Abstract
Aluminum (Al) toxicity, which is caused by the solubilization of Al3+ in acid soils resulting in inhibition of root growth and nutrient/water acquisition, is a serious limitation to crop production, because up to one-half of the world's potentially arable land is acidic. To date, however, no Al tolerance genes have yet been cloned. The physiological mechanisms of tolerance are somewhat better understood; the major documented mechanism involves the Al-activated release of Al-binding organic acids from the root tip, preventing uptake into the primary site of toxicity. In this study, a quantitative trait loci analysis of Al tolerance in Arabidopsis was conducted, which also correlated Al tolerance quantitative trait locus (QTL) with physiological mechanisms of tolerance. The analysis identified two major loci, which explain approximately 40% of the variance in Al tolerance observed among recombinant inbred lines derived from Landsberg erecta (sensitive) and Columbia (tolerant). We characterized the mechanism by which tolerance is achieved, and we found that the two QTL cosegregate with an Al-activated release of malate from Arabidopsis roots. Although only two of the QTL have been identified, malate release explains nearly all (95%) of the variation in Al tolerance in this population. Al tolerance in Landsberg erecta × Columbia is more complex genetically than physiologically, in that a number of genes underlie a single physiological mechanism involving root malate release. These findings have set the stage for the subsequent cloning of the genes responsible for the Al tolerance QTL, and a genomics-based cloning strategy and initial progress on this are also discussed.
Al toxicity is a major limiting factor for crop production worldwide. Al is the third most abundant element in the earth's crust and is toxic to plants when solubilized into the soil solution at acidic pH values (Kochian, 1995). When the soil pH drops below 5.0, Al is solubilized as the phytotoxic Al3+ species from nontoxic Al silicates and oxides. Al-intoxicated plants have limited root growth and development, and thus acquire water and nutrients from the soil poorly (Kochian, 1995). Root stunting directly translates into reduced plant vigor and yield. Acid soils make up approximately 30% of all land presently under cultivation and more than 50% of potentially arable lands (von Uexküll and Mutert, 1995). These soil types are not evenly distributed worldwide; Al toxicity is the primary limitation on crop production for 37.9% of farmland in Southeast Asia, 30.9% of Latin America, and approximately 20% in East Asia, SubSaharan Africa, and North America (Wood et al., 2000). Low tolerance to Al stress directly reduces food security in many areas where it is most tenuous. Furthermore, in developed countries such as the United States, high-input farming practices such as the extensive use of ammonia fertilizers are causing further acidification of agricultural soils, creating new acid soils from previously neutral ones (Jackson and Reisenauer, 1984). Although soil amendments such as lime can ameliorate soil acidity, this is neither an economic option for poor farmers nor an effective strategy for alleviating subsoil acidity (Rao et al., 1993).
The genetic analysis of Al tolerance has been an active area of research. Significant intraspecific variation for Al tolerance is well known in many plant species (Foy, 1988). The genetic architecture of tolerance is variable, being monogenic in some species but polygenic in others. In wheat (Triticum aestivum), crosses between multiple cultivars suggest that the majority of variability for Al tolerance apparently occurs at a single locus on chromosome 4DL (Kerridge and Kronstad, 1968; Camargo, 1984; Delhaize et al., 1993b). Other loci may contribute to Al tolerance in certain wheat cultivars, but variability in Al tolerance in wheat and other members of the Triticeae (e.g. rye [Secale cereale] and barley [Hordeum vulgare]) appears to be genetically very simple and centered on the long arm of chromosome 4 (Camargo, 1981; Berzonsky, 1992; Tang et al., 2000; Miftahudin et al., 2002). In other species, such as maize (Zea mays) and rice (Oryza sativa), Al tolerance appears to be quantitative and involves four to 10 QTL (Sawazaki and Furlani, 1986; Magnavaca et al., 1987; Prioli, 1987; Wu et al., 2000; Nguyen et al., 2001; Nguyen et al., 2002). Al tolerance has also recently been shown to be a quantitative trait in Arabidopsis (Kobayashi and Koyama, 2002). Despite the progress to date, no Al tolerance genes have yet been cloned from any plant species.
