Abstract
In wild-type Arabidopsis, levels of ASN1 mRNA and asparagine (Asn) are tightly regulated by environmental factors and metabolites. Because Asn serves as an important nitrogen storage and transport compound used to allocate nitrogen resources between source and sink organs, we tested whether overexpression of the major expressed gene for Asn synthetase, ASN1, would lead to changes in nitrogen status in the ultimate storage organ for metabolites—seeds. Transgenic Arabidopsis constitutively overexpressing ASN1 under the cauliflower mosaic virus 35S promoter were constructed (35S-ASN1). In seeds of the 35S-ASN1 lines, three observations support the notion that the nitrogen status was enhanced: (a) elevations of soluble seed protein contents, (b) elevations of total protein contents from acid-hydrolyzed seeds, and (c) higher tolerance of young seedlings when grown on nitrogen-limiting media. Besides quantitative differences, changes in the relative composition of the seed amino acid were also observed. The change in seed nitrogen status was accompanied by an increase of total free amino acids (mainly Asn) allocated to flowers and developing siliques. In 35S-ASN1 lines, sink tissues such as flowers and developing siliques exhibit a higher level of free Asn than source tissues such as leaves and stems, despite significantly higher levels of ASN1 mRNA observed in the source tissues. This was at least partially due to an enhanced transport of Asn from source to sink via the phloem, as demonstrated by the increased levels of Asn in phloem exudates of the 35S-ASN1 plants.
In higher plants, nitrogen is acquired from the environment via nitrate reduction (Crawford and Arst, 1993), ammonia uptake, or nitrogen fixation (Burris and Roberts, 1993). All inorganic nitrogen must first be reduced to ammonia by the action of nitrate and nitrite reductase before being assimilated into amino acids (Lea and Miflin, 1980; Miflin and Lea, 1980; Lea et al., 1990). In plants, the majority of ammonia is assimilated into organic form via the combined action of Gln synthetase and Gln 2-oxoglutarate aminotransferase. The products of Gln synthetase-Gln 2-oxoglutarate aminotransferase cycle, Gln, and Glu are highly reactive and are used in various anabolic pathways. Through the action of Asn synthetase (AS; EC 6.3.5.4), Gln will react with Asp to form Asn and Glu. Because all the substrates and products of the AS-catalyzed reaction are major nitrogen carriers in plant metabolism for transporting nitrogen in the phloem, the AS enzyme is believed to play an important role in regulating the flow of nitrogen into the organic nitrogen pool (Lam et al., 1994). Asn is an especially important nitrogen transport amino acid because it has a high nitrogen to carbon ratio (relative to the other amide amino acid Gln) and is relatively inert compared with the other nitrogen-transporting amino acids (Glu, Asp, and Gln). Therefore, Asn can be used for long-range nitrogen transport and storage, which is vital to physiological processes such as germination and nitrogen assimilation (Lea and Miflin, 1980; Sieciechowicz et al., 1988; Lea et al., 1990) and is also essential for organic nitrogen translocation between host and parasitic plants (Delavault et al., 1998). The relative importance of Asn in plant nitrogen assimilation and metabolism is reflected by the drastic changes of its levels under different physiological and environmental conditions (for review, see Lea and Miflin, 1980; Sieciechowicz et al., 1988; Lea et al., 1990; Lam et al., 1995, 1996).
To elucidate the functions of AS in plants using molecular approaches, several laboratories have begun their studies with the cloning of AS genes and examination of the corresponding gene expression patterns. In several species, it has been shown that the expression of AS genes is repressed by light and/or sugar, and this observation is consistent with the previous observed changes in levels of free Asn in light- versus dark-treated plants (Lam et al., 1995; Chevalier et al., 1996). Light repression of AS genes was shown to be at least partially regulated by phytochrome (Tsai and Coruzzi, 1990; Lam et al., 1994), and there is evidence that this light repression may be a calcium and cGMP-dependent process (Neuhaus et al., 1997). De novo synthesis of AS may be a consequence of the light/dark regulation of AS gene expression because the increase of AS activity in the dark is abolished by cycloheximide treatment (Dembinski et al., 1996b). However, there are reports showing that in some species, specific AS genes are insensitive to light (Waterhouse et al., 1996) or even induced by light under certain circumstances (Hughes et al., 1997; Lam et al., 1998). In asparagus (Asparagus officinalis) and broccoli (Brassica oleracea), AS gene expression increases during postharvesting period, and this increase is correlated to a depletion of carbon resources (Davis et al., 1996; Downs and Somerfield, 1997). Furthermore, environmental stresses such as salt and heavy metal also elevate the mRNA levels of AS genes (Chevalier et al., 1996). The complexity of gene regulation further suggests the importance of AS in intermediate metabolism.
To take advantage of using a genetic model to study the molecular/genetic aspects of Asn metabolism in plants, we previously cloned all members of the AS gene family from Arabidopsis (ASN1, ASN2, and ASN3). The ASN1 cDNA was cloned by heterologous hybridization to the pea (Pisum sativum) AS1 gene (Lam et al., 1994), whereas the ASN2 and ASN3 cDNAs were cloned by functional complementation of an AS-deficient yeast (Saccharomyces cerevisiae) mutant (Lam et al., 1998). Levels of ASN1 and ASN2 mRNA were shown to be reciprocally regulated by light and by organic carbon and nitrogen (Lam et al., 1998). Gene expression studies suggest that ASN1 is the major expressed gene controlling Asn synthesis in plants. This is evidenced by the fact that dark induction of ASN1 mRNA correlates with dark induction of Asn levels in plants. In contrast, ASN2 is expressed at low levels in the dark, and its mRNA accumulates preferentially in light-treated plants (Lam et al., 1994, 1995, 1998).
The above-cited studies provide circumstantial evidence for the importance of AS in controlling nitrogen status in higher plants. Previous studies from our laboratory examined the effects of altering the expression of the pea AS1 gene in transgenic tobacco (Nicotiana tabacum). Although expressing the pea AS1 gene ectopically in tobacco led to an increase in levels of free Asn in vegetative tissues, the effects on nitrogen status in sink tissues were not thoroughly analyzed (Brears et al., 1993).
In this report, we provide evidence showing that overexpression of ASN1 in Arabidopsis enhances the nitrogen status of seeds, the ultimate storage organs for metabolites.
