Abstract
Histone proteins play a central role in chromatin packaging, and modification of histones is associated with chromatin accessibility. SET domain [Su(var)3-9, Enhancer-of-zeste, Trithorax] proteins are one class of proteins that have been implicated in regulating gene expression through histone methylation. The relationships of 22 SET domain proteins from maize (Zea mays) and 32 SET domain proteins from Arabidopsis were evaluated by phylogenetic analysis and domain organization. Our analysis reveals five classes of SET domain proteins in plants that can be further divided into 19 orthology groups. In some cases, such as the Enhancer of zeste-like and trithorax-like proteins, plants and animals contain homologous proteins with a similar organization of domains outside of the SET domain. However, a majority of plant SET domain proteins do not have an animal homolog with similar domain organization, suggesting that plants have unique mechanisms to establish and maintain chromatin states. Although the domains present in plant and animal SET domain proteins often differ, the domains found in the plant proteins have been generally implicated in protein-protein interactions, indicating that most SET domain proteins operate in complexes. Combined analysis of the maize and Arabidopsis SET domain proteins reveals that duplication of SET domain proteins in plants is extensive and has occurred via multiple mechanisms that preceded the divergence of monocots and dicots.
Transcriptional regulation in eukaryotes is orchestrated by a combination of trans-acting factors that recognize cis-DNA elements acting in concert with temporal and spatial variation in the chromatin environment of a gene. Factors determining the expression potential of the chromatin environment include DNA modifications, histone modifications, and the composition of associated proteins (Pirrotta, 1998; Cheung et al., 2000; Strahl and Allis, 2000; Jenuwein and Allis, 2001). Chromatin states are important for determining gene expression potential in developmental regulation and for epigenetic silencing. For example, mutants in several plant chromatin proteins have been identified on the basis of their effect on plant development (Goodrich et al., 1997; Grossniklaus et al., 1998; Luo et al., 1999; Ohad et al., 1999; Gendall et al., 2001; Kaya et al., 2001; Yoshida et al., 2001; Wagner and Meyerowitz, 2002). Mutations in chromatin proteins have also been shown to affect epigenetic silencing in plants (Finnegan et al., 1996; Ronemus et al., 1996; Jeddeloh et al., 1998; Lindroth et al., 2001). Although numerous examples exist of chromatin level control of gene expression in plants, the specific details of molecular mechanisms controlling plant chromatin states remain poorly understood. Histone modification is emerging as a central theme in the control of chromatin states across organisms.
The observation that a complex system of histone modifications is important in controlling chromatin state has led to the histone code hypothesis (Strahl and Allis, 2000; Jenuwein and Allis, 2001). The N-terminal tails of the core histone proteins are highly conserved in eukaryotes and contain Lys, Arg, and Ser residues that are targets for posttranslational modifications including acetylation, methylation, phosphorylation, and ubiquitination (Strahl and Allis, 2000; Jenuwein and Allis, 2001). These posttranslational modifications can influence other modifications, directly influence the chromatin structure, or alter the composition of chromatin-associated proteins at a locus (Wu and Grunstein, 2000; Jenuwein and Allis, 2001; Zhang and Reinberg, 2001). There are examples of interactions between histone modifications such as ubiquitination leading to methylation (Dover et al., 2002; Sun and Winston, 2002) and methylation influencing histone acetylation (Bernstein et al., 2002). The histone tails serve as a repository for temporary information storage in the form of modifications. One group of proteins that are involved in histone modification is the SET domain proteins.
SET domain proteins have been found in chromatin-associated complexes that play a role in either promoting or inhibiting gene expression (Francis and Kingston, 2001). In Drosophila melanogaster, the Polycomb group (PcG) proteins maintain transcriptionally silent states throughout development (Simon, 1995), whereas Trithorax group (trxG) proteins maintain a transcriptionally active state (Francis and Kingston, 2001). The E(Z) (ENHANCER OF ZESTE) SET domain protein is a PcG protein that is present in a complex with another PcG protein, ESC (EXTRA SEX COMBS) and a histone deacetylase, RPD3 (van der Vlag and Otte, 1999; Tie et al., 2001). Two trxG SET domain proteins, TRX (TRITHORAX) and ASH1 (ABSENT OR SMALL HOMEOTIC DISCS1) have been found in a complex with the histone acetylase CBP (Bantignies et al., 2000; Petruk et al., 2001).
Biochemical evidence from a number of studies indicates that the SET domain proteins can methylate histones. Homologs of the SU(VAR)3-9 SET domain protein in mammals and Schizosaccharomyces pombe methylate the Lys-9 residue of histone H3 (Rea et al., 2000; Nakayama et al., 2001), creating a specific binding site for the chromodomain of HP1-like proteins (Bannister et al., 2001; Lachner et al., 2001; Nakayama et al., 2001). A related SET domain protein from animals, G9a, methylates histone H3 at Lys-9 and Lys-27 (Tachibana et al., 2001). The Neurospora crassa DIM-5 protein, which is similar to SU(VAR)3-9 and G9a, also methylates Lys-9 of histone H3 (Tamaru and Selker, 2001). Interestingly, the dim-5 mutation causes reductions in cytosine DNA methylation, providing a link between histone modification and DNA modification. Other examples of SET proteins that function as histone methyltransferases include: yeast (Saccharomyces cerevisiae) SET1, which methylates Lys-4 of histone H3 (Briggs et al., 2001; Roguev et al., 2001; Nagy et al., 2002); yeast SET2, which methylates Lys36 of histone H3 (Strahl et al., 2002); D. melanogaster ASH1 protein, which methylates Lys-4 and Lys-9 of histone H3 (Beisel et al., 2002); mammalian ESET, which methylates histone H3 (Yang et al., 2002); and mammalian SET7, which methylates Lys-4 of histone H3 (Wang et al., 2001).
Genetic studies have provided evidence that SET domain proteins are important for developmental and epigenetic regulation of gene expression. The PcG and trxG proteins act to stabilize transcriptional states during development. Mutations in SET domain proteins produce both PcG [E(z), Jones and Gelbart, 1990] and trxG (ash1, Tripoulas et al., 1994; and trx, Ingham and Whittle, 1980) phenotypes. Mutations in the SET domain gene, Su(var)3-9, result in suppression of position effect variegation (Tschiersch et al., 1994). The yeast SET1 protein is required for rDNA silencing (Briggs et al., 2001; Bryk et al., 2002), but the modification catalyzed by SET1p is highly correlated with transcribed regions of the yeast genome (Bernstein et al., 2002). For several SET domain proteins, mutations that affect the histone methyltransferase activity are associated with the mutant phenotype, indicating that histone methylation is a required biological function of these proteins (Rea et al., 2000; Nakayama et al., 2001; Tamaru and Selker, 2001; Beisel et al., 2002).
In plants, two proteins containing an SET domain, CLF (CURLY LEAF) and MEA (MEDEA), were identified by the developmental phenotype associated with loss-of-function mutations (Goodrich et al., 1997; Grossniklaus et al., 1998). CLF and MEA are related to the PcG protein, E(Z) (Goodrich et al., 1997; Grossniklaus et al., 1998), and homologs of these proteins have been identified in maize (Zea mays; Springer et al., 2002). A third SET domain protein, KYP (KRYPTONITE), was identified as a second-site suppressor of epigenetic silencing of SUP (Jackson et al., 2002). Mutations in the KYP protein result in reductions in genomic DNA methylation levels, with CpNpG sites showing greater reductions than CpG sites (Jackson et al., 2002). The Arabidopsis genome contains an additional 29 proteins with an SET domain (Baumbusch et al., 2001). The Baumbusch et al. (2001) study performed a phylogenetic analysis of 28 Arabidopsis SET domain proteins along with sequences representing four types of animal SET proteins. Based on this analysis, they divided Arabidopsis SET domain proteins into four classes, each named for the most closely related D. melanogaster protein. Another recent study characterized the relationship of several subgroups of Arabidopsis SET domain proteins with each other and with SET domain proteins from animal species (Alvarez-Venegas and Avramova, 2002).
The objective of this study was to analyze SET domain-containing proteins from maize and Arabidopsis using phylogenetic analysis and interpretations based on protein organization. In our analysis, we included sequences representing all orthology groups of D. melanogaster, mouse (Mus musculus), and yeast SET domain proteins and 22 SET domain proteins from maize. The addition of another plant species and additional proteins from non-plant species, together with a thorough analysis of all domains in these proteins, revealed additional classes of SET domains in plants.
RESULTS
BLASTP and TBLASTN searches identified 32 proteins containing an SET domain and five proteins containing an interrupted S-ET domain in the Arabidopsis genome (Table I). The Arabidopsis SET domain proteins that we identified were the same as those reported by Baumbusch et al. (2001). We have determined an alternative annotation relative to the predicted annotation available at the GenBank accession for 14 of the Arabidopsis proteins based on cDNA sequences or alignments to other SET domain proteins (Table I), which was confirmed using RT-PCR, EST, or ortholog alignments. Twenty-five maize SET domain genes were identified through searches of EST databases. The sequences of 21 of these genes have been extended or completed by further sequencing of EST clones or through RACE analysis. Each of the proteins was assigned a name, SDGX, with X being a number assigned based on the order each gene was discovered. The existing synonyms are listed in Tables I and II. The Arabidopsis SET-domain containing proteins are labeled SDG followed by a number less than 100, and the maize SET proteins are labeled SDG followed by a number greater than 100. Sequence, expression, and map information for the maize and Arabidopsis SET domain genes are located at the ChromDB Web site (www.chromdb.org) and are updated regularly.
Table I.