More progress has been made in recent years in understanding the physiological basis of Al tolerance. Exclusion of Al from the root tip is the most common tolerance mechanism. An exclusion mechanism based on root exudation of Al-chelating organic acids such as malate, citrate, or oxalate has been described in both monocots (for example, see Delhaize et al., 1993b; Pellet et al., 1995) and dicots (for example, see Miyasaka et al., 1991; Silva et al., 2001). The organic acid is released into the rhizosphere, where it acts as a ligand for Al3+ (the primary toxic Al species), forming a nontoxic complex that does not readily enter the root. It has been shown that Al-activated root malate release cosegregates with the Al tolerance locus in wheat, directly linking the genetic and physiological studies (Delhaize et al., 1993a, 1993b).
Quantitative trait locus (QTL) mapping allows one to statistically identify individual chromosomal regions containing genetic factors that contribute to variation in a complex trait (Alonso-Blanco and Koornneef, 2000). QTL mapping has been used widely in the dissection of complex traits, Al tolerance included, in crop plants and in model plants such as Arabidopsis (Burr and Burr, 1991; Doerge, 2002). QTL mapping is the first step in the positional cloning of genes underlying complex phenotypes. In recent years, positional cloning has become increasingly routine and is a particularly fruitful approach in a sequenced model organism such as Arabidopsis (Lukowitz et al., 2000).
Recombinant inbred lines (RIL) from a cross between two ecotypes or inbreds provide a convenient population type for QTL mapping. In the case of a selfing organism, each RIL is derived from an independent segregating individual (such as an F2) and is fixed by repeated generations of single-seed descent. As a result, each RIL is homozygous at (very nearly) every locus, and each line can be thought of as a unique mosaic of the paternal and maternal chromosome complements (Burr and Burr, 1991). An important property of an RIL population is that one can replicate the same genotype within and across conditions and experiments.
We used an RIL population derived from an F2 cross between the Arabidopsis ecotypes Landsberg erecta (Ler) and Columbia (Col) to identify factors contributing to Al tolerance (Lister and Dean, 1993). This population has been intensively characterized, both genotypically (http://nasc.nott.ac.uk/new_ri_map.html) and phenotypically (Yanovsky et al., 1997; Juenger et al., 2000; Lambrix et al., 2001; Kliebenstein et al., 2002; Perez-Perez et al., 2002; Quesada et al., 2002). Col is considerably more Al tolerant than Ler, requiring twice the level of Al to inhibit root growth to the same degree as seen in Ler (Larsen et al., 1996). Col, Ler, and 99 RIL were grown on an acidic medium ±Al to estimate Al tolerance by measuring root growth in both environments. Two significant QTL were detected for Al tolerance. In hydroponic culture, we then tested both parents and selected RIL to identify how Al tolerance was conferred by the two QTL. Both QTL correlated with a release of malate from Arabidopsis roots, suggesting that differences in malate release explain the differences in Al tolerance observed between Col, Ler, and their derivative RIL.
RESULTS
QTL Mapping and Analysis
Al tolerance was estimated by measuring the impact of Al stress on root growth compared with plants grown under identical conditions lacking Al. Plants were grown in solid, gellan gum media where the soluble, toxic Al was introduced by soaking the plates in a full-strength nutrient solution containing 1 mm AlCl3 (adapted from Larsen et al., 1996). The Ler × Col RIL set was chosen for our experiments because we already had established a difference in Al tolerance between these ecotypes and because of the availability of the densely genotyped mapping population (Lister and Dean, 1993; Larsen et al., 1996).
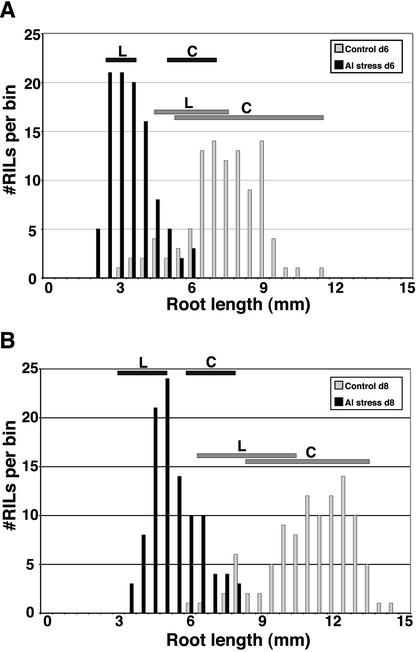
Ler and Col grew at approximately equal rates in the absence of Al but differed substantially in the presence of Al (Fig. 1). The distributions for root length in the RIL, under both control and Al stress conditions, were not obviously multimodal, which might have indicated a major gene. There was only minor transgressive segregation (no line was more than two sds more extreme than either parent), which indicates that Col contains most of the alleles that contribute to increased Al tolerance. Broad sense heritability was calculated for root growth under both control (h2b = 0.98) and Al-treated (h2b = 0.99) conditions, indicating that root growth is a highly heritable trait.