RESULTS
Construction of Transgenic Arabidopsis Overexpressing ASN1
The ASN1 cDNA previously isolated from Arabidopsis (Lam et al., 1994) was subcloned into a plant expression vector (pTEV; Brears et al., 1993) and was placed immediately downstream from the cauliflower mosaic virus (CaMV) 35S promoter. After transformation into Agrobacterium tumefaciens GV3101/pMP90 (Koncz and Schell, 1986), the target ASN1 gene was subsequently integrated into the genome of Arabidopsis ecotype Columbia-0 (Col-0) using vacuum infiltration techniques (Bechtold and Pelletier, 1993). Transgenic lines containing a single locus of the ASN1 transgene were identified by scoring the phenotypic ratio of kanamycin-resistant versus -sensitive seedlings (3:1; verified by chi square test) in the T2 generation resulting from self-pollination of T1 plants. Two 35S-ASN1 lines (362-D2-d6 and 362-4E-21) overexpressing the ASN1 gene obtained by independent transformation events and each carrying a single insertion locus were further propagated to obtain homozygous lines. As a control, a homozygous line (359-2C-8) containing a single insertion locus of the T-DNA of the vector (i.e. without the ASN1 gene) was also obtained.
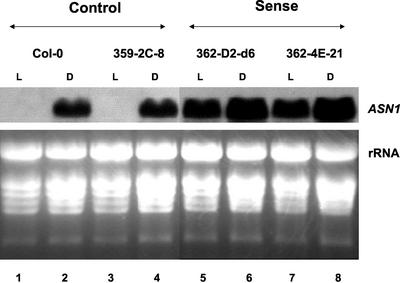
To determine the relative levels of the ASN1 mRNA in these transgenic lines, total RNA was extracted from leaves, and northern-blot analyses were performed (Fig. 1). In these studies, levels of ASN1 mRNA in the wild-type parent Col-0 and the vector-alone control 359-2C-8 were induced by continuous dark treatment and were repressed under continuous light treatment (Fig. 1, lanes 1–4), consistent with previous findings of the native ASN1 gene regulation (Lam et al., 1994). Notably, the levels of ASN1 mRNA were barely detectable in both controls grown under light conditions (Fig. 1, lanes 1 and 3). In contrast, ASN1 mRNA was constitutively overexpressed in 35S-ASN1 lines (362-D2-d6 and 362-4E-21) in both light-grown and dark-treated plants (Fig. 1, lanes 5–8). The ASN1 mRNA in light-grown 35S-ASN1 lines (Fig. 1, lanes 5 and 7) was found to be as abundant as levels detected in dark-adapted control plants (Fig. 1, lanes 2 and 4). Under dark conditions, where the ASN1 mRNA levels were generally high in the wild-type Col-0, the 35S-ASN1 lines (Fig. 1, lanes 6 and 8) still showed a higher expression than that of the controls (Fig. 1, lanes 2 and 4).
Figure 1.
Levels of ASN1 mRNA were dramatically elevated in 35S-ASN1 lines. Nine-day-old seedlings grown on Murashige and Skoog agar plates under a regular day/night cycle (16 h of light/8 h of dark) were transferred to soil for a further growth of 14 d. Plants were subsequently treated under continuous light (L) or continuous dark (D) conditions for 48 h. Total leaf RNA was extracted as described in “Materials and Methods.” An aliquot of 20 μg of total RNA from each line was loaded onto each lane. Northern-blot analysis was performed as described in “Materials and Methods.”
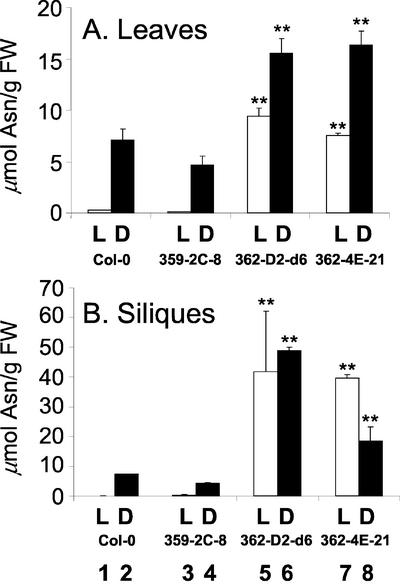
Precise study of ASN1 enzyme activities from plant extracts is hampered by instability of the AS enzyme in vitro (Sieciechowicz et al., 1988); copurification of a heat-stable, dialyzable inhibitor (Kern and Chrispeels, 1978; Joy et al., 1983); and the presence of contaminating asparaginase activity in plant extracts (Huber and Streeter, 1985) and the existence of multiple isoenzymes (Lam et al., 1998). Besides, our ultimate goal is to study the changes in amino acid metabolism and nitrogen status; therefore, we directly determined the steady-state levels of free Asn (product of AS enzymes) in leaves and siliques of 35S-ASN1 plants and compared them with control wild type (Col-0) or wild type transformed with an empty vector (359-2C-8; Fig. 2). For the experiment on leaf tissues, the plant materials were prepared using growth conditions identical to the northern-blot experiments described above. Nine-day-old seedlings grown under a normal day/night cycle were subject to 48 h of light or 48 h of dark treatment. As anticipated, the changes in the levels of free Asn in leaves of ASN1 transgenic and control plants (Fig. 2A) were found to parallel the changes in ASN1 mRNA levels (Fig. 1). The levels of free Asn in leaves of Col-0 and the vector-alone control were high in dark-adapted plants but nearly undetectable in light-grown plants (Fig. 2A, lanes 1 and 3). In contrast, levels of free Asn were elevated compared with the controls in 35S-ASN1 lines treated under both light- and dark-grown conditions (Fig. 2A, lanes 5–8). The levels of free Asn in the leaves of both 35S-ASN1 lines grown in light (Fig. 2A, lanes 5 and 7) were as high as (or higher than) that found in dark-adapted control plants (Fig. 2A, lanes 2 and 4).
Figure 2.
Elevation of amino acid levels in 35S-ASN1 lines grown under continuous light or continuous dark treatment. A, Leaves, plants were grown under the same treatment as in Figure 1. B, Siliques, 9-d-old seedlings were transferred to soil and grown under a regular day/night cycle (as described in Fig. 1) until most of the flower buds had turned into green siliques. One-half of the plants were subject to continuous light (L; white boxes) and the remaining one-half was subject to continuous dark treatment (D; black boxes), respectively, for 48 h. Leaves and siliques were harvested, and their amino acid levels were measured using amino acid analyzers (see “Materials and Methods”). Each bar represents an average of two samples. Error bars = ses. The data were analyzed by one-way ANOVA followed by lsd (Fisher's lsd) test. **, Corresponding lines were grouped distinctly from both control lines with P values less than 0.01. FW, Fresh weight.
