into the recombinant enzyme, which was utilized for affinity purification of the protein. SDS-PAGE of the recombinant protein before and after purification indicated that GMD1 could be highly purified via nickel-nitrilotriacetic acid agarose (Ni-NTA) chromatography and encodes a protein of approximately 43 kD (Fig. 4), which is in agreement with its predicted size based on the amino acid sequence including the His tag (42.1 kD).
Figure 4.
Coomassie Blue-stained SDS-polyacrylamide gel of crude and Ni-NTA purified GMD1 protein from Escherichia coli. Mr denotes a size marker.
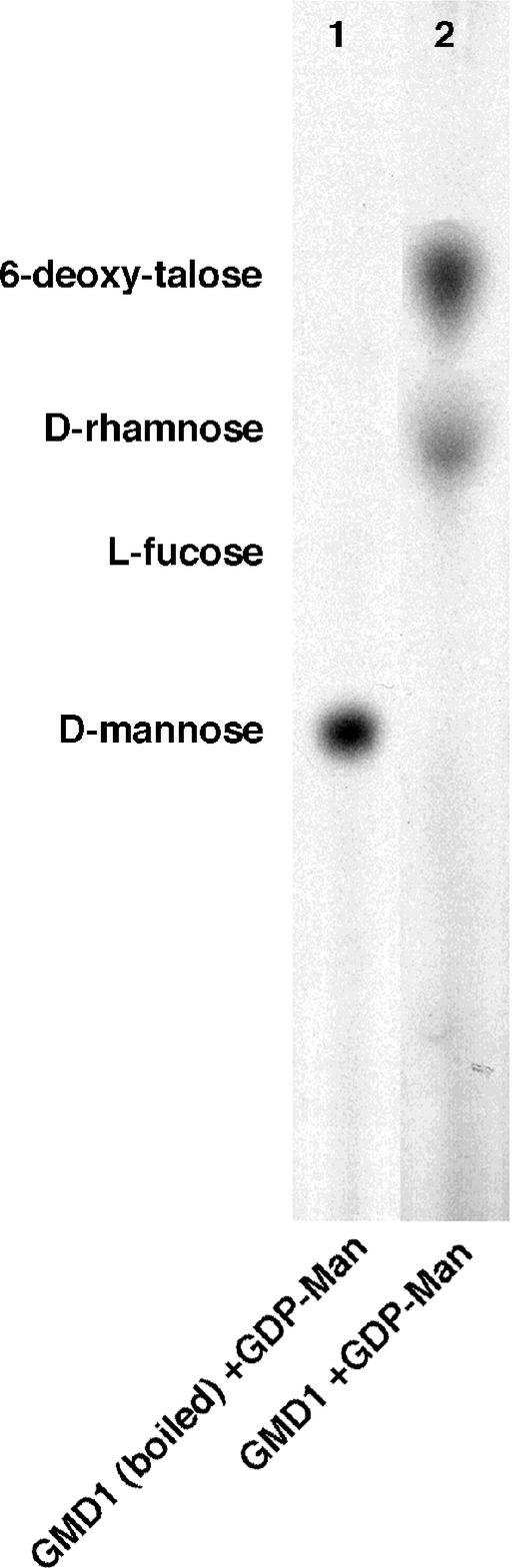
To assay GMD1 for 4,6-dehydratase activity, GDP-d-[14C]Man was incubated with purified recombinant GMD1 protein. As shown in Figure 5, lane 2, GDP-d-[14C]Man was converted to GDP-KDM (seen as a mixture of 6-deoxy-d-talose and d-Rha). These experiments demonstrate that, like the GMD2 protein, GMD1 exhibits GDP-d-Man 4,6-dehydratase activity in vitro.
Figure 5.
Autoradiograph of 14C-labeled sugars from assays of recombinant GMD1 protein for 4,6-dehydratase activity. Boiled (lane 1) and native (lane 2) GMD1 protein was incubated with radiolabeled GDP-d-Man. Nucleotide sugars were hydrolyzed to monosaccharides before loading. The sample in lane 2 was reduced with NaBH4 before hydrolysis, which leads to the formation of a mixture of d-Rha and 6-deoxy-d-talose.
GMD1 and GMD2 Are Differentially Expressed
To further explore the expression patterns of GMD1 and GMD2, DNA sequences upstream of the start codon were transcriptionally fused to β-glucuronidase (GUS) in the pCAMBIA 1391Z and 1381Z vectors, respectively, and transformed into Arabidopsis. Antibiotic-resistant T1 plants were transferred to soil and allowed to self. T2 plants collected at different developmental stages were then stained for GUS activity. Three independent lines sharing identical expression patterns were used for further analysis.