Figure 1.
Distribution of mean root lengths among RIL in the Ler × Col population. A, Day six. Mean root lengths for each RIL are organized into bins (0.5-mm increment) to show the distribution within the population. Control-treated plants are shown with vertical gray bars, whereas Al-treated plants are shown with vertical black bars. Average root lengths for Ler (L) and Col (C), including 1 sd above and below the mean, are given as horizontal bars. B, Day eight.
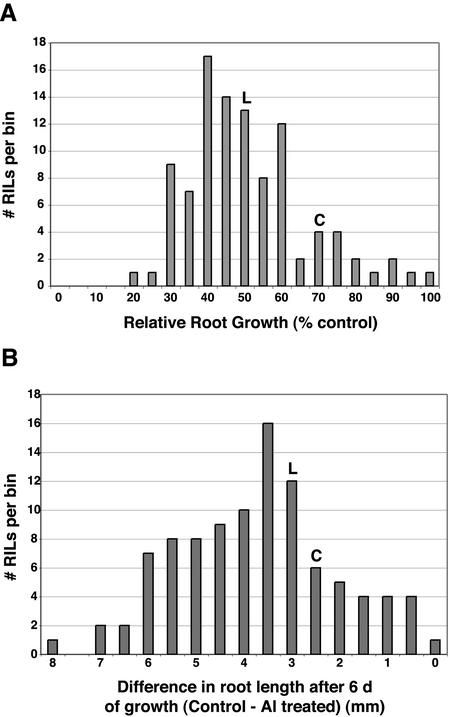
We investigated 10 variables related to root length to identify Al tolerance loci segregating in the RIL population. Four direct variables were used in the analysis, the mean root length at each time point (after d 6 or 8) and growth condition (RLAl6, RLAl8, RLC6, and RLC8) as well as six derived variables, which resulted from differences and ratios between mean root lengths for the ±Al-treated plants (D6, D8, R1 R2, DAl, and DC). The typical measure of Al tolerance is to consider a derived variable rather than a direct one in an attempt to scale root growth under Al stress to control conditions. For example, in Figure 2A, we present the distribution of mean relative root growth (RRG). As RRG approaches 100%, plants growth equally well under both conditions (highly tolerant when RLC6 = RLAl6), whereas RRG values that approach zero indicate high sensitivity to Al stress (when RLAl6 = 0). RRG due to Al stress in Ler was 47%, whereas RRG for Col was 71% (Fig. 2A). Individual RIL varied from 23% to 98% RRG. We also considered the simple difference in root growth D (D = RLcontrol - RLAl), because ratios can be problematic for statistical analyses (Fig. 2B; Sokal, 1995). As D approaches zero, root growth occurs at the same rate under both control and Al stress conditions (highly tolerant). As D approaches RLControl, root growth occurs at very different rates (highly sensitive). Ler and Col were more similar with regards to Al tolerance when the parameter D6 was used (3.4 and 2.5, respectively), but again, individual RIL varied from highly tolerant to highly sensitive (D6 of 0.1–8.1). For each of the 10 variables or traits we considered, empirical significance thresholds were determined from 1,000 permutations (data not shown).
Figure 2.
Distribution of RRG estimates between RIL in the Ler × Col population. A, RRG variable. Root growth inhibition was estimated using RRG for d 6: RLAl6/RLC6; RRG is a scaled variable and is a measure of RRG. RRG values are expressed as percentage values and organized into bins (5% increments) to show the distribution within the population. RRG values are shown for the Ler (L) and Col (C) parental varieties. B, D6 variable. Root growth inhibition was measured by D6 = RLC6 - RLAl6. Also shown are D6 values for Ler (L) and Col (C).