Because free Asn can be used for long-range nitrogen-transport and storage, we investigated whether ASN1 overexpression and the ensuing dramatic increases in Asn pools affected the nitrogen status of the plants during seed development. Mature plants with developing green siliques were grown in continuous light or continuous dark for 48 h before harvesting. Free amino acid levels in siliques were measured. Similar to the results observed in leaves, levels of free Asn were only detectable in dark-adapted siliques of the control plants (Fig. 2B, lanes 2 and 4). The level of free Asn in siliques of 35S-ASN1 lines was significantly higher than that of the controls under both light- and dark-grown conditions (Fig. 2B, lanes 5–8).
Overexpression of the ASN1 Gene Enhances Nitrogen Status in Seeds
The ultimate sink organ in Arabidopsis is the seed, and the bulk of free amino acids are eventually incorporated into storage proteins in seeds. To test our hypothesis that overexpression of ASN1 will lead to a change in seed protein contents in the 35S-ASN1 lines, both the soluble protein contents (buffer extractable) and total protein contents (deduced from amino acid contents of acid-hydrolyzed seeds) were measured. Each of these experiments was performed with two different batches of seeds, and similar results were obtained. Each batch of seeds (containing all four lines tested) was grown, collected, stored, and analyzed under exactly the same duration and conditions. The 35S-ASN1 lines shed seeds normally, and no growth defects were observed during the entire life cycle. Data from one batch of seeds were shown (Fig. 3; Table I).
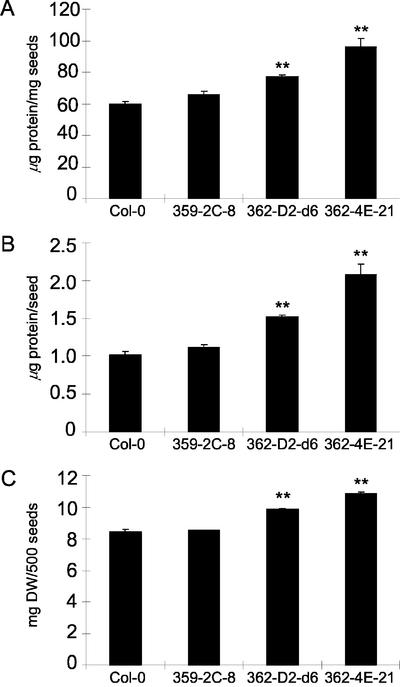
Figure 3.
Elevation of soluble protein content in seeds of 35S-ASN1 lines. Seeds were prepared, harvested, and then assayed for soluble seed protein contents and seed weight as described in “Materials and Methods.” Each data point represents an average value determined from six to eight aliquots of 500 seeds randomly sampled from two seed pools of 12 plants each for each line. Soluble seed protein contents were expressed both on per milligram seeds (A) and per seed (B) basis. Weight data for the 500-seed aliquots were also shown (C). Error bars = ses. The data were analyzed by one-way ANOVA followed by lsd test. **, Corresponding lines were grouped distinctly from both control lines with P values less than 0.01. DW, Air-dried weight.
Table I.
Total protein content and mol % of Asn + Asp (Asx) and Gln + Glu (Glx) in acid-hydrolyzed seeds
| Plant | Total Proteins | Asx | Glx |
|---|---|---|---|
| μg mg-1 seeds | mol % | ||
| Col-0 | 139.9 (1.2) | 7.2 (0.6) | 15.1 (1.4) |
| 359-2C-8 | 140.1 (7.2) | 7.4 (0.1) | 16.0 (0.5) |
| 362-D2-d6 | 161.3 (3.3)* | 10.7 (0.2)** | 15.8 (0.5) |
| 362-4E-21 | 169.3 (15.3)** | 9.3 (0.2)** | 15.7 (0.7) |
Plants were grown regularly as described in `Materials and Methods.' Seeds from about 10 plants for each line were pooled. Three aliquots of 70 to 100 mg of air-dried seeds were randomly sampled from each pool and then subjected to acid hydrolysis and amino acid determination (except for Col-0, where only two aliquots were used), as described in `Materials and Methods.' Total protein contents were deduced from total amino acid contents after acid hydrolysis of seeds. The relative mol % of Asx and Glx in seed proteins were also calculated. The data were analyzed by one-way ANOVA followed by LSD test* and **, Significant difference when compared with the wild type Col-0 with a P value less than 0.05 and 0.01, respectively. sds are shown in parentheses
Supporting our hypothesis, 35S-ASN1 lines exhibited an increase in seed-soluble protein contents compared with controls (Fig. 3). The increase in seed-soluble protein contents was observed both on per milligram of seeds (Fig. 3A) and per seed (Fig. 3B) basis. This result indicated that both the proportion and absolute amounts of seed-soluble proteins were increased. In fact, a light increase of seed weight was also observed (Fig. 3C). An aliquot of 500 seeds was used as one data point to circumvent the problem due to light weight of Arabidopsis seeds and to obtain a more homogenous sample.
To complement the data on seed-soluble protein contents, total protein contents were deduced from amino acid contents of acid-hydrolyzed seeds (without buffer extraction; Table I). Free amino acid contents in seeds were insignificant in this measurement because they were 2 orders of magnitude less than bound amino acids in seed proteins. Similar to the results of seed-soluble proteins, total protein contents in acid-hydrolyzed seeds in the two 35S-ASN1 lines were higher than the controls (Table I). Moreover, the mol % of Asx (representing major pool of four carbon amino acids) in total proteins was also increased. On the other hand, the mol % of Glx (representing major pool of five carbon amino acids) did not change. In fact, no drastic changes in the mol % of amino acids other than Asx were observed. It seemed that the increase in mol % of Asx was evenly balanced by subtle changes of other amino acids as a whole.
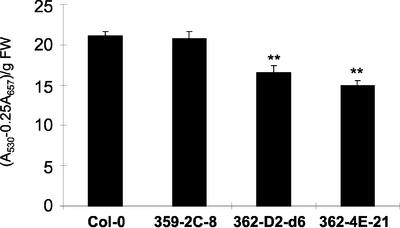
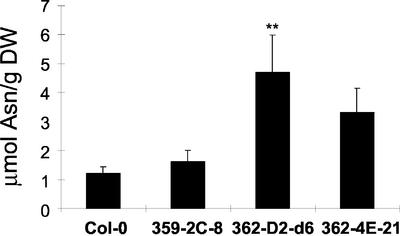
To test whether the increase of seed nitrogen in the 35S-ASN1 lines is of physiological significance, we determined the growth of young seedlings under nitrogen-limiting conditions. The growth of young seedlings under such conditions will be mainly dependent on the seed nitrogen storage. We previously showed that in high carbon and nitrogen-free media, young seedlings of Arabidopsis will accumulate anthocyanin. When exogenous Gln was added, nitrogen stress is only partially relieved due to nitrogen limitation (Hsieh et al., 1998). On nitrogen-free Murashige and Skoog medium containing 3% (w/v) Suc and 0.5 mm Gln, the levels of anthocyanin in young seedlings of 35S-ASN1 lines were found to be lower than that of the controls (Fig. 4), indicating that these transgenic lines are more tolerant to nitrogen limitation stress. On the other hand, when seedlings were grown on regular Murashige and Skoog medium containing 3% (w/v) Suc and 20 mm NH4NO3, the level of anthocyanin in all lines tested became undetectable (data not shown).