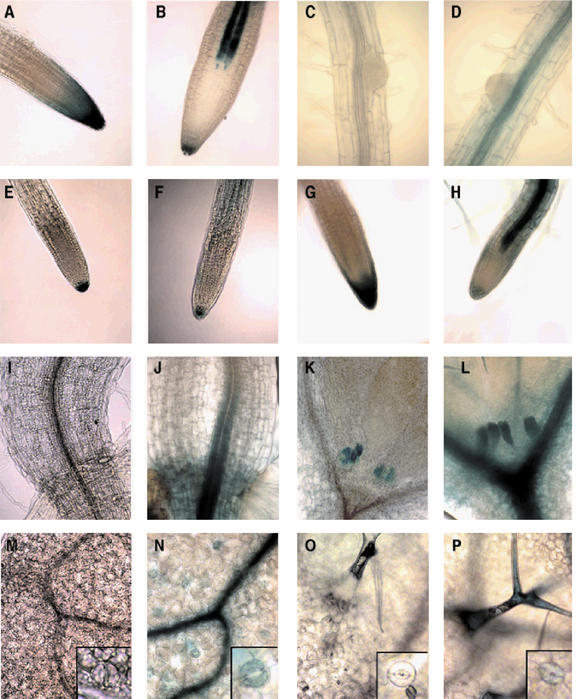
GMD1::GUS expression was seen a few days after germination at the tip of the developing primary root (Fig. 6A), whereas GMD2::GUS expression was found in all other regions of the plant except for the root meristem and the proximal part of the elongation zone (Fig. 6B). The expression of GMD1::GUS was primarily localized to the root meristem and columella root cap and showed some irregular expression through the lateral root cap and the epidermal cells close to the root tip at 3 d post-germination, and this expression pattern was maintained throughout growth (Fig. 6A). GMD2::GUS was expressed in a few columella root cap cells and the developing vasculature at 3 d post-germination (Fig. 6B) and also continued to be expressed throughout the growth of the primary root (Fig. 6D). Neither GMD1::GUS nor GMD2::GUS expression was detectable in emerging lateral roots (Fig. 6, C and D, respectively), but expression of both genes occurred at later stages of lateral root development (Fig. 6, E and F, respectively) and mimicked the expression observed within the primary root at maturity (Figs. 6, G and H, respectively). To determine whether GMD1 expression changes in response to the lack of GMD2 activity, we introduced the GMD1::GUS construct into the mur1 genetic background via standard crossing procedures. Staining for GUS activity indicated that the expression pattern of GMD1 was the same in wild-type and mur1 plants (data not shown).
Figure 6.
GMD1::GUS and GMD2::GUS expression patterns in roots, hypocotyls, cotyledons, and true leaves. GMD1::GUS (A) and GMD2::GUS (B) root tips 3 d post-germination. GMD1::GUS (C) and GMD2::GUS (D) emerging lateral roots. GMD1::GUS (E) and GMD2::GUS (F) immature lateral root tips. GMD1::GUS (G) and GMD2::GUS (H) mature lateral roots. GMD1::GUS (I) and GMD2::GUS (J) root-hypocotyl junctions. GMD1::GUS (K) and GMD2::GUS (L) apical meristems and stipules. GMD1::GUS (M) and GMD2::GUS (N) cotyledon surfaces and guard cells (insets). GMD1::GUS (O) and GMD2::GUS (P) true leaf surfaces with trichomes and guard cells (insets).
A more extensive analysis of promoter-GUS plants indicated that GMD1 was expressed in some aboveground tissues, specifically in stipules and in pollen grains. GMD1::GUS and GMD2::GUS expression could be seen in leaf stipules at 3 d post-germination (Fig. 6, K and L, respectively). GMD1::GUS expression was not seen at the root-hypocotyl junction or in cotyledons (Fig. 6, I and M, respectively), whereas GMD2::GUS was expressed in both of these regions, specifically within the vasculature and guard cells of the cotyledons (Fig. 6, J and N, inset, respectively). Inspection of true leaves revealed that GMD2::GUS was primarily expressed in trichomes and guard cells (Fig. 6P), whereas GMD1::GUS was not (Fig. 6O). Staining for GUS activity in whole developing florets indicated that GMD1 and GMD2 are transiently expressed in pollen just before anthesis (Fig. 7, A and B). Closer inspection of individual flowers at different stages of development confirmed this initial finding. At early stages of flower development, neither GMD1 nor GMD2 were expressed in pollen grains (Fig. 7, C and E, respectively), but were switched on at a slightly more developed stage (Fig. 7, D and F, respectively). This can be seen more clearly in a close-up of GMD1::GUS and GMD2::GUS anthers (Fig. 7, G and I, respectively). The expression of both GMD2 and GMD1 in only a subset of the pollen may be attributed to a different stage of development for pollen derived from single anthers. After inspecting several flowers, it seemed as if the development of pollen from individual anthers lacked synchronicity. A small percentage of flowers examined had anthers where it appeared as if all of the pollen grains were staining for GUS activity, suggesting that synchronous pollen development may be a rare event. After anthesis, expression of both GMD1 and GMD2 was no longer observed (Fig. 7, H and J). GMD1 transcription was not seen in any other floral organs, whereas GMD2 transcription was observed in portions of the inflorescence stem, the vasculature of flowers, the style, but not the stigma and the ovary (Fig. 7, B, E, and F).