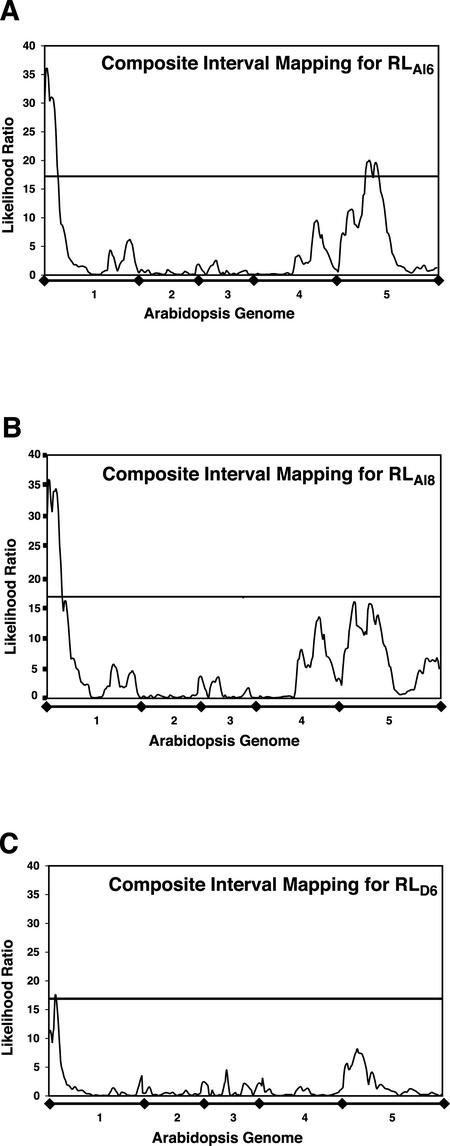
Composite interval mapping identified significant QTL for root growth with three of the 10 variables: RLAl6, RLAl8, and D6 (Fig. 3). No significant QTL were found for growth under control conditions. The two regions of the genome most strongly associated with Al tolerance were the north end of chromosome one (closest markers m488/apx1A) and the middle of chromosome five (closest marker TSL). Both QTL were identified as significant for the RLAl6 variable, whereas QTL no. 1 (chromosome one) was also identified by RLAl8 and D6. The closest linked markers, LR, estimated additive effect and percent variance explained for the significant QTL are listed in Table I. In all cases, the alleles from the Col parent conferred greater tolerance.
Figure 3.
Likelihood ratio (LR) plots for significant Al tolerance QTL. LR values for three traits plotted against a linear representation of the Arabidopsis genome. A, RLAl6. B, RLAl8. C, D6.
Table I.
Significant QTL: results from composite interval analysis
| Trait | Chromosome | cM | Marker | LR | Additive Effect | r2 |
|---|---|---|---|---|---|---|
| RLA16 | 1 | 7.74 | m488 | 36.03*** | 0.52 | 0.32 |
| RLA16 | 5 | 37.75 | TSL | 19.98** | 0.42 | 0.19 |
| RLA18 | 1 | 9.31 | apx1A | 34.40*** | 0.59 | 0.31 |
| D6 | 1 | 9.31 | apx1A | 17.60* | -0.70 | 0.16 |
The LR, additive effect, and proportion of the phenotypic variance explained by the QTL, conditioned on the background markers, are given at the point for which the LR is highest within each region. The location of each trait is indicated by the chromosome, the distance in cM from the first marker (position), and the name of the closest marker used in the analysis. LR maxima within 10 cM of a higher local LR maximum are not counted. The statistical significance of the LR is indicated by *, **, or ***, which denote a value in excess of the α = 0.1, 0.05, or 0.01 critical value, respectively. A positive additive effect indicates a longer root length in individuals homozygous for the Col allele for the first three traits. Due to the nature of the fourth trait (D6), a negative additive effect indicates Col contributed the superior allele
Using marker regression analysis (MRA; Weller, 1986), which does not control for factors segregating at unlinked loci, additional QTL were identified that surpassed empirical significance thresholds (Table II). Although MRA typically makes fewer type II errors (false negative QTL), it also tends to make more type I errors (false positive QTL). MRA suggests that there may be three (rather than one) QTL on chromosome five in the presence of Al. The previously identified locus near marker TSL has a stronger effect (Table II, b1 value) on d 6, whereas a second locus, near marker nga158, has a stronger effect on d 8. A third novel region of chromosome five, near marker ATR3, confers a greater change in root length between d 6 and d 8 in Al-treated plants in Col relative to Ler. Also, a locus on chromosome four near marker mi123 contributes to a larger difference in tolerance under Al treatment in Col than in Ler on both d 6 and 8. However, for this study, we will rely upon the more conservative QTL estimate from the composite interval analysis.