Figure 4.
Decrease of anthocyanin in young seedlings of 35S-ASN1 lines under high Suc and low nitrogen conditions. Anthocyanin contents (A530 - 0.25A657) of 10-d-old seedlings grown on nitrogen-free Murashige and Skoog agar plates containing 3% (w/v) Suc and 0.5 mm Gln were measured using a spectrometric method as described in “Materials and Methods.” Each data point represents an average value determined from four pools of 36 seedlings for each line. Seeds of different lines were sown onto the same agar plate to ensure homogeneity as described in “Materials and Methods.” Error bars = ses. The data were analyzed by one-way ANOVA followed by lsd test. **, Corresponding lines were grouped distinctly from both control lines with P values less than 0.01. FW, Fresh weight.
Overexpression of the ASN1 Gene Alters Amino Acid Composition in Sink Tissues of Seed-Shedding Plants
The above measurement of seed protein contents focused on the nitrogen status of mature seeds while the plant life cycle was completed. Mature seeds are developed from siliques that, in turn, come from flowers. During anthesis and silique development, vegetative tissues (leaves and stems) will act as source to supply nitrogen resources to the sink organs (flowers and developing siliques).
To study the effects of ASN1 overexpression on the sink-source relationships of amino acid metabolism, ASN1 mRNA and free amino acids in various source tissues (rosette leaves, stems, and cauline leaves) and sink tissues (flowers and siliques) were analyzed. Although the above results obtained under continuous light or continuous dark treatment (Figs. 1 and 2) amplified the contrast among the 35S-ASN1 lines and controls, analysis under regular day/night cycle will help to understand the physiological impact of such differences. The plants were grown under regular day/night cycle (16 h of light, 8 h of dark) for about 5 to 6 weeks until flowers and developing siliques emerged. Different tissues were harvested on the same day after 8 h into the light period (the difference between 35S-ASN1 lines and the wild type in light is easier to visualize than in the dark) so that the sink-source relationship could be compared.
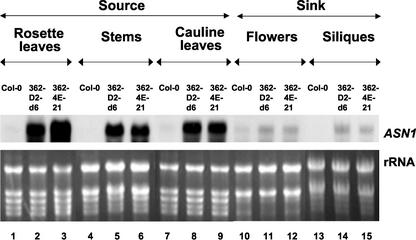
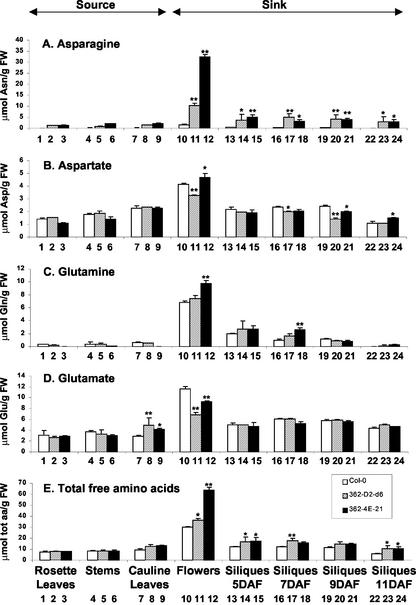
The levels of ASN1 mRNA in the 35S-ASN1 lines were higher than the wild-type Col-0 in all tissues tested (Fig. 5). However, the effect of ASN1 overexpression was much more prominent in vegetative tissues (rosette and cauline leaves and stems), which serve as source organs (Fig. 5, lanes 1–9), than in reproductive tissues (flowers and siliques), which serve as sink organs (Fig. 5, lanes 10–15). Steady-state free Asn levels in the same batch of samples were also measured. All tissues in the 35S-ASN1 lines exhibited a higher level of free Asn when compared with the wild-type Col-0 (Fig. 6A). However, contrasting to the level of ASN1 mRNA, the increase of free Asn levels in the sink tissues of these overexpressing lines (Fig. 6A, lanes 10–24) were much more drastic and significant than that of the source tissues (Fig. 6A, lanes 1–9). For instance, in flowers of the 35S-ASN1 lines, although there were only mild increases in the ASN1 mRNA levels (Fig. 5, lanes 10–12), the levels of Asn were most dramatically enhanced (Fig. 6A, lanes 10–12). Because free Gln (substrate of free Asn) levels in the sink tissues did not drop (Fig. 6C), it suggests that in addition to the possible increased de novo biosynthesis in the sink tissues (due to ASN1 overexpression), Asn accumulation in sink tissues such as flowers may be partially due to the transport of Asn from leaves, where ASN1 expression is the highest.
Figure 5.
Elevation of levels of ASN1 mRNA in sink versus source tissues of 35S-ASN1 lines grown under regular day/night cycle. Plants were grown under regular day/night cycle (16 h of light/8 h of dark) for about 5 to 6 weeks until flowers and development siliques emerged. Different tissues (rosette leaves, stems, cauline leaves, flowers, and siliques) were harvested on the same day after 8 h into the light period. The silique samples consisted of a mixture of siliques at the stage 5, 7, 9, and 11 d after flowering (DAF). Total RNA was extracted, and northern-blot analysis was performed using 20-μg total RNA aliquots as described in “Materials and Methods.”
Figure 6.
Changes in levels of free amino acid content in source versus sink tissues of 35S-ASN1 lines grown under regular day/night cycle. A to E, Free Asn, free Asp, free Gln, free Glu, and total free amino acids, respectively. The samples were harvested as described in Figure 5, except that siliques of different developmental stages were tagged, collected, and analyzed separately. Free amino acid analysis was performed as described in “Materials and Methods.” The contribution of Tyr was not included in the calculation of total free amino acids in E because the relative position of its peak recorded in the amino acid analyzer was very close to the internal standard nor-Leu and the absolute levels of Tyr were very low compared with other free amino acids. Each bar represents an average of two or three samples. Error bars = ses. The data were analyzed by one-way ANOVA followed by lsd test. * and **, Significant difference when compared with the wild-type Col-0, with a P value less than 0.05 or 0.01, respectively. FW, Fresh weight; Tot aa, total amino acids.