Arabidopsis SET domain proteins
| Gene | Other Names | Length | Accession No.a | AGI Locus No. | Expressionb | Class/Orthology Group |
|---|---|---|---|---|---|---|
| SDG1 | CLF | 902 | CAA71599 | At2g23380 | n/c | I/2 |
| SDG2 | ATXR3 | 2,283 | CAB10297c | At4g15180 | n/e/r | III/3 |
| SDG3 | SUVH2 | 651 | AAK28967 | At2g33290 | n/e/r | V/3 |
| SDG4 | ASHR3 | 497 | AAD10162*c | At4g30860 | e/c | II/2 |
| SDG5 | MEA | 689 | AAC39446 | At1g02580 | c | I/1 |
| SDG6 | SUVR5 | 1,114 | AAC17088*c | At2g23750 | e/r | V/7 |
| SDG7 | ASHH3 | 363 | AAC23419*c | At2g44150 | n/e/r | II/1 |
| SDG8 | ASHH2 | 1,792 | AAC34358c | At1g77300 | n/e/r | II/3 |
| SDG9 | SUVH5 | 794 | AAK28970 | At2g35160 | e/r | V/5 |
| SDG10 | EZA1 | 856 | AAD09108* | At4g02020 | c/n/e/r | I/3 |
| SDG11 | SUVH10 | 312 | AAC95167*c | At2g05900 | r | V/1 |
| SDG13 | SUVR1 | 734 | AAD10665 | At1g04050 | c/r | V/6 |
| SDG14 | ATX3 | 967d | CAB71104c | At3g61740 | e/r | III/2 |
| SDG15 | ATXR5 | 379d | CAB89351* | At5g09790 | c/e/r | IV/1 |
| SDG16 | ATX4 | 981d | CAB36760*c | At4g27910 | e/r | III/2 |
| SDG17 | SUVH7 | 693 | AAK28972* | At1g17770 | c/r | V/1 |
| SDG18 | SUVR2 | 717 | AAK92218* | At5g43990 | c/n/r | V/6 |
| SDG19 | SUVH3 | 669 | AAK28968 | At1g73100 | c/n/e/r | V/1 |
| SDG20 | SUVR3 | 354 | AAF00642 | At3g03750 | n/e/r | V/4 |
| SDG21 | SUVH8 | 755 | AAK28973 | At2g24740 | c/r | V/1 |
| SDG22 | SUVH9 | 650 | AAK28974 | At4g13460 | c/e/n/r | V/3 |
| SDG23 | SUVH6 | 790 | AAK28971* | At2g22740 | c/e/r | V/2 |
| SDG24 | ASHH4 | 352 | CAB75815 | At3g59960 | - | II/1 |
| SDG25 | ATXR7 | 1,421 | BAB10481 | At5g42400 | c/n/r | III/4 |
| SDG26 | ASHH1 | 492 | AAF04434*c | At1g76710 | e/r | II/3 |
| SDG27 | ATX1 | 1,062 | AAK01237 | At2g31650 | c/r | III/1 |
| SDG29 | ATX5 | 1,040 | BAA97320 | At5g53430 | n/e/r | III/2 |
| SDG30 | ATX2 | 1,063d | AAF29390*c | At1g05830 | c/n/e/r | III/1 |
| SDG31 | SUVR4 | 492 | AAF63769*c | At3g04380 | c | V/6 |
| SDG32 | SUVH1 | 670 | AAK28966 | At5g04940 | c/e | V/1 |
| SDG33 | KYP, SUVH4 | 624 | AAK28969 | At5g13960 | c/e | V/2 |
| SDG34 | ATXR6 | 349 | BAB10399 | At5g24330 | n/r | IV/1 |
| SDG35 | ATXR1 | 545 | AAF87042c | At1g26760 | e/r | N.A. (S-ET)e |
| SDG36 | ATXR2 | 559 | BAB02844c | At3g21820 | e | N.A. (S-ET)e |
| SDG37 | ASHR1 | 447 | AAD03568 | At2g17900 | - | N.A. (S-ET)e |
| SDG38 | ATXR4 | 325 | BAB11410*c | At5g06620 | e | N.A. (S-ET)e |
| SDG39 | ASHR2 | 341 | AAD10162 | At2g19640 | c/e | N.A. (S-ET)e |
For all accession nos. followed by an asterisk, we have chosen a model different from the predicted annotation. Our model is presented at http://www.chromdb.org
Any evidence for expression is indicated by n (northern blot available at chromdb.org), c (cloned cDNA available at Genbank), e (expressed sequence tag [EST]), or r (reverse transcriptase [RT]-PCR; C. Napoli, unpulished data)
The protein model presented at http://www.chromdb.org is distinct from that of Baumbusch et al., 2001
Evidence for alternative splicing resulting in two different lengths of proteins is presented at http://www.chromdb.org
The S-ET domain genes were not assigned to orthology groups
Table II.
Maize SET domain genes
| Gene | Synonyms | Lengtha | Accession No.b | Class/Orthology Group | Estimated Copy No.c | Map Position |
|---|---|---|---|---|---|---|
| SDG101 | 508* | AW091195b | V/1 | 1 | 7.02 | |
| SDG102 | 513 | AY122273 | II/3 | 4 | 2.04, 6.01, 6.02 | |
| SDG103 | 955* | AI987233b | V/2 | 1 | N.A. | |
| SDG104 | 501 | AY122272 | V/2 | 1 | 2.03 | |
| SDG105 | 678 | AY093419 | V/1 | 2 | 8.04 | |
| SDG106 | 248* | AI065600b | III/1 | 1 | 2.07 | |
| SDG107 | 783* | AI782865b | V/6 | 1 | 2.03 | |
| SDG108 | 469* | AI855041b | III/3 | 2 | 4.05, 10.03 | |
| SDG110 | 342 | AF545814 | II/1 | Complex | 7.02 | |
| SDG111 | 486 | AY187718 | V/2 | 2 | 6.06 | |
| SDG113 | 766 | AF545813 | V/1 | 1 | 3.06 | |
| SDG115 | 1,032* | BE225019b | III/2 | 2 | 3.08 | |
| SDG116 | 222* | BE575075b | V/4 | 5 | 2.07, 7.03 | |
| SDG117 | 1198 | AY187719 | V/7 | 2 | 3.07, 5.06 | |
| SDG118 | 696 | AY122271 | V/2 | 2 | 8.06 | |
| SDG119 | 418* | BG838020b | V/2 | 2 | 2.06 | |
| SDG122 | 109* | AI820207b | N.A. (S-ET)d | N.A. | 1.08 | |
| SDG123 | 303 | AY172976 | N.A. (S-ET)d | N.A. | 6.01 | |
| SDG124 | MEZ1 | 933 | AF443596 | I/2 | 1 | 6.01-6.02 |
| SDG125 | MEZ2 | 893 | AF443597 | I/3 | 2 | 9.04 |
| SDG126 | MEZ3 | 896 | AF443598 | I/3 | 2 | N.A. |
| SDG127 | 132* | BM500594b | III/4 | N.A. | N.A. | |
| SDG128 | 121* | BM501397b | III/1 | N.A. | N.A. | |
| SDG129 | 40* | BM736459b | IV/1 | N.A. | N.A. | |
| SDG130 | 410 | AAL75997 | N.A. (S-ET)d | N.A. | 5.02 |
The amino acid length of the protein sequence is indicated. All sequences denoted by an asterisk are partial sequences
The accession nos. in bold font represent full-length coding sequences. All accession nos. in italics represent a single EST representing this gene. For information about the other ESTs and additional sequence derived by RACE, see http://www.chromdb.org
The S-ET domain proteins were not assigned to orthology groups
The copy no. was for each gene was estimated based on DNA gel-blot analysis using two genotypes with six different restriction enzymes. In each case, we based our estimate upon the restriction enzymes showing the fewest no. of cross-hybridizing sequences
Plants Contain a Class of Proteins with Interrupted SET Domains
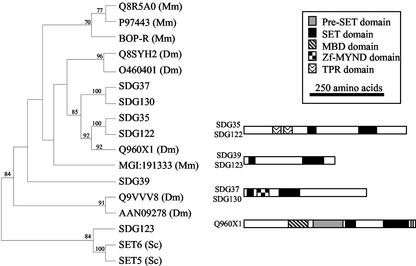
In addition to documenting a large family of plant proteins containing an intact SET domain, our analysis also revealed the presence of plant proteins containing a disrupted SET domain in which the N-terminal one-third of the SET domain is separated from the C-terminal two-thirds of the domain by 50 to 120 amino acids. Jenuwein and Allis (2001) referred to the disrupted SET domains as S-ET domains. The insertion of 50 to 120 amino acids in the plant S-ET domain sequences made it difficult to align these sequences with other SET domains. The S-ET domain proteins include the Arabidopsis SDG35, SDG36, SDG37, SDG38, and SDG39 proteins and the maize SDG122, SDG123, and SDG130 proteins. We were not able to investigate the relationship of SDG36 and SDG38 to other S-ET proteins because these proteins do not contain regions with significant similarity to the N-terminal portion of the SET domain. An alignment of the other plant S-ET proteins with animal S-ET proteins was used to determine the relationships between these proteins (Fig. 1). There is evidence for at least three orthology groups predating the divergence of monocots and dicots within the S-ET proteins. These orthology groups are SDG35/SDG122, SDG37/SDG130, and possibly SDG39/123, although there is less evidence for a close relationship of the last two proteins. The phylogenetic analysis indicates that the SDG35/SDG122 and SDG37/SDG130 have a single closest relative in animals. The domain architecture of the SDG35/SDG122, SDG37/SDG130 and closest D. melanogaster relative, Q960X1, proteins are quite distinct (Fig. 1). The SDG35/122 proteins contain an interrupted S-ET domain only. The SDG37/SDG130 proteins contain a Zf-MYND domain within the sequence that interrupts the SET domain. The D. melanogaster Q960X1 protein contains a methyl-binding domain pre-SET and S-ET domain. There is evidence that some S-ET domains retain the ability to catalyze protein methylation (Klein and Houtz, 1995; Zhang and Reinberg, 2001; Yang et al., 2002). The rest of the discussion will focus on the analysis of the SET domain proteins.
Figure 1.