Table II.
Significant QTL: results from marker regression analysis
| Trait | Chromosome | cM | Marker | LR | b1 |
|---|---|---|---|---|---|
| RLA16 | 1 | 7.74 | m488 | 34.10*** | 0.47 |
| RLA16 | 5 | 37.75 | TSL | 19.63*** | 0.40 |
| RLA18 | 1 | 7.74 | m488 | 33.88*** | 0.57 |
| RLA18 | 4 | 75.65 | mi123 | 12.48** | 0.37 |
| RLA18 | 5 | 18.12 | nga158 | 15.84** | 0.41 |
| D6 | 1 | 11.35 | ARR4 | 17.61*** | -0.70 |
| D8 | 1 | 11.35 | ARR4 | 16.35*** | -0.72 |
| DA | 5 | 118.44 | ATR3 | 14.44** | 0.19 |
The LR and additive effect (bl) are given at the point for which LR is highest within each region. Location and statistical significance are reported as in Table I
Both the m488/apx1A and TSL regions on chromosomes 1 and 5 were significant for trait RLAl6. The mean value of RLAl6 for each of the four two-loci genotypes is shown in Table III. A two-way ANOVA was used to test for a significant interaction between these two QTL. The results (Table IV) confirm the statistical significance of the trend apparent in Table III, whereby the gain in tolerance seen in the class containing both of the superior QTL alleles (Col, Col, or CC) is larger than would be expected based on the performance of CL and LC relative to LL (most sensitive; Ler at both QTL). The adjusted r2 is 0.398 for the joint model, whereas it is 0.260 and 0.196 for the single-classification ANOVAs including apx1A and TSL, respectively.
Table III.
Interaction between QTL on chromosome 1 and 5 for trait RLA16
| apx1A Genotype | TSL Genotype | Mean RLA16 | se |
|---|---|---|---|
| mm | |||
| Columbia | Columbia | 4.68 | 0.15 |
| Columbia | Landsberg | 3.71 | 0.14 |
| Landsberg | Columbia | 3.42 | 0.20 |
| Landsberg | Landsberg | 3.13 | 0.12 |
| Columbia (ecotype) | 6.10 | 0.21 | |
| Landsberg (ecotype) | 3.00 | 0.13 |
Mean and se of RLA16 for the six genotypic classes
Markers apx1A (chromosome 1) and TSL (chromosome 5) were used as factors in a two-way ANOVA for RLA16. Similar results were obtained when marker m488 was used in place of apx1A. The number of lines included in the analysis was 97, four lines lacked marker because information for m488, apx1A, or TSL. The adjusted r2 is 0.398 for the joint model. DF, degrees of freedom. SS, sum of squares
Table IV.
Analysis of interaction between QTL for trait RLA16
| Factor | DF | SS | F | P |
|---|---|---|---|---|
| apx1A | 1 | 17.49 | 35.21 | <0.0001 |
| TSL | 1 | 8.02 | 16.14 | 0.0001 |
| apx1A*TSL | 1 | 2.38 | 4.80 | 0.0309 |
| Error | 93 | 46.18 |
Markers apx1A (chromosome 1) and TSL (chromosome 5) were used as factors in a two-way analysis of variance for RLA16. Similar results were obtained when marker m488 was used in place of apx1A. The number of lines included in the analysis was 97, because four lines lacked marker information for m488, apx1A, or BL. The adjusted r2 is 0.398 for the joint model. DF, degrees of freedom. SS, sum of squares
Physiological Analysis of the Al Tolerance QTL
We identified two significant QTL for Al tolerance in the Ler × Col RIL population. These major QTL apparently act together in the same pathway, based on the evidence for epistasis, and together explain approximately 40% of the variation in tolerance among the RI lines. We attempted to determine the physiological mechanisms by which the QTL condition tolerance. This might reveal a phenotype more suitable for the fine-mapping and eventual positional cloning of the genes responsible for the QTL, if each QTL conferred Al tolerance by distinct mechanisms. Because Al-activated root release of organic acids is the best documented tolerance mechanism, we designed experiments to identify and quantify the components of Arabidopsis root exudates to test the potential involvement of the QTL.