Figure 7.
GMD1::GUS and GMD2::GUS expressional analysis of flowers. GMD1::GUS (A) and GMD2::GUS (B) whole florets. GMD1:: GUS (C) and GMD2::GUS (E) young flowers. GMD1::GUS (D) and GMD2::GUS (F) flowers just before anthesis. Close-up of GMD1::GUS (G) and GMD2::GUS (I) anthers just before anthesis. GMD1::GUS (H) and GMD2::GUS (J) stamens postanthesis. Arrows point to GUS-expressing anthers or pollen grains.
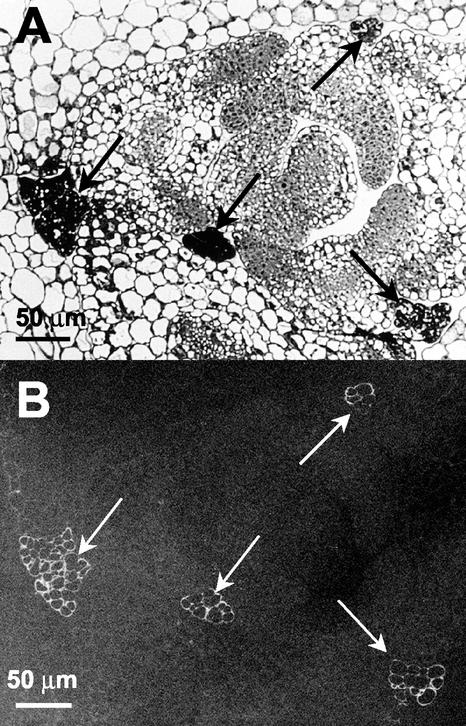
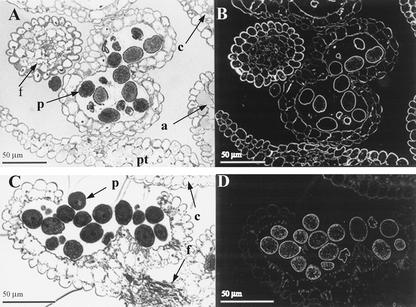
Because aboveground tissues of mur1 plants, which carry a mutation in GMD2, do not contain significant amounts of Fuc as determined by glycosyl composition analyses (Reiter et al., 1993), the expression of the GMD1::GUS fusion in stipules and pollen grains was unexpected and warranted closer examination using an independent method. The monoclonal antibody, CCRC-M1 (Puhlmann et al., 1994), detects fucosylated xyloglucans in Arabidopsis under the experimental conditions used in this study (Freshour et al., 2003). The CCRC-M1 antibody labeled the walls of all stipule cells but did not label any other cells in the rosette of mur1 plants (Fig. 8). In wild-type plants, all cell walls of the leaves and stipules were labeled by CCRC-M1 (data not shown). Immunolabeling with CCRC-M1 of anthers from wild-type and mur1 plants yielded staining of all floral cell walls in wild-type plants (Fig. 9B), whereas immunoreactivity was strictly confined to pollen grains in anthers from mur1 plants (Fig. 9D). All pollen grains stained with approximately equivalent intensities in wild-type and mur1 plants.
Figure 8.
Immunofluorescent labeling with CCRC-M1 of transverse sections through the rosette of a 21-d-old mur1 plant. A, Light micrograph of stipules (indicated by arrows) and portions of adjacent leaves. B, Adjacent section with immunofluorescent labeling with CCRC-M1.
Figure 9.
Immunofluorescent labeling with CCRC-M1 of transverse sections of stage 13 flowers of Arabidopsis. A and B, Wild-type plants. C and D, mur1 plants. A and C, Sections stained with toluidine blue. B and D, Immunofluorescent labeling with CCRC-M1. a, Anther; c, carpel; f, filament; pt, petal; p, pollen grain.
DISCUSSION
We have demonstrated that a gene with sequence similarity to the previously characterized GMD2 (MUR1) gene (Fig. 2) encodes a functional GDP-d-Man 4,6-dehydratase that catalyzes the committing step in the conversion of GDP-d-Man to GDP-l-Fuc (Fig. 5). The identification and characterization of a second gene involved in the biosynthesis of l-Fuc adds to our understanding of nucleotide sugar formation in Arabidopsis and raises new questions regarding the regulation of cell wall polysaccharide biosynthesis in plants.