To test whether the accumulation of free Asn in sink tissues of 35S-ASN1 lines is related to an enhancement of Asn transport from source (where the ASN1 transgene is strongly expressed) to sink tissues (where the ASN1 transgene is only weakly expressed), Asn levels in phloem exudates were analyzed. Because the excised leaves must be placed immediately into the EDTA solution for phloem analysis, fresh weight data could not be obtained accurately. Therefore, the amino acid contents in the phloem were presented in a per dry weight basis for comparison. At least one of the 35S-ASN1 lines showed a significant increase in the level of Asn transported in the phloem when compared with the controls (Fig. 7).
Figure 7.
Elevation of free Asn levels in phloem exudates of 35S-ASN1 lines. Plants were grown for 5 weeks under regular day/night cycle, and individuals that bore at least 10 to 15 developing siliques were used in the analysis. Phloem exudates were obtained using a scaled-down EDTA elution method, and free amino acid contents were determined as described in “Materials and Methods.” Each bar represents an average of three samples. Error bars = ses. The data were analyzed by one-way ANOVA followed by lsd test. **, Corresponding lines were grouped distinctly from both control lines with P values less than 0.01. DW, Dry weight after phloem exudate collection.
To investigate the impact of increasing levels of free transported Asn on the overall nitrogen sink-source relationship, the compositions of free amino acids in different tissues were analyzed (Fig. 6; Table II). The overall levels of free amino acids were generally higher in sink tissues (especially in flowers) than source tissues in all plant constructs tested (wild-type and 35S-ASN1 lines; Fig. 6E, lanes 10–12). There were no significant differences in the amount of total free amino acids in rosette leaves and stems when comparing the 35S-ASN1 lines with the wild type (Fig. 6E, lanes 1–6).
Table II.
Mol % of free Asx and Glx in different tissues
| Plant | Amino Acid | Rosette Leaf | Stem | Cauline Leaf | Flower | 5-DAF Silique | 7-DAF Silique | 9-DAF Silique | 11-DAF Silique |
|---|---|---|---|---|---|---|---|---|---|
| Col-0 | Asx | 19.4 (2.8) | 22.1 (0.1) | 24.5 (1.0) | 19.1 (0.3) | 21.8 (1.3) | 22.3 (0.8) | 22.9 (0.4) | 18.8 (1.2) |
| 362-D2-d6 | Asx | 35.4 (1.0)** | 32.4 (0.6)* | 30.8 (2.9) | 37.9 (5.8)** | 31.2 (11.5) | 38.1 (7.4)** | 37.1 (8.3)** | 35.0 (18.1)** |
| 362-4E-21 | Asx | 31.8 (2.1)* | 42.9 (4.2)** | 34.7 (0.4)* | 58.5 (0.8)** | 39.7 (5.2)** | 31.8 (4.8)* | 41.3 (2.3)** | 39.9 (11.8)** |
| Col-0 | Glx | 43.9 (11.6) | 48.7 (3.6) | 36.6 (2.4) | 61.3 (0.1) | 57.6 (1.3) | 58.8 (1.3) | 59.9 (1.1) | 72.9 (3.8) |
| 362-D2-d6 | Glx | 35.4 (3.2) | 44.2 (2.4) | 42.2 (7.9) | 40.0(5.5)** | 46.9 (6.7)* | 43.0 (4.9)** | 45.7 (6.6)** | 50.1 (13.2)** |
| 362-4E-21 | Glx | 36.0 (0.0) | 38.9 (7.3) | 32.6 (2.3) | 30.0 (1.3)** | 42.2 (3.9)** | 49.5 (4.4)* | 44.0 (3.1)** | 49.0 (11.4)** |
Relative mol % of Asx and Glx in total free amino acid pools of various plant tissues were measured. Details of this experiment were described in Figure 4 and `Materials and Methods.' The data were analyzed by one-way ANOVA followed by lsd test. * and **, Significant difference when compared with the wild-type Col-0 with a P value less than 0.05 or 0.01, respectively. sds are shown in parentheses
On the other hand, a slight increase of total free amino acids was observed in the sink tissues of the 35S-ASN1 lines (Fig. 6E). In flowers and siliques, the predominant increase of Asn (Fig. 6A, lanes 10–24) was one major factor attribute to the increase in levels of total free amino acids (Fig. 6E, lanes 10–24). For all plants tested, a drop of free amino acid pool was observed in 11-DAF siliques (Fig. 6E, lanes 22–24), suggesting that large amounts of free amino acids were required for the synthesis of seed proteins at this development stage.
A quality change in the total free amino acids was also observed in the 35S-ASN1 lines when compared with the wild type (Table II). Due to the increase in levels of Asn, the mol % of Asx relative to total amino acid was significantly increased in nearly all tissues tested. On the other hand, the mol % of Glx (two major 5-C amino acids) was found to drop, especially in sink tissues despite that the absolute amount of Glx did not drop (Fig. 6, C and D).
DISCUSSION
In current high-throughput agricultural practices, nitrogen-containing fertilizers are applied to soil to increase crop yield. Aside from the fact that the fertilizers themselves are costly, high fertilizer applications are deleterious to the environment, causing ground water contamination. Therefore, increasing the efficiency of either nitrogen assimilation or utilization in higher plants would have significant economical and environmental implications. One way to optimize nitrogen utilization is to allocate more nitrogen resources to the organ of interest, such as seeds (edible part of many crop plants).
The correlation between Asn and seed nitrogen status has been established in several previous reports. Because the efficiency of protein synthesis depends on the light/dark regulation of AS activities (Dembinski et al., 1996a), elevations of leaf AS activities and Asn levels have been used as parameters to screen for high grain protein cultivars in maize (Dembinski et al., 1995) and rye (Secale cereale; Dembinski and Bany, 1991). Moreover, Asn may also play a role in the formation of seed nitrogen reserves (Dilworth and Dure, 1978; Sieciechowicz et al., 1988). A current model on seed protein biosynthesis also suggests that free Asn in siliques may play a major role in supplying Asp and nitrogen resources to seed coats and cotyledons during seed development (Bewley et al., 2000). This may explain the dramatic changes in Asn levels that accompany seed development in several plant species (Karchi et al., 1994).
In this report, we demonstrated that it is possible to manipulate the relationship between Asn and seed nitrogen status by controlling the expression of the ASN1 gene. In the 35S-ASN1 lines, an increase of ASN1 mRNA (Fig. 1) is accompanied by an increase of free Asn (Fig. 2), suggesting that ASN1 may encode an AS enzyme that plays a key role in regulating free Asn pools in Arabidopsis. Both seed-soluble protein (Fig. 3) and total seed protein (Table I) are higher in 35S-ASN1 lines compared with the controls. It is worthwhile to note that previous attempts to manipulate seed storage protein genes have resulted in altered protein composition but not in overall nitrogen storage in seed (Tabe et al., 2002). Our data suggest that quantitative changes in seed nitrogen reserves may require enhanced transporting nitrogen resources.