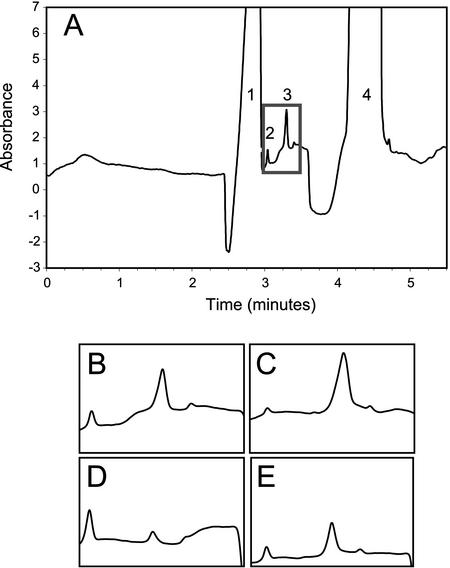
Previous studies have used high-pressure liquid chromatography (HPLC) for the analysis of root exudates (Pellet et al., 1995; Ryan et al., 1995; Larsen et al., 1998). To discriminate between inorganic and organic anions, HPLC requires the use of a very simple salt solution as a hydroponic growth medium for collection of root exudates. Arabidopsis is highly sensitive to hydroponic growth conditions; the roots of seedlings neither grow well nor release substantial amounts of organic acids when grown in simple salt solution (Larsen et al., 1998). Thus, a different analytical technique, capillary electrophoresis, was employed so that seedlings could be grown and root exudates collected from a nutrient solution more optimal for root growth. Using capillary electrophoresis, organic anions (e.g. citrate and malate) released from the plant can be discriminated from inorganic anions (e.g. sulfate, phosphate, and nitrate) in nutrient solution (Piñeros et al., 2002). Preliminary experiments demonstrated that Arabidopsis does alter the composition of root exudates in response to Al stress and that differences exist between Col and Ler consistent with the difference in tolerance (Fig. 4). A representative electropherogram is shown, which contains four principal peaks (Fig. 4A). Both of the major peaks derive from the nutrient solution. The early major peak (1) is composed of Cl-, NO3-, and SO42- anions, all of which essentially comigrate. The trailing major peak (4) is composed of Homopipes, a pH buffer that keeps the small volume of the exudation solution in the relevant pH range to maintain Al3+ toxicity during the exudation component of the experiment. The early minor peak (2) is citrate and the slightly later minor peak (3) is composed of malate and phosphate. Representative traces are shown for exudates collected from Col and Ler, grown in control and Al-treated conditions, with a focus on the citrate/malate/phosphate region (Fig. 4, B–E). Both ecotypes exhibit the same general response to Al stress—citrate release decreases while malate/phosphate release increases. However, the malate/phosphate peak for Col is approximately eight times as large as the one seen in Ler, indicating that Col releases more protective ligands in response to Al stress than does Ler, which is consistent with the difference in tolerance observed.
Figure 4.
Representative capillary electrophoresis traces for hydroponic media solutions. A, Representative complete electropherogram for root exudates collected from Col seedlings grown under control conditions. B, Col, control sample for time window 3.0 to 3.6 min, which brackets the elution of citrate (first peak) and malate/phosphate (second peak). C, Col, Al treated, for 3.0- to 3.6-min elution period. D, Ler, control, for 3.0- to 3.6-min elution period. E, Ler, Al treated, for 3.0- to 3.6-min elution period.
To test whether the two major Al tolerance QTL condition organic acid release, 10 RIL for each of the four genotypic classes at the two loci (Col, Col-CC; Col, Ler-CL; Ler, Col-LC; Ler, Ler-LL) were selected to be representative of those genotypes. The observations for each of the four genotypic classes were pooled to focus on the effects of the major QTL and to randomize the effects of the rest of the genome. One limitation of the capillary electrophoresis technique is that malate and phosphate comigrate due to similarities in size and charge density. Therefore, phosphate levels were also determined using a spectrophotometric assay, and the joint malate/phosphate peaks were deconvoluted accordingly (Baykov et al., 1988).