GMD1 transcription appears to be essentially root specific (Fig. 3), although it was also detected in stipules and in pollen grains (Figs. 6 and 7). The expression in roots was expected based on our initial hypothesis that a gene other than GMD2 must be responsible for the presence of l-Fuc in mur1 roots, which is approximately 60% that of wild-type roots (Reiter et al., 1993). This transcriptional expression pattern correlated with a decreased 4,6-dehydratase activity in mur1 roots as compared with wild-type roots, suggesting that the GMD1 protein is responsible for synthesis of the remaining l-Fuc found within this tissue. An evaluation of the entire Arabidopsis genome (Arabidopsis Genome Initiative, 2000) did not reveal any candidate genes for GDP-d-Man 4,6-dehydratase other than GMD1 and GMD2. Accordingly, the production of all l-Fuc in Arabidopsis appears to depend on the activity of these two genes.
We used GMD1::GUS and GMD2::GUS plants to more closely examine the transcriptional expression patterns of these genes in Arabidopsis (Figs. 6 and 7). These experiments have provided us with compelling evidence that GMD1 is not only transcribed within specific cells of the root but is temporally expressed within stipules and pollen. As expected from the presence of l-Fuc in mur1 roots and our GMD1 northern data (Fig. 3), GMD1::GUS expression was found to be localized mainly in the root but appeared to be confined to the tips of both the primary root and lateral roots with particularly strong expression in the meristematic and columellar root cap zones (Fig. 6, A, E, and G). These results are in remarkable agreement with immunolocalization studies carried out with the CCRC-M1 antibody on roots of the mur1 mutant, where only the walls of meristematic and columellar cells of mature roots stained (Freshour et al., 2003).
Interestingly, neither GMD1 nor GMD2 appeared to be expressed in the lateral root primordia (Fig. 6, C and D), yet the walls of wild-type root primordia contain fucosylated xyloglucans as demonstrated by CCRC-M1 immunolabeling (Freshour et al., 2003). Furthermore, GMD1::GUS expression was undetectable in root hairs both in the wild-type and mur1 genetic backgrounds even though root hairs of mur1 plants react strongly with the CCRC-M1 antibody (Freshour et al., 2003). These results suggest that the sensitivity of GMD1::GUS and GMD2::GUS expression assays is not always sufficient to predict the presence of the CCRC-M1 epitope presumably because of posttranscriptional events that are not detectable by reporter gene studies. On the other hand, we never encountered an instance where the presence of the CCRC-M1 epitope was expected based on reporter gene expression but not observed by immunocytochemistry.
GMD2 appears to be expressed in all other parts of the plant, which was expected based on our previous characterization of the mur1 mutant and the omnipresence of l-Fuc in Arabidopsis. Aside from the columella root cap, GMD2 expression is switched on within the elongation zone of the root where differentiation of specialized cell types occurs. Specifically, GMD2 is expressed to a high degree in the developing stele (Fig. 6B) and to a lesser degree within surrounding cells. Because specialized transport cells within the vasculature require a strong cell wall to resist conductive forces, the production and insertion of fucosylated polymers may aid in their integrity. Immunolocalization data using the CCRC-M1 antibody did not show significant differences in labeling intensity of the vasculature versus surrounding cells (Freshour et al., 1996), suggesting that if the high levels of GMD2 expression observed in the stele lead to increased Fuc production, this Fuc is incorporated into glycoconjugates other than xyloglucan. Examples of other fucosylated glycoconjugates that are not recognized by CCRC-M1 include glycoproteins and rhamnogalacturonan II, of which the latter has been shown recently to play a significant role in controlling growth characteristics of plant cells (O'Neill et al., 2001). The strong expression of GMD2 in guard cells may similarly reflect a high demand for GDP-l-Fuc for glycoconjugates needed to strengthen or otherwise influence the properties of these specialized cell walls. Intense labeling of wild-type guard cell walls by CCRC-M1 suggests that these walls contain higher levels of xyloglucan than walls of other leaf cells (Fig. 10).
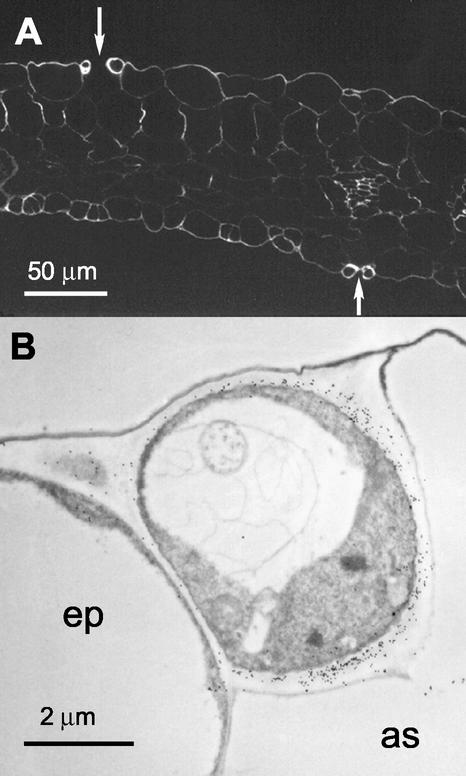
Figure 10.