One direct impact of nitrogen resources in seeds is on the development of young seedlings, especially when exogenous nitrogen resources are limited. Using anthocyanin accumulation as an indicator of nitrogen limitation stress, we showed that the young seedlings of 35S-ASN1 lines are less stressful under nitrogen-limiting conditions (Fig. 4). This result further demonstrates the enhanced nitrogen status in seeds of the 35S-ASN1 lines.
Besides quantitative changes, the composition of amino acid in seeds of the 35S-ASN1 lines also differs from the controls. In acid-hydrolyzed samples, mol % Asx was increased (Table I), consistent with the results in developing siliques (Table II). Asx provides the substrate for the Asp amino acid biosynthetic pathway (products including essential amino acids such as Lys, Met, Thr, and Ile; Galili and Hofgen, 2002). An alternation in the relative percentage of free Asx may have strong implications concerning the amino acid metabolism in the sink tissues.
On the other hand, the result of mol % Glx in acid-hydrolyzed proteins (no change in 35S-ASN1 lines; Table I) was different from that in free amino acid pools of developing siliques (decreased in 35S-ASN1 lines; Table II). The observation that a decrease in mol % of Glx in developing siliques does not result in a corresponding decrease in mol % of Glx in seed proteins suggests that the free Glx pool in developing siliques may be saturated in supplying Glx to seed proteins. The main function of the remaining free Glx may be used as nitrogen donors to make other nitrogen-containing compounds in seeds, including other amino acids (Bewley et al., 2000). The explanation is still tentative and needs to be verified.
It will be just a zero sum game for overall nitrogen status in sink tissues (flowers and developing siliques) if no additional nitrogen resource is being transported in from source tissues (leaves and stems) during seed development. To obtain a better understanding of the changes in nitrogen sink-source relationship in 35S-ASN1 lines, the relative changes in ASN1 mRNA level and free amino acid composition in various source and sink tissues were analyzed. Apparently, the ASN1 mRNA levels in the sink tissues were lowered than that of the source tissues in 35S-ASN1 lines. This empirical result suggests that either the CaMV 35S promoter used in constitutive expression of the ASN1 gene actually expresses preferentially in vegetative tissues (discrepancy of the “constitutiveness” of constitutive promoters has been reported previously; Holtorf et al., 1995), or a tissue-specific mRNA degradation mechanism occurs in sink tissues. We cannot distinguish these two possibilities at this moment. In 35S-ASN1 lines, although the source tissues contained a much higher ASN1 mRNA level than the sink tissues, the levels of free Asn were higher in sink tissues than that of source tissues (Figs. 5 and 6). This result suggests that at least some of the increased free Asn produced in the source tissues may have been transported to the sink tissues. This hypothesis is supported by the results of phloem exudate analysis (Fig. 7) in which an increase of free Asn was observed in 35S-ASN1 lines.
In summary, this report shows that by controlling the expression of the ASN1 gene in source tissues, both the quality and quantity of nitrogen resources in the sink tissues can be altered. To apply this result (increase of Asx) to seed quality improvement, further manipulation is needed. In particular, deregulation of end production feedback inhibition and other transcriptional and posttranscriptional control mechanisms in the biosynthesis of the Asp family amino acids should be achieved (Galili and Hofgen, 2002). In addition, the presence of effective sinks must be engineered (e.g. Lys-rich [Sun et al., 1999] or Met-rich [Sun et al., 1987]) seed storage proteins) to entrap these essential amino acids. The impact of such alteration of nitrogen status on the composition of other vital metabolic components should also be analyzed.
MATERIALS AND METHODS
Plant Growth and Lighting Conditions
Seeds were sown on Murashige and Skoog (Murashige and Skoog, 1962) basal medium (pH adjusted to 5.7 with KOH; Invitrogen, Carlsbad, CA) containing 3% (w/v) Suc and 0.9% (w/v) agar. The seeds were then allowed to imbibe on Murashige and Skoog agar plates at 4°C in the dark for 2 d before being transferred to environmentally controlled growth chambers. For soil growth tests, 7- to 10-d-old seedlings were transferred to Metro-Mix 200 soil (Hummert, St. Louis) for further growth. Different lines were transferred to separate pots and subirrigated using a common tray. Water and nutrients were added to the tray but not to the individual pots to ensure even distribution of nitrogen resources. Regular day/night cycle was set to a day length (illumination of 80 μE at 22°C) of 16 h and a dark period (at 20°C) of 8 h. In several experiments (as indicated), plants were grown in a regular day/night cycle before transferred to continuous light (light grown) or continuous dark (dark adapted) for 48 h.
Generation of 35S-ASN1 Lines
The ASN1 cDNA (Lam et al., 1994) was subcloned into a pTEV vector (Brears et al., 1993) and expressed under the control of the CaMV 35S promoter. Binary vector constructions were transferred into the disarmed Agrobacterium tumefaciens strain GV3101/pMP90 (Koncz and Schell, 1986) by electroporation. Six- to 8-inch-height Col-0 plants with abundant flowering buds were subject to vacuum infiltration transformation (Bechtold and Pelletier, 1993). The transformed plants were allowed to continue to grow and set seed. Positive transformants were screened by sowing the resulting seeds on Murashige and Skoog agar plates supplemented with 50 μg mL-1 kanamycin. Single-locus insertions were scored (using chi square test) by observing a 3:1 ratio (kanamycin-resistant:kanamycin-sensitive seedlings) in T2 progeny resulted from self-pollination of T1.
RNA Analyses
Tissue samples were freshly collected and frozen immediately in liquid nitrogen. Total RNA was obtained by a phenol extraction protocol (Jackson and Larkins, 1976). Digoxigenin-labeled probes were generated by a commercial kit (Roche, Mannheim, Germany) according to the manufacturer's instructions. Northern-blot analyses were performed at 42°C in 50% (v/v) formamide hybridization solution for at least 16 h. Post-hybridization washing and chemiluminescent detection were done according to the Genius System User's Guide (Roche).
Amino Acid and Phloem Exudate Analyses
For free amino acid analysis, plant tissues were harvested and frozen immediately in liquid nitrogen. The samples were ground in 175 μL of protein grinding buffer (0.1 m Tris-HCl [pH 8.0] and 0.5% [v/v] 2-mecaptoethanol) supplemented with a total of 100 to 300 nmol of nor-Leu as the internal standard for amino acid analyses. One hundred sixty-five microliters of each sample extract was mixed with 510 μL of methanol: chloroform (6:2.5 [v/v]) followed by vortexing and incubation on ice for 30 min. After addition of 450 μL of water and 300 μL of chloroform, the samples were vortexed again before spun for 30 s using a microfuge. The top layer of each sample was collected and dried in a speed vacuum system. The dried pellets were resuspended in 400 to 500 μL of lithium buffer and filtered through 0.45-μm nylon filters before analyzed with amino acid analyzers (System 6300, Beckman, Fullerton, CA; and L8800, Hitachi, Tokyo).