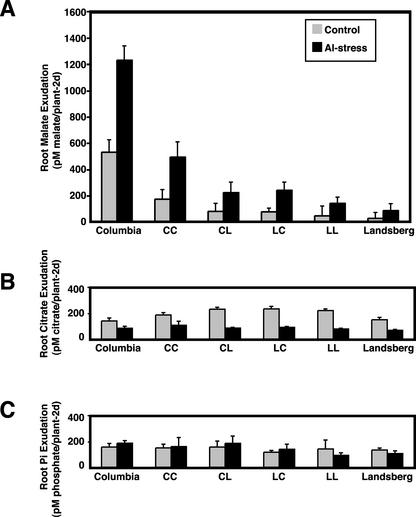
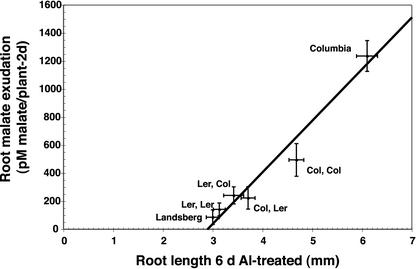
Al stress increased the release of malate from Arabidopsis roots (Fig. 5A). All of the genotypes tested (both parents and the four QTL classes) demonstrated approximately a 3-fold increase in malate release in the Al stress treatment (Fig. 5, black bars) compared with the control treatment (Fig. 5, gray bars). However, the absolute magnitude of the malate flux varied according to genotype in a pattern consistent with the pattern of Al tolerance. The CC RIL genotypes, as a class, released more malate than the other three genotypes, the CL and LC classes were indistinguishable from each other, and the LL class was equivalent to the Ler parent (Fig. 3A; Table V). An ANOVA confirmed that the six genotypic classes (two parents and four QTL classes) differ in malate release (F = 15.96, P = 5.7e-11). The almost perfect linear relationship (r = 0.975) between malate release and root length under Al stress for the six genotypic classes is shown in Figure 6. This suggests that the two major Al tolerance QTL both affect tolerance, as measured by root length, via regulation of malate release. Furthermore, the fact that the linear relationship holds even with inclusion of the parental genotypes is suggestive evidence that the remaining (undetected) QTL may also act through this same mechanism.
Figure 5.
Root organic ligand exudation by six Arabidopsis genotypes. A, Root malate exudation. Mean malate release (picomoles of malate per plant per 2 d) from hydroponically grown Arabidopsis in the presence (black) and absence (gray) of Al. Vertical error bars depict se. Six varieties were tested: ecotypes Col and Ler and the four QTL classes that derive from two QTLs (CC, CL, LC, and LL). B, Root citrate exudation. C, Root phosphate exudation.
Table V.
Effect of genotype and environmental conditions on root exudates
| Genotype/Treatment 1 | Mean Flux 1 | Treatment/Genotype 2 | Mean Flux 2 | P | DF |
|---|---|---|---|---|---|
| pM plant-12 d-1 | pM plant-12 d-1 | ||||
| Malate Exudation | |||||
| Control | 94 | Al-treated | 280 | 9E-06 | 157 |
| QTL 1-Col | 365 | QTL 1-Ler | 192 | 0.014 | 77 |
| (Al) | (Al) | ||||
| QTL 2-Col | 383 | QTL 2-Ler | 185 | 0.004 | 76 |
| (Al) | (Al) | ||||
| Citrate Exudation | |||||
| Control | 218 | Al-treated | 93 | 1.9E-20 | 157 |
| QTL 1-Col | 99 | QTL 1-Ler | 87 | 0.476 | 77 |
| (Al) | (Al) | ||||
| QTL 2-Col | 102 | QTL 2-Ler | 85 | 0.294 | 76 |
| (Al) | (Al) | ||||
| Phosphate Exudation | |||||
| Control | 143 | Al-treated | 141 | 0.896 | 29 |
| QTL 1-Col | 161 | QTL 1-Ler | 121 | 0.18 | 29 |
| (all) | (all) | ||||
| QTL 2-Col | 140 | QTL 2-Ler | 142 | 0.916 | 29 |
| (all) | (all) | ||||
| Columbia | 170 | Landsberg | 118 | 0.02 | 19 |
Mean malate exudation was calculated for three comparisons: effect of Al-treatment versus control (all varieties); effect of QTL 1 allele (Col versus Ler) on malate exudation in Al-treated plants; effect of QTL 2 allele (Col versus Ler) on malate exudation in Al-treated plants. All three comparisons are significantly different. Mean citrate exudation was calculated for three comparisons as malate above. Only one comparison (control versus Al-treated) was significantly different. Mean phosphate exudation was calculated for four comparisons, the three used for malate and citrate and a comparison of mean phosphate exudation from the parental types (Col and Ler, control and Al-treated combined together, because Al treatment has no effect on phosphate exudation). Col and Ler have different patterns of phosphate exudation
Figure 6.