Immunolabeling with CCRC-M1 of thin sections taken from cotyledons of 5-d-old wild-type seedlings. A, Immunofluorescent labeling with CCRC-M1. Arrows point to pairs of heavily labeled guard cells. B, Immunogold labeling of a single guard cell located between stomatal air space (as) and an epidermal cell (ep).
The expression of GMD1 in stipules and pollen was unexpected but could be confirmed by l-Fuc immunolocalization studies in these tissues from mur1 plants. It appears clear that both GMD1 and GMD2 are expressed in pollen to provide Fuc for the synthesis of the pollen grain wall (Fig. 9). In addition, expression of these two genes just before anthesis might provide sufficient amounts of GDP-d-Man 4,6-dehydratase to permit l-Fuc synthesis during pollen tube growth after pollen germination. Labeling of mur1 pollen tubes by the CCRC-M1 antibody has been observed (Freshour et al., 2003), suggesting the persistence of GMD1 activity through pollen germination and pollen tube growth.
Stipules represented the only vegetative shoot organ where both GMD1 and GMD2 were strongly expressed, presumably reflecting a high demand for GDP-l-Fuc in these leaf-like structures. Stipules from Arabidopsis are characterized by elaborate endomembrane systems (Bowman, 1994) that may play a role in the synthesis of fucosylated glycans.
Our examination of the biochemical function and expression patterns of the GMD1 and GMD2 genes raises the question of why Arabidopsis maintains two enzymes with the same catalytic function in a nucleotide sugar interconversion pathway. Although both GMD1 and GMD2 are GDP-d-Man 4,6-dehydratases, the two proteins may differ in terms of protein stability, kinetics, and regulatory properties such as feedback inhibition. Differences in these properties may make one isoform more suitable for specific cell types than the other one. Although l-Fuc can be recycled via a salvage pathway, plants cannot convert this 6-deoxysugar into other monosaccharides, making it an end product destined to be incorporated into cell wall polymers and glycoproteins. Accordingly, the de novo synthesis of GDP-l-Fuc needs to be tightly regulated, presumably in part at the posttranscriptional and/or posttranslational levels.
The final steps in the conversion of GDP-d-Man to GDP-l-Fuc are catalyzed by GDP-l-Fuc synthase, an enzyme encoded by the GER1 gene in Arabidopsis (Bonin and Reiter, 2000). Like GMD2, the GER1 gene is expressed in all major plant organs and appears to account for virtually all GDP-l-Fuc synthase activity, at least in leaf material. The Arabidopsis genome contains a close homolog to GER1 that is transcribed and likely to encode an isoform of GDP-l-Fuc synthase (Bonin and Reiter, 2000). It will be interesting to determine the precise expression patterns of the GER1 and “GER2” genes to see whether they are coordinately regulated with GMD2 and GMD1, respectively.
MATERIALS AND METHODS
Plant Material
Plants were grown in environmental chambers at 23°C and 60% to 70% humidity under continuous fluorescent light (60–70 μ mol m–2 s–1). Wild-type plants of the Columbia ecotype were used for the isolation of DNA and RNA.
Isolation of the GMD1 Gene
GMD1 genomic clones were isolated from a λEMBL3 library constructed by CLONTECH Laboratories Inc. (Palo Alto, CA) using DNA from the Columbia ecotype of Arabidopsis. Synthesis of a digoxigenin-labeled hybridization probe was accomplished by random primer labeling of the EcoRI/BamHI insert fragment of the cDNA isolated previously (Bonin et al., 1997). The λEMBL3 library was screened with the digoxigenin-labeled probe using the colorimetric detection system supplied by Boehringer Mannheim (Indianapolis). Lambda clones hybridizing to the DNA probe were grown in liquid media using standard procedures (Sambrook et al., 1989), and DNA was purified using the lambda DNA purification kit (Qiagen Inc., Chatsworth, CA). Two hybridizing SalI fragments that corresponded to 5′ and 3′ portions of the gene due to a SalI site within the coding sequence were isolated from the largest lambda clone and ligated separately to SalI-digested pBluescript KS+ vector. The ligation products were transformed into Escherichia coli XL1-Blue supercompetent cells (Stratagene, La Jolla, CA).
Nucleic Acid Sequence Determination and Analysis
All sequencing reactions were done by the enzymatic method with approximately 10 μg of template DNA using the Cy5 Auto Read Labeling kit (Pharmacia, Piscataway, NJ) and an automated laser fluorescent sequencer (Pharmacia). Analysis of nucleic acid and derived amino acid sequences of the GMD1 gene was carried out with MacVector software (Oxford Molecular Group, Oxford). All DNA sequences were determined from both strands.