Proteins containing an interrupted SET domain were aligned using ClustalW, and the alignment was analyzed by parsimony using PHYLIP. The resulting phylogenetic tree is displayed with all bootstrap values >70% indicated at the nodes. The domain organization of representative plant proteins and the most closely related animal protein were investigated using National Center for Biotechnology Information (NCBI)-conserved domain database (CD) and SMART searches. All proteins are displayed as scaled schematic diagrams with the N terminus at the left. Shaded boxes within the protein schematics indicate recognizable domains. The accession numbers for the sequences used in the alignment are Q8R5A0-Mm (AAH23119), P97443-Mm (BAB26947), BOP-R-Mm (NP_081464), Q8SYH2-Dm (AAL49177), O46040-Dm (O46040), Q960X1-Dm (AAK93223), MGI:191333-Mm (XP_134310), Q9VVV8-Dm (AAF49199), AAN09278-Dm (AAN09278), ScSET5 (P38890), and ScSet6 (NP_015160).
Plants Contain Five Classes of SET Domain Proteins
We sought to classify the SET domain proteins of maize and Arabidopsis on the basis of phylogenetic analyses and domain organization. Thirty-one Arabidopsis, 19 maize, eight D. melanogaster, 12 mouse, and four yeast proteins were included in our phylogenetic analysis (SDG11 from Arabidopsis was not included because we could not align the full SDG domain; SDG108, SDG128 and SDG129 from maize were not included because we do not have sequence for the entire SET domain; none of the S-ET sequences were included from any species; in the first alignment, we used an additional six D. melanogaster and five mice genes that did not cluster with any of the plant groups within the analysis, and these sequences were removed for the final analysis). The SET domain of each protein, bounded by GWG on the N terminus and TYDY on the C terminus, was aligned using ClustalW (see Supplementary Fig. 1 at http://www.plantphysiol.org). The structure of four different SET domain proteins has been determined (Min et al., 2002; Trievel et al., 2002; Wilson et al., 2002; Zhang et al., 2002). The majority of the plant SET domain proteins show conservation to regions of the structures important for substrate interactions and secondary structure (see Supplementary Fig. 1 at http://www.plantphysiol.org). This alignment was then analyzed to find a parsimonious tree using PHYLIP (Fig. 2). This phylogenetic tree supported the existence of five distinct classes of SET domain proteins in plants. Four of the classes identified by our analysis agree with the classes identified by Baumbusch et al. (2001), whereas our class IV represents sequences not included in the phylogenetic analysis performed by Baumbusch et al. (2001).
Figure 2.
The SET domains from maize and Arabidopsis SET domain proteins were aligned with the SET domain of yeast, D. melanogaster, and mouse proteins using ClustalW (see Supplementary Fig. 1 at www.plantphysiol.org). All bootstrap values >70% are indicated at the nodes. The accession numbers for the plant SET domain sequences are shown in Tables I and II. The D. melanogaster proteins used for this alignment were E(Z) (AAC46462), CG4976 (AAF56762), CG17396 (AAF45425), ASH1 (AAF49140), MES-4 (AAK84931), TRX (AAF55041), TRR (AAF45684), G9a-like (AAF45487), and SU(VAR3-9) (CAB93768). The mouse proteins used for the alignment are EZH1 (AAC50778), EZH2 (Q61188), NSD1 (AAC40182), ASH1 (AAK26242), HRX (AAA62593), MLL3-like (AAK70214), G9a (AAC84164), MLL2-like (BAB27589), Q91Z33 (AAH10250) SET7 (Q9NQR1), SUV39h1 (AF193862), and Suv39h2 (AAG09134). The yeast sequences used for the alignment are ScSET1 (AAB68867), ScSET2 (NP_012367), ScSET3 (NP_012954), and ScSET4 (NP_012430).
We divided the SET domain proteins of plants into five classes on the basis of this phylogenetic analysis and the domain organization of plant proteins within a clade (indicated in Fig. 2). Some classes, such as class I, contain plant and animal proteins that are conserved across all domains of the protein (data not shown). Other classes, such as class II, are conserved only in the SET domain, whereas the organization of domains outside of the SET domain in the plant proteins and the overall length of the protein are quite different from the most closely related animal proteins (Fig. 3). We have defined the presence of 19 putative orthology groups of SET domain genes in plants. The term orthology group will be used to refer to a group of proteins that are likely to have evolved from a single progenitor present in the last common ancestor of maize and Arabidopsis. These groups were inferred based upon the phylogenetic analysis in this study and relationships with sequences from other plant species. This more detailed level of evolutionary interpretation was possible relative to previous studies (Baumbusch et al., 2001) because of the inclusion of both maize and Arabidopsis proteins and additional proteins from non-plant species.
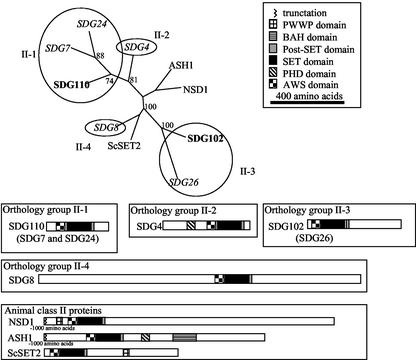
Figure 3.
Class II SET domain proteins. The SET domain protein sequence from the plant class II SET domain proteins and several well-characterized animal class II SET domain proteins were aligned using ClustalW. The relationships of these sequences were investigated using PHYLIP using bootstrap analysis. All bootstrap values >70% are shown. The circles indicate the putative orthology groups. The Arabidopsis proteins are indicated by italic text, and the maize proteins are indicated by bold text. The domain organization of the plant proteins was investigated using NCBI-CD and SMART searches. All proteins are displayed as scaled schematic diagrams with the N terminus at the left. Shaded boxes within the protein schematics indicate recognizable domains. Due to their longer lengths, the animal NSD1 and ASH1 protein schematics are truncated at the N terminus; the sequence not shown does not contain any recognizable domains.
Class I SET Domain Proteins
The class I SET domain proteins, which include the D. melanogaster PcG protein E(Z) and the Arabidopsis CLF (SDG1) and MEA (SDG5) proteins, have been well characterized in plants and animals. A limited expansion of class I proteins has occurred in plants, with two orthology groups of class I proteins present in both maize and Arabidopsis. A third type of class I SET domain protein, represented by MEDEA in Arabidopsis, has only been found in dicots to date (Springer et al., 2002).
The sequence characteristics of the plant class I SET domain proteins have been previously described (Goodrich et al., 1997; Grossniklaus et al., 1998; Springer et al., 2002). The class I SET domain proteins contain five domains that have been conserved between plants and animals (Springer et al., 2002). The EZD1 (Enhancer of zeste domain1) and EZD2 (Enhancer of zeste domain2) are present only in E(z)-like proteins and do not have a known function. The class I proteins also contain a SANT (SWI3, ADA2, N-CoR, and TFIIIB DNA-binding domains), Cys-rich, and SET domain. The SANT domain is a nonspecific DNA-binding domain (Aasland et al., 1996). The Cys-rich region of E(z)-like proteins contains 15 invariant Cys residues with a conserved spacing pattern (Springer et al., 2002).
Class II SET Domain Proteins
On the basis of domain organization outside the SET domain, the proteins included in class II are a structurally diverse group, both between plant orthology groups and between plants and animals (Fig. 3). The animal proteins in class II include the D. melanogaster ASH1 (Tripoulas et al., 1994, 1996) and mouse NSD1 (Huang et al., 1998). Many of the animal proteins in this class are long proteins (1,000 amino acids) and contain other domains including PWWP (domain containing Pro-Trp-Trp-Pro motif), PHD (plant homeodomain), Bromodomain, and BAH domains in addition to the conserved AWS, SET, and PostSET domains.
Although the support for the clustering of all class II sequences was much lower than the support for other classes, there are several common features of the plant and animal class II SET domain proteins that make it logical to consider all of these sequences as a single class. All class II SET domain proteins (except SDG4) contain an AWS domain located just N terminal of the SET domain. The AWS domain is a subdomain of the PreSET domain that contains several highly conserved Cys residues. Another common feature of the plant and animal class II proteins is the location of the SET domain. In all other classes, the SET domain is found very near the C terminus of the protein, whereas the SET domain of class II proteins is more centrally located. The relationship of the SET domain location and common presence of AWS domains in plant and animal class II SET domain proteins suggests that they are likely to be related based on origin and function.
The plant class II proteins were characterized based on overall structure and phylogenetic relationships generated from the SET domain (Fig. 3). Based on this analysis, there are four orthology groups of plant class II proteins. Orthology group II-1 proteins are relatively short (approximately 350 amino acids) and contain an SET domain along with AWS and PostSET domains (Fig. 3). The SDG7 and SDG24 genes are located in collinear duplicated regions of the Arabidopsis genome on chromosomes 2 and 3. The maize gene, Sdg110, is most closely related to SDG7 and SDG24 and contains a similar organization of domains (Fig. 3).
A single Arabidopsis sequence, SDG4, represents the second orthology group (II-2) of class II SET domain proteins. The SET domain of SDG4 is similar to the SET domains of orthology group II-1 proteins, but the N-terminal and C-terminal regions of the proteins are different. The N-terminal extension contains a PHD zinc finger domain. PHD domains are found in a number of chromatin-associated proteins and are thought to be involved in protein-protein interactions important in the assembly of multiprotein complexes (Aasland et al., 1995). The SDG4 protein is the only class II SET domain protein from plants that does not contain the AWS domain.
Orthology group II-3 of the plant class II SET domain proteins includes SDG26 from Arabidopsis and SDG102 from maize. The SET and PostSET domains of orthology group II-3 proteins are located near the N terminus or in the middle of the protein (Fig. 3). The alignment of SDG8 and SDG102 shows significant conservation in an approximately 80-amino acid Cys-rich region located on the N-terminal side of the SET domain.
The final orthology group of class II proteins is represented by SDG8, which is a long protein (1,767 amino acids) with both C- and N-terminal extensions relative to SDG26 and SDG102 (the C-terminal extension has been supported by EST data, whereas the N-terminal extension is based upon a predicted annotation). We were able to find a rice genomic sequence, AP004876, which is more closely related to SDG8 than it is to SDG102 or SDG26. It is likely that there is a maize gene belonging to orthology group II-4 that has not yet been detected by EST sequencing projects.