Analysis and preparation of acid-hydrolyzed seeds were performed by the Australian Proteome Analysis Facility (Macquarie University, Sydney, Australia). About 70 to 100 mg of air-dried seeds underwent liquid phase acid hydrolysis in 6 m HCl at 110°C. After being dried and derivatized with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate, samples were analyzed by RP-HPLC (Waters 2690 Alliance, Waters, Milford, MA). Total protein was deduced by the summation of amino acid contents minus the water used in peptide bonds.
For phloem exudate collections, 5-week-old plants were grown under a regular day/night cycle, and plants that bore 10 to 15 developing siliques were used. At this stage of development, the sink is well established. An EDTA elution method (King and Zeevaart, 1974) was adopted and scaled down to fit the experimental need. The plants were allowed to enter the light cycle for 8 h (the differences between the 35S-ASN1 lines and the controls are easier to visualize in light than in the dark) before four rosette leaves from each plant were excised, and the petioles were placed immediately into a 500-μL microfuge tube filled with 100 μL of 20 mm EDTA (disodium salt of ethylenediamine-tetraacetic acid [pH 7.0]). The tubes were left opened in a sealed transparent container in which the atmosphere was water saturated to prevent uptake of the EDTA solution by the leaves. The excised leaves were allowed to complete their light cycles, and the phloem exudates were vacuum dried before being resuspended in 1 mL of lithium buffer, filtered, and analyzed as free amino acids (see above). The remaining leaf tissues were oven dried at 95°C for 48 h to obtain the dry weight.
Determination of Soluble Seed Protein Contents
Seeds were sown directly onto soil. Ten-day-old seedlings were transferred to new soil pots (three seedlings per pot) and allowed to grow under regular day/night cycle. The plants were allowed to shed seeds without further addition of fertilizers. For each line, all thoroughly air-dried seeds from 12 plants were pooled to form a homogenous seed pool. Two seed pools were made for each line, and three to four aliquots of 500 seeds were randomly sampled from each pool. The mean for each data point was calculated from a total of six to eight samples. Seed-soluble proteins were extracted as described previously (Sun et al., 1987). Each aliquot of 500 thoroughly air-dried seeds was first weighed using the AD-4 Autobalance (accuracy down to the microgram level; Perkin Elmer, Norwalk, CT) before being ground in 1 mL of seed extraction buffer (0.25 m sodium chloride and 0.05 m potassium phosphate [pH 7.0]) using mortar and pestle. Acid-washed sand was added to aid the grinding step. The homogenates were transferred to microfuge tubes and centrifuged for 4 min at 12,000g. About 0.4 mL of each supernatant was aspirated and placed in a new microfuge tube. After a second round of centrifugation, 0.2 mL of each supernatant was recovered. Protein content was then measured using the Bradford assay (Bradford, 1976).
Anthocyanin Measurement in Young Seedlings
Anthocyanin determination was largely based on a protocol described previously (Mita et al., 1997). Forty seeds of each line were sown onto the same nitrogen-free Murashige and Skoog agar plate supplemented with 3% (w/v) Suc and 0.5 mm Gln. After imbibition, the seeds were allowed to germinate and grow under a regular day/night cycle. Four sets of each setting were made. For each line, 36 germinated 10-d-old seedlings from each agar plate (constituting one data point) were extracted with 500 μL of methanol containing 1% (v/v) HCl at 4°C with continuous shaking in a cold room for 1 d. After centrifugation, the absorbance of the supernatant at 530 and 657 nm was measured by a Microplate Spectrophometer (SPECTRA max 250, Molecular Devices, Sunnyvale, CA), and the content of anthocyanin was calculated as A530 - 0.25A657.
Acknowledgments
The authors would like to thank Miss Iris Tong and Mr. Hung-Kong Lau (The Chinese University of Hong Kong) for their professional assistance in generating transgenic plants. Amino acid analysis was performed with the technical help of Mr. Thomas Tang (The Chinese University of Hong Kong) and the Australian Proteome Analysis Facility (Macquarie University, Sydney, Australia).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.020123.
This work was supported by the Hong Kong Research Grant Council (earmarked grant no. CUHK4292/98M to H.-M.L.) and by the U.S. Department of Energy (grant no. DEFG01–92–20071 to G.M.C.).