Isolation and Analysis of Arabidopsis RNA
RNA was isolated from different organs of Arabidopsis plants as described previously (Bonin et al., 1997). Root RNA was extracted from 2-week-old plants grown vertically under axenic conditions on agar plates containing a nutrient mixture as described by Haughn and Somerville (1986). Leaf RNA was extracted from 2-week-old plants, and RNA from stems, flowers, and siliques was extracted from 4-week-old plants. Northern blotting and transcript size determination was performed as described by Bonin et al. (1997) using 32P-labeled GMD1 or GMD2 probes. An NcoI/NotI fragment derived from pET28b/GMD1 and an NcoI/XhoI fragment derived from pET28b/GMD2, which correspond to the coding region of each gene, were used for random primer 32P labeling. Hybridizations and filter washings were done at 70°C using the buffer system described by Church and Gilbert (1984). To verify equal loading of the lanes, filters were stripped according to the protocol of the manufacturer (Amersham Pharmacia Biotech, Piscataway, NJ) and reprobed with a 32P-labeled EcoRI fragment derived from plasmid CD3–196 (obtained from the Arabidopsis Biological Resource Center, Ohio State University, Columbus), which contains most of an 18S rDNA gene from Arabidopsis.
Expression of the GMD1 Gene in E. coli
The sequences of oligonucleotide primers used for cloning of the GMD1 coding region into the pET28b expression vector (Novagen, Madison, WI) were as follows: GMD1/pET28b-upper, 5′-TCCCATGGCCTCCAGATCTCTCAATGGCG-3′; and GMD1/pET28b-lower, 5′-TAAGCGGCCGCCGGTTGCTGCTGAGCGTCC-3′, engineering an NcoI site into the upper primer and an NotI site into the lower primer. The PCR was performed in a volume of 30 μL with approximately 50 ng of template (wild-type Columbia DNA) using a model 2400 GeneAmp thermocycler and PCR Core Reagents (Perkin Elmer, Foster City, CA) with the following conditions: denaturation of the DNA at 95°C for 3 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 1 min, extension at 72°C for 1 min, and final extension at 72°C for 7 min. One unit of Taq DNA polymerase and 1 μg of each primer were used in the reaction, and the final concentration of MgCl2 was 2 mm.
The GMD1 PCR product was cloned into the pET28b expression vector as follows: A PCR fragment of the expected size was purified from an agarose gel using the Qiaquick gel extraction kit (Qiagen), digested with NcoI and NotI, and purified with the Qiaquick PCR purification kit. The pET28b vector was digested with NcoI and NotI and purified as above. Ligation of vector to insert DNA was followed by transformation of the construct into E. coli BL21 (DE3) competent cells according to the manufacturer's protocol (Stratagene).
Purification of the recombinant protein via Ni-NTA columns was done according to the manufacturer's protocol (Qiagen) with slight modifications of induction conditions. Culture flasks containing 50 mL of Luria-Bertani broth and 50 μg mL–1 of kanamycin were inoculated with 1 mL of an overnight culture and grown to an OD600 of approximately 0.6 at 37°C in a rotary shaker at 250 rpm. Isopropyl β-d-thiogalactoside was then added to a final concentration of 5 to 10 μm, and cultures were grown for an additional 3 h at ambient temperature (approximately 25°C).
Enzyme Extraction and Assay
Crude enzyme extracts of Arabidopsis were prepared as described earlier (Bonin et al., 1997). For assays of 4,6-dehydratase activity, 20 μL of these extracts containing approximately 0.2 mg of total protein was mixed with 926 Bq of GDP-d-[14C]Man contained in 10 μL of 20 mm Tris-HCl (pH 7.6) and incubated at 25°C overnight. NADPH was added to a final concentration of 5 mm for assaying the entire pathway from GDP-d-Man to GDP-l-Fuc. The reactions were terminated by boiling for 3 min, and precipitated protein was removed by centrifugation. All experiments using recombinant GMD1 protein (approximately 3 μg per reaction) were pre-incubated overnight at 25°C with 1 mm NAD+ (final concentration) in 20 mm Tris-HCl (pH 7.6) in a final volume of 30 μL. Enzymatic activity of the GMD1 protein was assayed by adding the above pre-incubation reaction to a tube containing 926 Bq of dried GDP-d-[14C]Man and incubating at 37°C for 90 min. Reactions were boiled, reduced with 1 μmol of NaBH4 at 37°C for 1 h, and hydrolyzed by addition of 100 μL of 2 m trifluoroacetic acid and incubation at 95°C for 20 min. Samples were dried under vacuum and resuspended in 15 μL of 70% (v/v) ethanol. Resultant monosaccharides were separated by thin-layer chromatography either on Baker-flex cellulose plates (J.T. Baker Inc., Phillipsburg, NJ), developed in 1-butanol:acetic acid:water (12:3:5 [v/v]), dried, and developed further by ethyl acetate:pyridine:water (8:2:1 [v/v]) in the same dimension. Radioactivity was visualized by phosphor imaging (Bio-Rad Laboratories, Hercules, CA), and authentic sugar standards run in parallel were stained with aniline-hydrogen phthalate (Fry, 1988). Quantification was performed with Molecular Analyst Software (Bio-Rad Laboratories). Control experiments were performed using transformed E. coli cells carrying the pET28b vector without an insert.