Class III SET Domain Proteins
The class III SET domain proteins include the D. melanogaster TRX (TRITHORAX) and TRR (TRITHORAX-RELATED) proteins, the mouse HRX and MLL3-like proteins, and the yeast SET1 protein. Two Arabidopsis homologs of Trx were previously identified and named ATX1 (SDG27) and ATX2 (SDG30; Alvarez-Venegas and Avramova, 2001). The authors documented the presence of a conserved DAST (Domain Associated with SET in Trithorax; referred to as FYR [Phe-Tyr-rich domain] by the SMART database) domain found in all plant and animal TRX proteins. The findings of Baumbusch et al. (2001) and our study show that plants contain additional proteins similar to TRX.
Analysis of the plant class III SET domain proteins supports the existence of four orthology groups (Fig. 4). Orthology group III-1 includes SDG27 and SDG30, which both contain a similar arrangement of domains including a PWWP domain, an FYR domain (named DAST by Alvarez-Venegas and Avramova, 2001), and two PHD domains (Fig. 4). SDG27 and SDG30 are found in regions of Arabidopsis chromosomes 1 and 2 that are collinear duplicated regions. The domain structure and expression pattern of these genes was characterized by Alvarez-Venegas and Avramova (2001). The PWWP domain is predicted to be involved in mediating protein-protein interactions in proteins that are regulators of cell growth and differentiation (Stec et al., 2000). The FYR domain is composed of an FYR-C terminal portion and an FYR-N terminal portion that often occur near each other but can be separated (Schultz et al., 2000). The only FYR domains present in the mouse and D. melanogaster genome are present in class III SET domain proteins. In plants, two types of proteins contain FYR domains, the SDG27/30 proteins and a group of jumonji-domain proteins (Balciunas and Ronne, 2000; Alvarez-Venegas and Avramova, 2001). The FYR domain is not found in the other SET proteins in the class III group. The absence of the FYR domain and the finding that the domain organization of the other class III proteins differs from that of TRX suggests that the remaining class III proteins may function differently than trithorax (Fig. 4).
Figure 4.
Class III SET domain proteins. The SET domains of the plant class III SET domain proteins and several related animal proteins were aligned using ClustalW. The relationship of these sequences was examined using PHYLIP, and a parsimonious tree is shown with bootstrap values >70%. Circles are used to indicate putative maize-Arabidopsis orthology groups. The Arabidopsis proteins are indicated by italic text, and the maize proteins are indicated by bold text. Several maize proteins, which are currently only partially sequenced, were placed within the orthology group that they are most closely related to. The domain organization of the class III SET domain proteins was analyzed by NCBI-CD and SMART searches. Schematic diagrams show the domain organization of these proteins with the N terminus on the left side. For several of the longer proteins, a region of the protein that did not contain any recognizable domains was truncated.
We have documented the presence of a maize gene, Sdg128, which encodes a class III orthology group III-1 protein. Although the sequence is not currently complete, it does provide evidence for a maize member of group III-1.
A second orthology group (III-2) of the class III plant SET domain proteins includes SDG14, SDG16, SDG29, and SDG115. The domain organization of these proteins is similar; they all contain a PWWP domain, two PHD domains, and a PostSET domain in addition to the SET domain (Fig. 4). SDG16 and SDG29 are located in collinear duplicated regions of Arabidopsis chromosomes 4 and 5. The maize gene Sdg106 is currently represented by a partial sequence. This sequence is closely related to both groups III-1 and III-2, and it is not currently possible to assign this gene to one orthology group.
The final two orthology groups (III-3 and III-4) of class III SET domain proteins found in plants are represented by SDG2 and SDG25 from Arabidopsis and SDG108 and SDG127 from maize. The yeast ScSET1 catalyzes histone H3 Lys-4 methylation (Briggs et al., 2001; Roguev et al., 2001) and is closely related SET domain protein to the III-4 orthology group. The Arabidopsis proteins SDG2 and SDG25 both have similar domain architecture. NCBI-CD searches reveal that SDG2 and SDG25 contain two partial GYF domains near the N terminus and an SET domain near the C terminus (Fig. 4). GYF domains are involved in binding Pro-rich regions of other proteins (Freund et al., 1999). Both maize genes (Sdg108 and Sdg127) are represented by partial sequences and do not include the regions expected to contain the GYF domains.
Class IV SET Domain Proteins
Our phylogenetic analysis supports the existence of a class of SET domain proteins only present in yeast and plants. The class IV SET domain proteins include two proteins from Arabidopsis and two proteins from yeast (Fig. 2). These four SET domain proteins all contain an SET domain and a PHD domain (Fig. 5) but lack a PreSET or PostSET domain. A maize gene, Sdg129, which is related to the Arabidopsis SDG15 and SDG34 has been identified. The partial sequence obtained for SDG129 does not include the SET domain; therefore, this protein is not included in the phylogenetic analysis.
Figure 5.
Class IV SET domain proteins. The amino acid sequences of the class IV SET domain proteins were aligned using ClustalW. This alignment was shaded using Boxshade such that identical amino acids are shaded black, and conserved resides are shaded in gray. The locations of the PHD (Λ) and SET (*) domains are indicated above the alignment. Schematic diagrams of these proteins are shown below the alignment.
Null mutants for either ScSET3 or ScSET4 and the double mutant are viable (Pijnappel et al., 2001). The ScSET3 protein is found in a large multiprotein complex including two histone deacetylases and does not possess detectable histone methyltransferase activity in vitro (Pijnappel et al., 2001). The alignment of the SET domain sequence reveals that several of the amino acids determined to be critical for SET domain histone methyltransferase activity are not conserved in the class IV SET domain proteins (see Supplementary Fig. 1 at http://www.plantphysiol.org). Alignments of the plant and yeast proteins do not reveal other regions of significant conservation between these proteins. The phylogeny supports independent duplication of class IV SET domain proteins in both Arabidopsis and yeast.
Class V SET Domain Proteins
The class V proteins are the largest group of SET domain proteins in plants. The D. melanogaster, mouse, and human genomes each contain two or three class V SET domain proteins compared with 15 in the Arabidopsis genome (Fig. 6). This is the only class of SET domain proteins that contains both PreSET and PostSET domains.
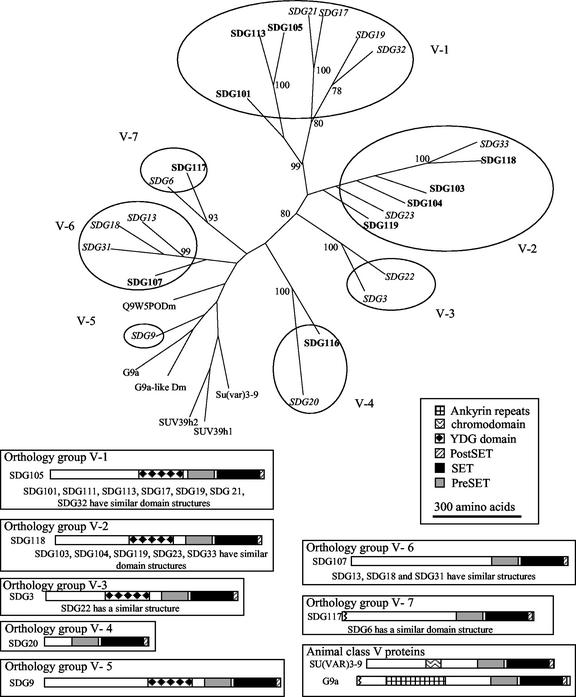
Figure 6.
Class V SET domain proteins. The SET domain of all class V SET domain proteins was aligned using ClustalW. The relationship of these sequences was examined using PHYLIP, and a parsimonious tree is shown with bootstrap values >70%. Circles are used to indicate putative maize/Arabidopsis orthology groups. The Arabidopsis proteins are indicated by italic text, and the maize proteins are indicated by bold text. Several maize proteins, which are only partially sequenced, are placed within the orthology group that they are most closely related to. We searched for recognizable domains in these proteins using NCBI-CD and SMART searches. Schematic diagrams indicate the domain organization for each of the full-length proteins, with the N terminus on the left.
The PreSET domain is a Cys-rich putative Zn+-binding domain that is only found associated with SET domains. A partial PreSET domain (the AWS domain) is found in class II SET domain proteins, including ASH1 and NSD1. The PostSET domain is a small Cys-rich region often found at the C terminus of SET domains. To date, at least one member of each class of animal proteins containing both PreSET and- PostSET domains has been shown to be functional histone methyltransferase enzymes (Rea et al., 2000; Tachibana et al., 2001; Yang et al., 2002).
Domain Organization of Class V SET Domain Proteins
The animal class V SET domain proteins can be divided into two groups based on domain structure. The SU(VAR)3-9 protein and mammalian homologs all contain a chromodomain near the N terminus. The G9a protein and a related D. melanogaster sequence (AAF45487) both contain ankyrin repeats. The domain organization of the plant class V SET domains is distinct from that of the animal proteins. None of the plant class V SET domain proteins contain a chromodomain or ankyrin repeats.
The orthology groups V-1, V-2, V-3, and V-5 are all YDG/PreSET/SET/PostSET domain proteins, whereas the orthology groups V-4, V-6, and V-7 all lack the YDG domain. The YDG domain is also referred to as a SET and RING-finger associated domain (SRA) (Baumbusch et al., 2001). Ten Arabidopsis class V SET domain proteins, which fall into four orthology groups, all contain YDG, PreSET, SET, and PostSET domains (Fig. 6). The remaining Arabidopsis class V SET proteins, which fall into three orthology groups, do not contain YDG domains (Fig. 6). The maize class V proteins identified to date fall into five orthology groups (Fig. 6). The N-terminal portion of these proteins does not contain any recognizable domains.