References
- Bechtold N, Pelletier G (1993) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In J Martinez-Zapater, J Salinas, eds, Arabidopsis Protocols. Humana Press Inc., Totowa, NJ, pp 259-266 [DOI] [PubMed]
- Bewley JD, Hempel FD, McCormik S, Zambryski P (2000) Reproductive development. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 988-1043
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254 [DOI] [PubMed] [Google Scholar]
- Brears T, Liu C, Knight TJ, Coruzzi GM (1993) Ectopic overexpression of asparagine synthetase in transgenic tobacco. Plant Physiol 103: 1285-1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burris RH, Roberts GP (1993) Biological nitrogen fixation. Annu Rev Nutr 13: 317-335 [DOI] [PubMed] [Google Scholar]
- Chevalier C, Bourgeois E, Just D, Raymond P (1996) Metabolic regulation of asparagine synthetase gene expression in maize (Zea mays L.) root tips. Plant J 9: 1-11 [DOI] [PubMed] [Google Scholar]
- Crawford NM, Arst HNJ (1993) The molecular genetics of nitrate assimilation in fungi and plants. Annu Rev Genet 27: 115-146 [DOI] [PubMed] [Google Scholar]
- Davis KM, Seelye JF, Irving DE, Borst WM, Hurst PL, King GA (1996) Sugar regulation of harvest-related genes in asparagus. Plant Physiol 111: 877-883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delavault P, Estabrook E, Albrecht H, Wrobel R, Yoder JI (1998) Host-root exudates increase gene expression of asparagine synthetase in the roots of a hemiparasitic plant Triphysaria versicolor (Scrophulariaceae). Gene 222: 155-162 [DOI] [PubMed] [Google Scholar]
- Dembinski E, Bany S (1991) The amino acid pool of high and low protein rye inbred lines (Secale cereale L.). J Plant Physiol 138: 494-496 [Google Scholar]
- Dembinski E, Bany S, Raczynska-Bojanowska K (1995) Asparagine and glutamine in the leaves of high and low protein maize. Acta Physiol Plant 17: 361-365 [Google Scholar]
- Dembinski E, Wisniewska I, Raczynska-Bojanowska K (1996a) The efficiency of protein synthesis in maize depends on the light regulation of the activities of the enzymes of nitrogen metabolism. J Plant Physiol 149: 466-468 [Google Scholar]
- Dembinski E, Wisniewska I, Zebrowski J, Raczynska-Bojanowska K (1996b) Negative regulation of asparagine synthetase in the leaves of maize seedling by light, benzyladenine and glucose. Physiol Plant 96: 66-70 [Google Scholar]
- Dilworth MF, Dure L (1978) Developmental biochemistry of cotton seed embryogenesis and germination: X. Nitrogen flow from arginine to asparagine in germination. Plant Physiol 61: 698-702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs CG, Somerfield SD (1997) Asparagine synthetase gene expression increases as sucrose declines in broccoli after harvest. N Z J Crop Hortic Sci 25: 191-195 [Google Scholar]
- Galili G, Hofgen R (2002) Metabolic engineering of amino acids and storage proteins in plants. Metab Eng 4: 3-11 [DOI] [PubMed] [Google Scholar]
- Holtorf S, Apel K, Bohlmann H (1995) Comparison of different constitutive and inducible promoters for the overexpression of transgenes in Arabidopsis thaliana. Plant Mol Biol 29: 637-646 [DOI] [PubMed] [Google Scholar]
- Hsieh M-H, Lam H-M, van de Loo FJ, Coruzzi G (1998) A PII-like protein in Arabidopsis: putative role in nitrogen sensing. Proc Natl Acad Sci USA 95: 13965-13970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber TA, Streeter JG (1985) Purification and properties of asparagine synthetase from soybean root nodules. Plant Sci 42: 9-17 [Google Scholar]
- Hughes CA, Beard HS, Matthews BF (1997) Molecular cloning and expression of two cDNA encoding asparagine synthetase in soybean. Plant Mol Biol 33: 301-311 [DOI] [PubMed] [Google Scholar]
- Jackson AO, Larkins BA (1976) Influence of ionic strength, pH, and chelation of divalent metals on isolation of polyribosomes from tobacco leaves. Plant Physiol 57: 5-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy KW, Ireland RJ, Lea PJ (1983) Asparagine synthesis in pea leaves, and the occurrence of an asparagine synthetase inhibitor. Plant Physiol 73: 165-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchi H, Shaul O, Galili G (1994) Lysine synthesis and catabolism are coordinately regulated during tobacco seed development. Proc Natl Acad Sci USA 91: 2577-2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern R, Chrispeels MJ (1978) Influence of the axis in the enzymes of protein and amide metabolism in the cotyledons of mung bean seedlings. Plant Physiol 62: 815-819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Zeevaart JAD (1974) Enhancement of phloem exudation from cut petioles by chelating reagents. Plant Physiol 53: 96-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204: 383-396 [Google Scholar]
- Lam H-M, Coschigano K, Schultz C, Melo-Oliveira R, Tjaden G, Oliveira I, Ngai N, Hsieh M-H, Coruzzi G (1995) Use of Arabidopsis mutants and genes to study amide amino acid biosynthesis. Plant Cell 7: 887-898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H-M, Coschigano KT, Oliveira IC, Melo-Oliveira R, Coruzzi G (1996) The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47: 569-593 [DOI] [PubMed] [Google Scholar]
- Lam H-M, Hsieh M-H, Coruzzi G (1998) Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana. Plant J 16: 345-353 [DOI] [PubMed] [Google Scholar]
- Lam H-M, Peng S, Coruzzi G (1994) Metabolic control of asparagine synthetase gene expression in Arabidopsis thaliana. Plant Physiol 106: 1347-1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea PJ, Miflin BJ (1980) Transport and metabolism of asparagine and other nitrogen compounds within the plant. In PK Stumpt, EE Conn, eds, The Biochemistry of Plants, Vol 5. Academic Press, New York, pp 569-607 [Google Scholar]
- Lea PJ, Robinson SA, Stewart GR (1990) The enzymology and metabolism of glutamine, glutamate, and asparagine. In BJ Miflin, PJ Lea, eds, The Biochemistry of Plants: Amino Acids and Derivatives, Vol 16. Academic Press, New York, pp 121-159 [Google Scholar]
- Miflin BJ, Lea PJ (1980) Ammonia assimilation. In BJ Miflin, ed, The Biochemistry of Plants: Amino Acids and Derivatives, Vol 5. Academic Press, New York, pp 169-202 [Google Scholar]
- Mita S, Murano N, Akaike M, Nakamura K (1997) Mutants of Arabidopsis thaliana with pleiotropic effects on the expression of the gene for betaamylase and on the accumulation of anthocyanin that are inducible by sugars. Plant J 11: 841-851 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for the growth and bioassay with tobacco tissue culture. Physiol Plant 15: 473-497 [Google Scholar]
- Neuhaus G, Bowler C, Hiratsuka K, Yamagata H, Chua NH (1997) Phytochrome-regulated repression of gene expression requires calcium and cGMP. EMBO J 16: 2554-2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieciechowicz KA, Joy KW, Ireland RJ (1988) The metabolism of asparagine in plants. Phytochemistry 27: 663-671 [Google Scholar]
- Sun SS-M, Leung FW, Tomic JC (1987) Brazil nut (Bertholletia excelsa) protein: fractionation, composition and identification of a sulfur-rich protein. J Agric Food Chem 35: 232-235 [Google Scholar]
- Sun SS-M, Wang M-L, Tu HM, Zuo W-N, Xiong L, Cheng M-K (1999) Transgenic approach to improve protein, starch and taste quality of food plants. In JR Whitaker, NF Haard, CF Shoemaker, RP Singh, eds, Food for Health in the Pacific Rim. Food & Nutrition Press Inc., Trumbull, CT, pp 560-563
- Tabe L, Hagan N, Higgins TJV (2002) Plasticity of seed storage protein composition in response to nitrogen and sulfur availability. Curr Opin Plant Biol 5: 212-217 [DOI] [PubMed] [Google Scholar]
- Tsai FY, Coruzzi GM (1990) Dark-induced and organ-specific expression of two asparagine synthetase genes in Pisum sativum. EMBO J 9: 323-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RN, Smyth AJ, Massonneau A, Prosser IM, Clarkson DT (1996) Molecular cloning and characterization of asparagine synthetase from Lotus japonicus: dynamics of asparagine synthesis in N-sufficient conditions. Plant Mol Biol 30: 883-897 [DOI] [PubMed] [Google Scholar]