Construction of GMD1::GUS and GMD2::GUS Plants
To obtain regulatory sequences upstream of the GMD1 and GMD2 coding regions, bacterial artificial chromosomes (BACs) were screened using gene-specific probes. Texas A&M University and Institut für Genbiologische Forschung BAC filters provided by the Arabidopsis Biological Resource Center were screened with 32P-labeled GMD1 and GMD2 probes, respectively, leading to the identification of four Texas A&M University BACs hybridizing to the GMD1 probe and seven Institut für Genbiologische Forschung BACs hybridizing to the GMD2 probe. Restriction analysis and Southern hybridizations were performed with these BACs to determine the amount of sequence upstream of the start codon. A 2.2-kb PstI/XhoI fragment corresponding to GMD1, which contains 2.1 kb of sequence upstream of the start codon, and a 5.0-kb BglII/ClaI fragment corresponding to GMD2, which contains 3.5 kb of sequence upstream of the start codon, were cloned separately into pBluescript KS+ and transformed into E. coli XL1-Blue MRF1 as described above. To clone part of the 2.2-kb GMD1 fragment into pCAMBIA 1391Z (Center for the Application of Molecular Biology to International Agriculture, Canberra, Australia), PCR was performed using a T7 primer located in the vector and a GMD1-specific primer. The sequence of the oligonucleotide primer used for cloning of the GMD1 upstream region into the pCAMBIA 1391Z vector was as follows: PGDgus, 5′-AAGGATCCATCAGAAGAAATGATTGG-3′, incorporating a BamHI site. The last 18 nucleotides of this primer corresponded to positions –28 to –45 relative to the GMD1 translational initiation codon. The PCR product and vector were then cleaved with BamHI and PstI, purified, ligated, and transformed first into E. coli XL1-Blue MRF1, followed by electroporation into Agrobacterium tumefaciens GV3101 using a Bio-Rad Genepulser. One of several kanamycin-resistant colonies was then used to transform Arabidopsis via vacuum infiltration (Bechtold et al., 1993). T1 plants were selected on plates containing Murashige and Skoog salts (Murashige and Skoog, 1962), 1% (w/v) Suc, 25 μg mL–1 hygromycin, and 500 μg mL–1 vancomycin. These were then transferred to soil after 10 d and allowed to self. T2 plants were again selected on hygromycin, transferred to soil, and allowed to self.
The regulatory sequences of the GMD2 gene were cloned in a similar fashion except that a GMD2-specific primer and the pCAMBIA 1381Z vector were used. The sequence of the oligonucleotide primer used for cloning of the GMD2 upstream region into the pCAMBIA 1381Z vector was as follows: PMRgus, 5′-AATTGTCGACGGATCTGGGATTTCAGAG-3′ incorporating a SalI site. The last 19 nucleotides of this primer corresponded to positions –13 to –31 relative to the GMD2 translational initiation codon. The remaining steps for cloning were the same as for the GMD1 construct described above except that SalI and BamHI were used.
Staining for GUS Activity
Promoter::GUS plants were stained for GUS activity as described by Jefferson et al. (1987) using X-Gluc CHA salt (Inalco Pharmaceuticals, San Luis Obispo, CA) at 37°C for 1 to 2 d. Plants were then dehydrated in an ethanol series (15%, 30%, 50%, then 70% [v/v] ethanol; 30 min for each step) and stored in this solution at room temperature.
Immunocytochemistry
Immunolocalization of the Fuc-containing CCRC-M1 epitope was carried out as described by Freshour et al. (1996, 2003).
Acknowledgments
We would like to thank J. Peter Gogarten for the λEMBL3 genomic library, the Arabidopsis Biological Research Center for providing DNA clones, and the Center for the Application of Molecular Biology to International Agriculture (Canberra, Australia) for plant transformation vectors.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.022368.
This work was supported by the Department of Energy's Energy Biosciences Program (grant nos. DE–FG02–95ER20203 and DE–FG02–96ER20220).