Evolution of Class V SET Domain Proteins in Plants
Comparison of the maize and Arabidopsis class V SET domain proteins suggests that the amplification of class V proteins in plants occurred through duplication events both before and after the divergence of monocots and dicots (Fig. 6). The parsimonious analysis of class V SET domain proteins shown in Figure 6 supports the presence of at least seven class V orthology groups. In our analysis, we have chosen the minimum number of orthology groups, and it is likely that some of the groups we have designated as a single group may actually represent multiple orthology groups. The orthology groups V-1, V-2, V-3, and V-5 are all YDG/PreSET/SET/PostSET domain proteins, whereas the orthology groups V-4, V-6, and V-7 all lack the YDG domain.
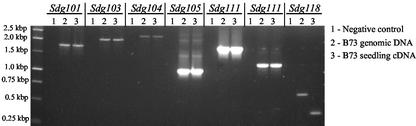
Baumbusch et al. (2001) noted that the majority of the Arabidopsis SET domain proteins that also contain an YDG domain do not contain introns. We tested the coding sequence of several YDG-SET domain genes from maize for the presence of introns (Fig. 7). Introns were detected within the coding sequence of only one maize YDG-SET domain gene, SDG118. The fact that both maize and Arabidopsis proteins lack introns indicates that this class was amplified before the divergence of maize and Arabidopsis, possibly by an ancient retrotransposition-like event. SDG33 (KYP) is the only Arabidopsis protein from orthology group V-1, V-2, V-3, and V-5 that contains introns within the coding sequence. The most closely related maize sequence, Sdg118, also contain introns within the coding sequence (Fig. 7). We also investigated the genomic sequence of class V YDG-SET domain genes present in the rice genome and found that only one, the homolog of SDG33/SDG118, contained introns within the coding sequence (data not shown).
Figure 7.
Maize contains intron-less class V SET domain genes. PCR was used to test for the presence of introns in the sequence of several class V maize genes, Sdg101, Sdg103, Sdg104, Sdg105, Sdg111, and Sdg118. A segment of the coding region for each of these genes was amplified from B73 genomic DNA and 10-d-old B73 seedling cDNA.
Expression of Maize SDG Genes
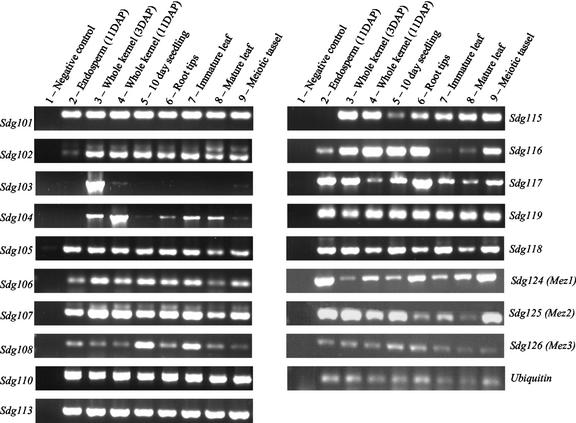
The majority of the maize SDG genes are constitutively expressed (Fig. 8). We tested the expression of 18 SDG genes by PCR of cDNA from eight different tissue sources. In all cases, one of the primers used was located in the 3′-untranslated region, which is expected to be more divergent than coding sequences and should allow for specific amplification of the target gene. Genomic controls were performed for all primers pairs and in every case except Sdg101, Sdg104, Sdg105, Sdg106, and Sdg113, the product amplified from genomic DNA was larger than that amplified from cDNA, indicating that the primers used flanked introns (data not shown). We did not detect any amplification products when two primer pairs specific for genomic DNA (one primer located within an intron) were used to test for genomic contamination of our cDNA (data not shown). Sdg101, Sdg102, Sdg105, Sdg106, Sdg107, Sdg108, Sdg110, Sdg113, Sdg116, Sdg117, Sdg118, Sdg119, Sdg124, Sdg125, and Sdg126 transcripts were detected in all tissues tested. Sdg103 transcripts were only detected in 3-DAP whole-kernel and 11-DAP whole-kernel tissues. The absence of products in 11-DAP endosperm tissue suggests that Sdg103 might be expressed specifically in embryo tissue. Sdg104 and Sdg115 transcripts were not detected in endosperm tissue.
Figure 8.
Expression patterns of maize SET genes. PCR was performed on cDNA from eight different tissue sources to test for expression of maize SET genes. The sequence amplified is indicated next to the image of the gel, and the source of the RNA for each lane is indicated above the pictures (1, blank; 2, endosperm [11 d after pollination {DAP}]; 3, whole kernel [3 DAP]; 4, whole kernel [11 DAP]; 5, 10-d-old seedling; 6, root tips; 7, immature leaf; 8, mature leaf; 9, meiotic tassel).
DISCUSSION
We have characterized 25 expressed SET domain genes from maize and compared these sequences with the 32 SET domain proteins present in the Arabidopsis genome, 30 of which are expressed. Our phylogenetic analysis suggests that the plant SET domain proteins form five classes, and further domain analysis suggests these can be subdivided into 19 orthology groups. The presence of a larger number of SET domain proteins in plants relative to non-plant species results from SET domain protein duplication that occurred via multiple mechanisms. Importantly, the domains outside of the SET domain are often quite different from those found in animal SET domain proteins. The domains present in many of the plant SET domain proteins are predicted to play roles in mediating protein-protein interactions, indicating that the plant putative histone methyltransferases may act in complexes quite distinct from those found in animals and yeast. The significant difference between plant and animal SET proteins indicates that detailed biochemical characterization of plant chromatin remodeling complexes will be necessary to fully understand their unique function.
Duplication of SET Domain Proteins in Plants
Plant SET domain genes show an increased degree of duplication relative to other organisms. For example, Arabidopsis contains 32 SET domain proteins, whereas D. melanogaster contains 14, mouse contains 17, and yeast contains four. The plant proteins have been divided into three class I orthology groups, four class II orthology groups, four class III orthology groups, one class IV orthology group, and seven class V orthology groups. The 19 orthology groups of SET domain proteins identified in plants are much larger than the nine orthology groups present in between D. melanogaster and mouse. This indicates that there was significant duplication and divergence of SET domain proteins in the plant lineage before the divergence of monocots and dicots.
We identified at least one maize gene in 15 of the 19 orthology groups and detected a monocot homologs for two of the four other orthology groups. The barley (Hordeum vulgare) EST BG345006 belongs to orthology group II-2, and the barley ESTs AV915295 and AV920392 represent orthology group V-3. The presence of ESTs from monocot species for these orthology groups indicates that it is likely that an as yet uncharacterized maize representative for these orthology groups exists. We did not detect any ESTs from other plant species or any genomic sequences from rice representing the other two orthology groups, I-1 (represented by SDG5/MEA in Arabidopsis) and V-5 (represented by SDG9 in Arabidopsis). This could reflect the fact that these genes are expressed at very low levels or in specific tissues, or it could indicate that these are genes specific to Arabidopsis and close relatives.
Phylogenetic analyses of SET domain genes indicate that there have been numerous gene duplication events in plants. One type of duplication event that has occurred in both maize and Arabidopsis is the result of polyploidization or chromosome addition. In Arabidopsis, duplications consistent with ancient polyploid or chromosome duplication events include the SDG7/24, SDG27/30, and SDG16/29 pairs of genes found in collinear duplicated genomic regions (Baumbusch et al., 2001). An example of duplication in maize is Sdg125/Sdg126, which are predicted to be duplicate genes resulting from the ancient allopolyploid origin of this species (Springer et al., 2002).
A second type of duplication event is represented by related genes found in non-collinear regions, such as SDG15/34, SDG3/22, SDG17/21, SDG19/32, and SDG13/18 from Arabidopsis. These gene pairs are found in regions of the Arabidopsis genome not classified as collinear regions (http://mips.gsf.de/proj/thal/db/gv/rv/rv_frame.html). These duplications may have arisen from the same mechanisms that gave rise to the duplications found in the collinear region, followed by successive reorganizations. Alternatively, these duplications may have occurred via small-scale transposition or illegitimate recombination events.
The third type of duplication event of the plant SDG genes has occurred via a putative retrotransposition-like event (Baumbusch et al., 2001). Our data show that the intron-less class V SET domain proteins are found in four orthology groups, three of which include at least one maize sequence. We tested five of the maize orthologs of the intron-less Arabidopsis class V proteins, Sdg101, Sdg103, Sdg104, Sdg105, and Sdg111 and found that the maize genes also lacked introns within the coding sequence. The closest relative to Sdg33 (Kyp), the only intron-containing YDG/SET domain gene from Arabidopsis, is Sdg118, which also contains introns. Together, these findings suggest that a retrotransposition-like event occurred before the divergence of monocots and dicots.
The large number of conserved SET domain proteins in plants suggests that many of the products of gene duplication events have adopted distinct functions. When a gene duplication event occurs, both products must adopt at least partially nonoverlapping function or one will tend to be lost by mutation (Lynch and Conery, 2000). The function of the two genes can be nonoverlapping either by having different expression patterns or by having distinct biochemical functions. The majority of SET domain proteins are expressed in most tissues tested (Fig. 8; Baumbusch et al., 2001; http://www.chromdb.org), although more detailed analysis will be necessary to detect any temporal or spatial expression patterns that were not revealed using pooled tissue samples. If the plant SET domain genes are constitutively expressed, it would suggest that the SET domain proteins of plants have adopted at least partially nonoverlapping biochemical functions.
Plant SET Domain Proteins Are Likely to Encode Histone Methyltransferases with Distinct Substrate Specificities
Several studies have documented that the SET domain is a histone methyltransferase motif in yeast and animals and that different SET domain proteins often display substrate preferences for specific Lys residues within histones H3 and H4 (Rea et al., 2000; Briggs et al., 2001; Nakayama et al., 2001; Roguev et al., 2001; Tachibana et al., 2001; Wang et al., 2001; Beisel et al., 2002; Fang et al., 2002; Nishioka et al., 2002; Strahl et al., 2002; Yang et al., 2002). On the basis of the conservation within the SET domain, we predict that many of the plant SET proteins are likely to encode functional histone methyltransferase enzymes. Evidence from animals suggests that each class of SET domain proteins is likely to have distinct substrate specificities. On the basis of homology between the plant SDG proteins and the animal proteins for which biochemical analysis has been performed, we can speculate about potential substrate specificities for each class of plant SET domain proteins.