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bacic A, Harris PJ, Stone BA (1988) Structure and function of plant cell walls. In PK Stumpf, EE Conn, eds, The Biochemistry of Plants, Vol 14. Academic Press, New York, pp 297–371 [Google Scholar]
- Baldo BA, Reid AL, Boniface PA (1983) Lectins as cytochemical probes of the developing wheat grain: IV. Demonstration of mucilage containing l-fucose associated with roots in ungerminated grain. Aust J Plant Physiol 10: 459–470 [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci 316: 1194–1199 [Google Scholar]
- Bonin CP, Potter I, Vanzin GF, Reiter W-D (1997) The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-d-mannose-4,6-dehydratase catalyzing the first step in the de novo synthesis of GDP-l-fucose. Proc Natl Acad Sci USA 94: 2085–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin CP, Reiter W-D (2000) A bifunctional epimerase-reductase acts downstream of the MUR1 gene product and completes the de novo synthesis of GDP-l-fucose in Arabidopsis. Plant J 21: 445–454 [DOI] [PubMed] [Google Scholar]
- Bowman J (1994) Arabidopsis. An Atlas of Morphology and Development. Springer Verlag, New York
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feingold DS, Avigad G (1980) Sugar nucleotide transformations in plants. In PK Stumpf, EE Conn, eds, The Biochemistry of Plants, Vol 2. Academic Press, New York, pp 101–170 [Google Scholar]
- Freshour G, Bonin CP, Reiter W-D, Alberheim P, Darvill AD, Hahn MG (2003) Distribution of fucose-containing xyloglucans in cell walls of the mur1 mutant of Arabidopsis. Plant Physiol 131: 1602–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freshour G, Clay RP, Fuller MS, Albersheim P, Darvill AG, Hahn MG (1996) Developmental and tissue-specific structural alterations of the cell wall polysaccharides of Arabidopsis thaliana roots. Plant Physiol 110: 1413–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC (1988) The growing plant cell wall: chemical and metabolic analysis. John Wiley and Sons, New York
- Greenland DJ (1979) The physics and chemistry of the soil-root interface: some comments. In JL Harley, RS Russel, eds, The Soil-Root Interface. Academic Press, London, pp 83–89
- Haughn GW, Somerville C (1986) Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet 204: 430–434 [Google Scholar]
- Jefferson RA, Kavanaugh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 13: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulichak AM, Bonin CP, Reiter W-D, Garavito RM (2002) Structure of the MUR1 GDP-mannose 4,6-dehydratase from Arabidopsis thaliana: implications for ligand binding and specificity. Biochemistry 41: 15578–15589 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Ohyama C, Smith PL, Angata K, Fukuda MN, Lowe JB, Fukuda M (1998) Molecular cloning and expression of GDP-d-mannose-4,6-dehydratase, a key enzyme for fucose metabolism defective in Lec13 cells. J Biol Chem 273: 14582–14587 [DOI] [PubMed] [Google Scholar]
- O'Neill MA, Eberhard S, Albersheim P, Darvill AG (2001) Arabidopsis growth requires borate cross-linking of the cell wall pectic polysaccharide rhamnogalacturonan II. Science 294: 846–849 [DOI] [PubMed] [Google Scholar]
- Puhlmann J, Bucheli E, Swain MJ, Dunning N, Albersheim P, Darvill AG, Hahn MG (1994) Generation of monoclonal antibodies against plant cell wall polysaccharides: I. Characterization of a monoclonal antibody to a terminal (1→2)-linked fucosyl-containing epitope. Plant Physiol 104: 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter W-D, Chapple CCS, Somerville CR (1993) Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science 261: 1032–1035 [DOI] [PubMed] [Google Scholar]
- Reiter W-D, Chapple CCS, Somerville CR (1997) Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J 12: 335–345 [DOI] [PubMed] [Google Scholar]
- Reiter W-D, Vanzin GF (2001) Molecular genetics of nucleotide sugar interconversion pathways in plants. Plant Mol Biol 47: 95–113 [PubMed] [Google Scholar]
- Rougier M (1981) Secretory activity of the root cap. In W Tanner, FA Loewus, eds, Encyclopedia of Plant Physiology, New Series, Vol 13B, Plant Carbohydrates II. Springer-Verlag, Berlin, pp 542–574 [Google Scholar]
- Sambrook J, Fritsch EF, Maniatas T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sturla L, Bisso A, Zanardi D, Benatti U, De Flora A, Tonetti M (1997) Expression, purification and characterization of GDP-d-mannose-4,6-dehydratase from Escherichia coli. FEBS Lett 412: 126–130 [DOI] [PubMed] [Google Scholar]
- Sullivan FX, Kumar R, Kriz R, Stahl M, Xu G-Y, Rouse J, Chang X-j, Boodhoo A, Potvin B, Cumming DA (1998) Molecular cloning of human GDP-mannose 4,6-dehydratase and reconstitutions of GDP-fucose biosynthesis in vitro. J Biol Chem 273: 8193–8202 [DOI] [PubMed] [Google Scholar]
- Tonetti M, Sturla L, Bisso A, Benatti U, DeFlora A (1996) Synthesis of GDP-l-fucose by the human FX protein. J Biol Chem 271: 27274–27279 [DOI] [PubMed] [Google Scholar]