Several of the animal class I proteins (Enhancer of zeste and homologs) have been shown to methylate predominately Lys-27 of histone H3 with a lower affinity for Lys-9 of histone H3 (Cao et al., 2002; Czermin et al., 2002; Kuzmichev et al., 2002; Muller et al., 2002). These studies have suggested that the activity of class I SET domain proteins requires interaction with other proteins. Based on these studies and the similarity between plant and animal class I SET domain proteins, it is likely that the plant class I SET domain proteins, including SDG1 (CLF), SDG5 (MEA), SDG10 (EZA1), SDG124 (MEZ1), SDG125 (MEZ2), and SDG126 (MEZ3), are likely to encode H3K27 methyltransferases.
Animal class II proteins that have been shown to encode functional histone methyltransferase enzymes include ScSET2 and ASH1 (Beisel et al., 2002; Strahl et al., 2002). ScSET2 methylates H3-Lys-36, whereas ASH1 methylates H3-Lys-4 and Lys-9 and H4-Lys-20. To date, there has not been a specific biochemical activity associated with the class II proteins as a group. The plant class II SET domain proteins, including SDG7, SDG8, SDG24, and SDG26, contain related SET domains but do not display similarity in other regions of the proteins. Based on the current literature, it is difficult to predict a substrate specificity that will be common to all class II proteins.
Several class III proteins, including ScSET1 and HRX, have been shown to encode functional histone methyltransferases (Briggs et al., 2001; Roguev et al., 2001; Milne et al., 2002; Nakamura et al., 2002) that methylate H3-Lys-4. The H3-Lys-4 methylation has been correlated with transcriptional activity in animals. In general, the class III proteins from animals have been correlated with transcriptional activity, and it has been proposed the H3-Lys-4 is an epigenetic mark of active chromatin. Published results are consistent with the idea that the plant class III proteins are likely to encode proteins capable of methylating H3-Lys-4, thereby promoting the formation of active chromatin.
The animal class V SET domain proteins, including SU(VAR)3-9 (Rea et al., 2000; Nakayama et al., 2001) and G9a (Tachibana et al., 2001, 2002), possess histone H3-Lys-9 methyltransferase activity. The plant SET domain protein KRYPTONITE (SDG33) also methylates H3-Lys-9 (Jackson et al., 2002). The remaining class V SET domain proteins from plants are proposed to methylate Lys-9 of histone H3 also. The methylation of H3-Lys-9 is generally correlated with the presence of silent chromatin.
The significant conservation within the SET domain suggests that many of the plant SET domain proteins will encode functional histone methyltransferase enzymes and that like the animal proteins, they may display substrate specificity for the modification of specific Lys residues present in histone tails. Further studies on the function of the SET domain and associated regions will provide a more detailed model for the exact biochemical modifications catalyzed by the plant SET proteins.
SET Domain Proteins Are Likely to Function in Complexes
The majority of SET domain proteins characterized in animals are present in large protein complexes. The domains present in many of the plant SET domain proteins, such as the PHD, PWWP, and YDG domains, suggest that they are likely to be present in protein complexes also. The PHD domain is a putative zinc finger that is involved in mediating protein-protein interactions (Aasland et al., 1995). The PWWP domain is also involved in mediating protein-protein interactions (Stec et al., 2000). The domains present in the N-terminal portion of SET domain proteins may be important for determining interactions with other proteins. In addition, several studies have indicated that the SET domain itself may also play a role in mediating protein-protein interactions (Cui et al., 1998; Rozenblatt-Rosen et al., 1998; Rozovskaia et al., 2000).
Many of the SET domain proteins in animals are present in large protein complexes. Although it is expected that some of these complexes will be conserved in plants, it is likely that many of the plant SET domain proteins will exist in complexes that are specific to plants. The class I and several class III plant proteins contain a domain structure very similar to related animal proteins, and these are predicted to exist in similar complexes as in animals. Other plant SET domain proteins do not contain any similarity to animal proteins outside of the SET domain; these will probably exist in complexes that are plant specific.
The duplication of SET domain proteins in plants may have required duplications of other interacting proteins, or it could be that the SET domain protein determines the specificity of a complex and a single complex can interact with multiple SET domain proteins. In some cases, there is evidence that the associated proteins have not undergone duplication. All three of the class I SET domain proteins from Arabidopsis, CLF, MEA, and EZA1, physically interact with the same protein, FIE (Luo et al., 2000; Spillane et al., 2000; Yadegari et al., 2000). If this is true for other SET domain protein complexes, it would suggest that the SET domain protein is important for determining the specificity of the complex.
Although many of the basic mechanisms of chromatin-based regulation are conserved in plants and animals, the flexibility of these systems and the ability of these systems to respond to developmental and environmental cues is likely to be quite different in plants and animals. In animals, developmental decisions regarding gene expression and differentiation are complete at an early stage of development. Plants often switch developmental fates throughout their life cycle, especially to respond to environmental stimuli such as light, temperature, and water availability. The presence of a much larger family of SET domain proteins may allow plants more specific control of developmental decisions. The Su(z)12 homologs of Arabidopsis provide an example of amplification of a chromatin protein that has adopted specific functions in regulation of development. Arabidopsis encodes three Su(z)12 homologs, Fis2 (Fertilization independent seed 2), Emf2 (Embryonic flower 2), and Vrn2 (Vernalization 2; Luo et al., 1999; Gendall et al., 2001; Yoshida et al., 2001). The proteins regulate distinct developmental transitions including endosperm development (Fis2), floral development (Emf2), and floral development in response to temperature treatments (Vrn2). By analogy, the different SET domain proteins may be important for regulation of different groups of genes or different chromatin types.
This study has further characterized the SET domain proteins of plants. Our analysis has suggested functional relationships between plant SET domain proteins that will be important for the interpretation of data from a model system, such as Arabidopsis, to other economically important crops, such as maize. The analysis presented in this paper will serve as a framework for ongoing functional analysis of this diverse group of proteins.
MATERIALS AND METHODS
SET Domain Gene Discovery and Annotation in Arabidopsis
The Arabidopsis SET domain group (SDG) protein sequences used in this study were identified by nucleic acid and protein BLAST analysis using E(Z) (AAC46462), ASH1 (AAF49140), TRX (AAF55041), TRR (AAF45684), G9a like (AAF45487), and SU(VAR3-9) (CAB93768) as queries. The resulting Arabidopsis SDG domain proteins were then used to query the Arabidopsis genome to find other Arabidopsis proteins. These proteins are the same proteins identified by Baumbusch et al. (2001). Our gene models predicted different splice sites relative to the model available at the GenBank accession number listed in Table I for 10 of the 37 proteins, which were confirmed by RT-PCR, EST, or ortholog alignment. Gene models were updated using EST data, targeted PCR analysis, and alignments with other plant ESTs. The gene models used for this study and expression data for many of the maize (Zea mays) and Arabidopsis SDG genes are available at http://www.chromdb.org. Collinear genome localization of the most closely related pairs of Arabidopsis sequences was assessed using the Munich Information Center for Protein Sequences Arabidopsis thaliana database redundancy viewer (http://mips.gsf.de/proj/thal/db/gv/rv/rv_frame.html). Gene pairs that did not fall into previously described collinear regions were, by default, considered to be duplications that did not occur by large-scale genome events.
SET Domain Gene Discovery and Sequencing in Maize
The SET domain protein sequences from Arabidopsis were used to search all maize ESTs present in GenBank (last searched August 5, 2002). Putative SET domain proteins, identified by automated searching, were arbitrarily named SDG101 to SDG130. In some cases, further sequencing revealed that two ESTs actually corresponded to the same gene, and one name was dropped. We obtained full-length cDNA sequence for Sdg102 (BE345442) and Sdg105 (AW216196) by sequencing EST clones. Full-length sequence for Sdg104, Sdg110, Sdg113, and Sdg118 was obtained by RACE. RACE reactions were performed using the Marathon cDNA kit (CLONTECH, Palo Alto, CA) on cDNA produced from 10-day-old B73 seedlings. Advantage2 polymerase (CLONTECH) was used in the RACE reactions. The primers used in the RACE reactions were Set104R1 (5′-CCT CTG ATT GAC TGC AAC AGC CAC C-3′) and Set104R2 (5′-GTG CGC ATG ACA CGA TAC TAA CAG CC-3′) for Sdg104, Set110R1 (5′-CCA CAA TGA CAA ACC TGA GCT GCT CC-3′) and Set110R2 (5′-TCC AAC CCT GGT CTC TCC ATC AAC AG-3′) for Sdg110, Set113R1 (5′-GCT TTG CTC CCC TAT CAA TTC AGG TCC-3′) and Set113R2 (5′-ATG AAC CAG CCC GTA TAG CGT CCC-3′) for Sdg113, and Set118R1 (5′-CTG CCC AAG CGA TAA CCG TAG CC-3′) and Set118R2 (5′-GGA GCT CAT GAC GCA CTG GAC G-3′) for Sdg118. RACE products were gel purified and cloned into pCR-BluntII (Invitrogen, Carlsbad, CA). For Sdg101, Sdg103, Sdg106, Sdg107, Sdg108, Sdg114, Sdg115, Sdg116, and Sdg117 we have extended the EST sequence either by sequencing of EST clones or through RACE analysis. Because these sequences are not full length, they have not yet been submitted to GenBank but are publicly available at http://www.chromdb.org.
PCR Analysis of Genomic Structure of Maize Class V Genes
Many of the Arabidopsis class V SET domain genes are intron-less as first described by Baumbusch et al. (2001). We used PCR to determine whether maize genes Sdg101, Sdg103, Sdg104, Sdg105, Sdg111, and Sdg 118 contain introns. The presence or absence of introns was determined by running the products amplified from B73 seedling cDNA adjacent to the products amplified from B73 genomic DNA. Conditions of the PCR were as follows: 94°C for 2 min, 35 cycles of 94°C for 30 s, 63°C for 30 s, 72°C for 2 min, followed by 72°C for 7 min. Amplified products were separated in a 1% (w/v) agarose Tris-borate/EDTA buffer gel and visualized by ethidium bromide staining. The primers used for the PCR reactions were Set101F3 (5′-CCC AAA CGT TTG CAG GAT AGT TCA G-3′) and Set101R6 (5′-CTA CAC TTC GGG GAC CAA CAT AAG C-3′) for Sdg101; Set103F1 (5′-GGA AAC CGT ACG CGA AAG GTG G-3′) and FlSet103R1 (5′-CAG CAG CAT CTC GTG TCA TCA TCT AGG-3′) for Sdg103; Set104F2 (5′-GCT CGC ACC CAG GAA TTC AGG-3′) and FlSet104R1 (5′-CCC ATT GGC AAC TAA AAA CAC TGA TG-3′) for Sdg104; Set105F2 (5′-GCG GCT TCA AGG ATC CAT TTT GC-3′) and Set105R3 (5′-ATC CCC TGC AGT TTT GTG ATC CAC-3′) for Sdg105; FLSet111F1 (5′-GTG CCA AGG TCC GCA TAT TCG-3′) and Set111R2 (5′-GAT ACA TAT GTG CTA GCT TCA CC-3′) and Set111F1 (5′-GGT GCC ATT GAT GTG CTG GTA TAC AG-3′) and Set111R1 (5′-ATA GTC CAC GGC AGT TTT GTG ATC C-3′) for Sdg111; and Set118F3 (5′-GAG GAG GAC TGA AGA TCT GGA TGG-3′) and ZmKrpR1 (5′-CTG CCC AAG CGA TAA CCG TAG CC-3′) for Sdg118.
Domain Predictions
The protein sequences of all SET domain proteins were analyzed for additional recognizable domains using NCBI-CD searches (http://ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The low-complexity filter was turned off, and the expect value was set at 1 to detect short domains or regions of less conservation in this analysis. Domains were not considered significant unless the alignment included more than 70% of the domain. All domains were referred to using the names present in the SMART domain database (Schultz et al., 2000). The domain characteristics and number of times a particular domain occurred in a species was determined using the SMART domain database (http://smart.embl-heidelberg.de/; Schultz et al., 2000).
Phylogenetic Analysis
The complete group of nonredundant yeast (Saccharomyces cerevisiae), mouse (Mus musculus), and Drosophila melanogaster SET domain proteins were obtained using the SMART database (Schultz et al., 2000). For all proteins analyzed, the region of the SET domain used for the alignment began at the conserved GWG motif and ended at the conserved TYDY motif (matched amino acids 296–409 of SUV39h1 [AF193862]). The selected SET domain sequences were aligned using ClustalW. This alignment was then submitted to the PHYLIP server (http://bioweb.pasteur.fr/seqanal/phylogeny/phylip-uk.html). The protpars feature was used with bootstrapping performed before analysis. One hundred replicates were examined to determine bootstrap values. The consensus tree was then displayed with bootstrap values.
DNA Gel-Blot Analysis
Genomic DNA was purified from young leaves of the inbred lines B73 and Mo17 by CsCl centrifugation as described previously (Cone et al., 1986). DNAs were digested overnight with the restriction endonucleases BamHI, DraI, EcoRI, EcoRV, HindIII, and XbaI according to the manufacturers' specifications, and digests were fractionated on agarose gels, blotted to nylon membranes (Magnagraph; Osmonics, Inc., Westborough, MA), and hybridized as previously described (Cone et al., 1986). To prepare probes, inserts were liberated from cDNA clones by digesting with restriction enzymes; then, the inserts were fractionated and excised from low-melting point agarose gels and labeled by random priming. Probes were derived from cDNA clones obtained from Virginia Walbot (Stanford University, Stanford, CA). Clone numbers and corresponding SDG genes were: 687009G06 (Sdg101), 946034H07 (Sdg102), 660002D05 (Sdg103), 486039C05 (Sdg104), 683028B05 (Sdg105), 687017H04 (Sdg106), 614005G10 (Sdg107), 707050F11 (Sdg108), 606074E04 (Sdg110), 660018D02 (Sdg113), 945053G08 (Sdg115), 946091A05 (Sdg116), 947006D12 (Sdg117), 618069F07 (Sdg118), and Zm10_04e12_A (Sdg119). Digital images of these survey DNA gel blots are available on the gene information Web pages at www.chromdb.org.
RT-PCR Analysis
RT-PCR was used to assess expression patterns because of the relatively low expression of the maize PcG homologs and because most of the genes were duplicated. Total RNA was extracted with Trizol (Invitrogen) from 10 tissues from the inbred B73 (endosperm [11 DAP], whole kernel [3 DAP], whole kernel [11 DAP], 10-d-old seedling [whole plant included], root tips, immature leaf [leaves three–five], mature leaf [fully expanded leaf 10], and meiotic tassel). One microgram of total RNA was used to make cDNA with the SMART cDNA synthesis kit according to the manufacturer's instructions (CLONTECH). PCR reactions were performed in a 25-μL total volume containing approximately 0.5 ng of cDNA, 5 pmol of each primer, 1 unit of Taq polymerase (Promega, Madison, WI), 2.5 μL of 10× reaction buffer, 2 μL of 25 mm MgCl2, and 0.3 μL of 25 mm dNTPs. Primers used for the RT-PCR reactions were Set101F1 (5′-CGC GGA CGA CCT AGG AAA ATT GAT ACC-3′) and Set101R1 (5′-CAG CAA TTC CGG TGC ATA GTT CCA TC-3′) for Sdg101, FLSet102F1 (5′-GTT CAG TCT TCA GAG CTG GGT TCG G-3′) and Set102R2 (5′-GCT CTC CGT TTG GCT TCC TTC C-3′) for Sdg102, Set103F2 (5′-GGA GCA GCG TTC ATT GAA GAT GAG-3′) and FLSet103R1 (5′-CAG CAG CAT CTC GTG TCA TCA TCT AGG-3′) for Sdg103, Set104F1 (5′-TGG GAC CAA CGT TTT CCG AGA CG-3′) and Set104R1 (5′-CCT CTG ATT GAC TGC AAC AGC CAC C-3′) for Sdg104, Set105F2 (5′-GCG GCT TCA AGG ATC CAT TTT GC-3′) and Set105R2 (5′-GCA AGC AAA CGC TCT GGC ATC C-3′) for Sdg105, Set106F1 (5′-CTT TTA TGG GCG ATG CGT GTC TC-3′) and Set106R1 (5′-GCA GGG CTT TGA ACC ATT TAT GCG-3′) for Sdg106, Set107F1 (5′-CTC TTA GAT GCT GGT TGG GGT CCT G-3′) and Set107R2 (5′-GGA CCC CAA CCA GCA TCT AAG AGC AC-3′) for Sdg107, Set108F1 (5′-GCA TGG AAA AAC AGG CAC AGA GAC C-3′) and FLSet108R1 (5′-CTC CGC AAG GTA TGT AGG GAC TGG-3′) for Sdg108, FLSet110F2 (5′-CGT CAC CCT TCG CCT AAA TCA CC-3′) and Set110R1 (5′-CCA CAA TGA CAA ACC TGA GCT GCT CC-3′) for Sdg110, Set113F3 (5′-GAT GGG GTT GCA ATC TGG AAG ATG-3′) and Set113R2 (5′-ATG AAC CAG CCC GTA TAG CGT CCC-3′) for Sdg113, Set115F1 (5′-GAG TAT CGC GGT GAG CTG GTC AG-3′) and FLSet115R1 (5′-ACT GGC CGT AGT GAA TAC AAC TGT GG-3′) for Sdg115, Set116F1 (5′-GAA GCG CGG AGA CGA CAC AAG G-3′) and FLSet116R1 (5′-CTG TAA GCA GGA AAC ACA TGT CCA GC-3′) for Sdg116, Set117F1 (5′-CAT GTA TTT GTG ACT CGT CCT GCC AG-3′) and FLSet117R1 (5′-CTC GCC TAC GAA CAG AGC AGC C-3′) for Sdg117, Set118F3 (5′-TGA GGA GGA CTG AAG ATC TGG ATG G-3′) and FLSet118R1 (5′-ATC AAA ATG GAA ACA CAC TGC AGG TC-3′) for Sdg118, Set119F1 (5′-GAA GTG TTG GAA TGT TGG CAA GAA GG-3′) and Set119R1 (5′-GTC CGA GCA GCC GTT TGT ACA GTT G-3′) for Sdg119, Mez1F1 (5′-GGG TGT GGT GAT GGT ACA TTG G-3′) and Mez1R1 (5′-CGG GAC CTA ACT CTA CGG ATG G-3′) for Sdg124, Mez2F8 (5′-CCC CTG TTT TGC AGC CAG TCG TGA-3′) and Mez2R8 (5′-GGT GAG AGA AGG ATG CCT CGT CC-3′) for Sdg125, Mez3F3 (5′-AGT ATG TGT TGG ATG CTT ATC GCA AGG-3′) and Mez3R2 (5′-GGT TGT CAG TTT GTC ACC TTC CGA CC-3′) for Sdg126, and Ubi1F1 (5′-TAA GCT GCC GAT GTG CCT GCG TCG-3′) and Ubi1R1 (5′-CTG AAA GAC AGC ACA TAA TGA GCA CAG GC-3′) for Ubiquitin. Conditions of the PCR were as follows: 94°C for 2 min, 35 cycles of 94°C for 30 s, 63°C for 30 s, 72°C for 2 min, followed by 72°C for 7 min. Amplified products were separated in a 1% (w/v) agarose Tris-borate/EDTA buffer gel and visualized by ethidium bromide staining.
Supplementary Material
Acknowledgments
We would like to thank Dean Bergstrom, Erin Guthrie, Sarah Kerns, Laura Schmitt, and Lyudmila Sidorenko for help with cloning and sequencing; Dean Bergstrom and Miriam Hankins for generating DNA gel-blot data; and Lewis Lukens for helpful discussions about phylogenetic analysis.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.013722.
This work was supported by the National Science Foundation (grant no. 9975930).
The online version of this article contains Web-only data. The supplemental material is available at http://www.plantphysiol.org.
References
- Aasland R, Gibson TJ, Stewart AF (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci 20: 56-59 [DOI] [PubMed] [Google Scholar]
- Aasland R, Stewart AF, Gibson T (1996) The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci 21: 87-88 [PubMed] [Google Scholar]
- Alvarez-Venegas R, Avramova Z (2001) Two Arabidopsis homologs of the animal trithorax genes: a new structural domain is a signature feature of the trithorax gene family. Gene 271: 215-221 [DOI] [PubMed] [Google Scholar]
- Alvarez-Venegas R, Avramova Z (2002) SET-domain proteins of the Su(var)3-9, E(z) and trithorax families. Gene 285: 25-37 [DOI] [PubMed] [Google Scholar]
- Balciunas D, Ronne H (2000) Evidence of domain swapping within the jumonji family of transcription factors. Trends Biochem Sci 25: 274-276 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120-124 [DOI] [PubMed] [Google Scholar]
- Bantignies F, Goodman RH, Smolik SM (2000) Functional interaction between the coactivator Drosophila CREB-binding protein and ASH1, a member of the trithorax group of chromatin modifiers. Mol Cell Biol 20: 9317-9330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB (2001) The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res 29: 4319-4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C, Imhof A, Greene J, Kremmer E, Sauer F (2002) Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419: 857-862 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL (2002) Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA 99: 8695-8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD (2001) Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharamyces cerevisiae. Genes Dev 15: 3286-3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk M, Briggs SD, Strahl BD, Curcio MJ, Allis CD, Winston F (2002) Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr Biol 12: 165-170 [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039-1043 [DOI] [PubMed] [Google Scholar]
- Cheung WL, Briggs SD, Allis CD (2000) Acetylation and chromosomal functions. Curr Opin Cell Biol 12: 326-333 [DOI] [PubMed] [Google Scholar]
- Cone KC, Burr FA, Burr B (1986) Molecular analysis of the maize anthocyanin regulatory locus C1. Proc Natl Acad Sci USA 83: 9631-9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, De Vivo I, Slany R, Miyamoto A, Firestein R, Cleary ML (1998) Association of SET domain and myotubularin-related proteins modulates growth control. Nat Genet 18: 331-337 [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell 111: 185-196 [DOI] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A (2002) Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277: 28368-28371 [DOI] [PubMed] [Google Scholar]
- Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y (2002) Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol 12: 1086-1099 [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA 93: 8449-8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE (2001) Mechanisms of transcriptional memory. Nat Rev Mol Cell Biol 2: 409-421 [DOI] [PubMed] [Google Scholar]
- Freund C, Dotsch V, Nishizawa K, Reinherz EL, Wagner G (1999) The GYF domain is a novel structural fold that is involved in lymphoid signaling through proline-rich sequences. Nat Struct Biol 6: 656-660 [DOI] [PubMed] [Google Scholar]
- Gendall AR, Levy YY, Wilson A, Dean C (2001) The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107: 525-535 [DOI] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44-51 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB (1998) Maternal control of embryogenesis by Medea, a polycomb group gene in Arabidopsis. Science 280: 446-450 [DOI] [PubMed] [Google Scholar]
- Huang N, vom Baur E, Garnier JM, Lerouge T, Vonesch JL, Lutz Y, Chambon P, Losson R (1998) Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J 17: 3398-3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, Whittle (1980) Trithorax: a new homeotic mutations of Drosophila melanogaster causing transformations of abdominal and thoracic imaginal segments. Mol Gen Genet 179: 607-614 [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556-560 [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Bender J, Richards EJ (1998) The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes Dev 12: 1714-1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074-1080 [DOI] [PubMed] [Google Scholar]
- Jones RS, Gelbart WM (1990) Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics 126: 185-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T (2001) FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104: 131-142 [DOI] [PubMed] [Google Scholar]
- Klein RR, Houtz RL (1995) Cloning and developmental expression of pea ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit N-methyltransferase. Plant Mol Biol 27: 249-261 [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893-2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116-120 [DOI] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE (2001) Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077-2080 [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A (2000) Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA 97: 10637-10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM (1999) Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 296-301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151-1155 [DOI] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL (2002) MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 10: 1107-1117 [DOI] [PubMed] [Google Scholar]
- Min J, Zhang X, Cheng X, Grewal SI, Xu RM (2002) Structure of the SET domain histone lysine methyltransferase Clr4. Nat Struct Biol 9: 828-832 [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA (2002) Histone methyltransferase activity of a Drosophila polycomb group repressor complex. Cell 111: 197-208 [DOI] [PubMed] [Google Scholar]
- Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML (2002) A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci USA 99: 90-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E (2002) ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell 10: 1119-1128 [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110-113 [DOI] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R et al. (2002) PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell 9: 1201-1213 [DOI] [PubMed] [Google Scholar]
- Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL (1999) Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11: 407-416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, Nakamura T, Canaani E, Croce CM, Mazo A (2001) Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science 294: 1331-1334 [DOI] [PubMed] [Google Scholar]
- Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, Rigaut G, Seraphin B, Aasland R, Stewart AF (2001) The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev 15: 2991-3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V (1998) Polycombing the genome: PcG, trxG, and chromatin silencing. Cell 93: 333-336 [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD et al. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593-599 [DOI] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WWM, Wilm M, Aasland R, Stewart AF (2001) The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J 20: 7137-7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus MJ, Galbiati M, Ticknor C, Chen J, Dellaporta SL (1996) Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273: 654-657 [DOI] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, Nakamura T, Croce CM, Mazo A, Canaani E (1998) The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci USA 95: 4152-4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozovskaia T, Rozenblatt-Rosen O, Sedkov Y, Burakov D, Yano T, Nakamura T, Petruck S, Ben-Simchon L, Croce CM, Mazo A et al. (2000) Self-association of the SET domains of human ALL-1 and of Drosophila TRITHORAX and ASH1 proteins. Oncogene 19: 351-357 [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P (2000) SMART: a Web-based tool for the study of genetically mobile domains. Nucleic Acids Res 28: 231-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J (1995) Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol 7: 376-385 [DOI] [PubMed] [Google Scholar]
- Spillane C, MacDougall C, Stock C, Kohler C, Vielle-Calzada JP, Nunes SM, Grossniklaus U, Goodrich J (2000) Interaction of the Arabidopsis polycomb group proteins FIE and MEA mediates their common phenotypes. Curr Biol 10: 1535-1538 [DOI] [PubMed] [Google Scholar]
- Springer NM, Danilevskaya O, Hermon P, Helentjaris T, Phillips RL, Kaeppler HF, Kaeppler SM (2002) Sequence relationships, conserved domains, and expression patterns for Zea mays homologs of the Drosophila polycomb group genes E(z), esc, and E(Pc). Plant Physiol 128: 1332-1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stec I, Nagl SB, van Ommen GJ, den Dunnen JT (2000) The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett 473: 1-5 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403: 41-45 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF et al. (2002) Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol 22: 1298-1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Winston F (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104-108 [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y (2001) Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem 276: 25309-25317 [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H et al. (2002) G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16: 1779-1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H, Selker EU (2001) A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414: 277-283 [DOI] [PubMed] [Google Scholar]
- Tie F, Furuyama T, Prasad-Sinha J, Jane E, Harte PJ (2001) The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development 128: 275-286 [DOI] [PubMed] [Google Scholar]
- Trievel R, Beach B, Dirk L, Houtz R, Hurley J (2002) Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell 111: 91-103 [DOI] [PubMed] [Google Scholar]
- Tripoulas N, LaJeunesse D, Gildea J, Shearn A (1996) The Drosophila ash1 gene product, which is localized at specific sites on polytene chromosomes, contains a SET domain and a PHD finger. Genetics 143: 913-928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripoulas NA, Hersperger E, La Jeunesse D, Shearn A (1994) Molecular genetic analysis of the Drosophila melanogaster gene absent, small or homeotic discs1 (ash1). Genetics 137: 1027-1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G (1994) The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J 13: 3822-3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vlag J, Otte AP (1999) Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet 23: 474-478 [DOI] [PubMed] [Google Scholar]
- Wagner D, Meyerowitz EM (2002) SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr Biol 12: 85-94 [DOI] [PubMed] [Google Scholar]
- Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y (2001) Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell 8: 1207-1217 [DOI] [PubMed] [Google Scholar]
- Wilson J, Jing C, Walker P, Martin S, Howell S, Blackburn G, Gamblin S, Xiao B (2002) Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell 111: 105-115 [DOI] [PubMed] [Google Scholar]
- Wu J, Grunstein M (2000) 25 years after the nucleosome model: chromatin modifications. Trends Biochem Sci 25: 619-623 [DOI] [PubMed] [Google Scholar]
- Yadegari R, Kinoshita T, Lotan O, Cohen G, Katz A, Choi Y, Katz A, Nakashima K, Harada JJ, Goldberg RB et al. (2000) Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12: 2367-2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Xia L, Wu D, Wang H, Chansky HA, Schubach WH, Hickstein DD, Zhang Y (2002) Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene 21: 148-152 [DOI] [PubMed] [Google Scholar]
- Yoshida N, Yanai Y, Chen L, Kato Y, Hiratsuka J, Miwa T, Sung ZR, Takahashi S (2001) EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13: 2471-2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15: 2343-2360 [DOI] [PubMed] [Google Scholar]
- Zhang X, Tamaru H, Khan S, Horton J, Keefe L, Selker E, Cheng X (2002) Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell 111: 117-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.