Correlation analysis of root malate exudation in Al-treated plants versus root growth in Al-treated plants. Error bars depict se for both root length and malate release.
What role do citrate and phosphate release play in Al tolerance in this cross? Al stress decreased the release of citrate from Arabidopsis roots (Fig. 5B). Although differences existed between the genotype classes under control conditions, the six genotypes had nearly identical levels of citrate release in the presence of Al. Thus, although Al exposure clearly influences citrate release, the two QTL do not appear to contribute to tolerance via this mechanism (Table V). Al did not affect the release of phosphate from Arabidopsis roots (Fig. 5C). However, Col and Ler did exhibit a small but significant difference in phosphate release (Table V). The trend for slightly greater phosphate release in Col is echoed in comparisons between the QTL 1-Col and QTL 1-Ler genotype classes, but the difference is not significantly different from zero (P = 0.18).
Identification of Candidate Tolerance Genes
One of the advantages to working in Arabidopsis is the wealth of genomic resources available to researchers. The ultimate goal of this research project is to isolate and characterize the Arabidopsis genes responsible for Al tolerance. The results of the QTL analysis conducted here identified regions of the Arabidopsis genome on chromosomes 1 and 5 associated with Al tolerance that each span approximately 3-Mb physical regions that contain approximately 700 predicted genes (Arabidopsis Genome Initiative, 2000). To begin to narrow down this list of genes to a manageable number for identification of possible candidate genes, we identified Al-inducible genes via microarray hybridization in collaboration with the Arabidopsis Functional Genomics Consortium (AFGC; Wisman and Ohlrogge, 2000). This gene expression-profiling experiment used the 11,500 spotted cDNA array produced by the AFGC for two experiments (dye-swap replicates). We used a pooling strategy for probe labeling combining RNA isolated from Col plants at three different time points during the Al stress response (3, 8, and 24 h ±Al) to maximize the possibility of gene discovery. Using permissive criteria, we identified 234 genes that increased in mRNA abundance in response to the Al treatment (Supplemental Table I). A subset of the Al-inducible genes were found to be located in the QTL containing regions and are listed in Table VI based on their locus name (e.g. At1g05430). Seven of these genes map to the portion of chromosome 1 consistent with QTL 1, whereas eight map to chromosome 5 consistent with QTL 2. These genes were further analyzed using the Cereon polymorphism database for ecotype Ler (Jander et al., 2002) to identify which genes may encode polymorphic alleles between the parents of the RIL set. Thirteen of the 15 genes have nucleotide differences identified by Cereon either within the predicted coding sequences or in closely flanking (<500 nucleotides) regions, which may contribute to differences in expression or function. On the basis of their annotation, a number of the genes (seven) encode putative proteins of unknown function. It is interesting to note that three of the other genes may encode proteins involved in organic acid transport or metabolism: MNJ7.15, a putative dicarboxylate transporter; F24B9.2, an ascorbate oxidase; and F14D16.13, a putative oxalate oxidase (although this gene contained no polymorphisms between Col and Ler, at least according to the Cereon database).
Table VI.
Al stress stimulated genes within QTL-containing regions
| Position | Gene Name | Annotation | Polymorphism within CDS | Polymorphism near CDS |
|---|---|---|---|---|
| kb | ||||
| chr 1 | ||||
| 630 | F14D16.13 | Putative oxalate oxidase | None listed | CER474838&9 |
| 630 | F14D16.17 | Similar to PSUVII | CER474909-913 | CER474915 |
| 1,020 | F5M15.22 | cor47 | None listed | CER475172-5 |
| 1,500 | yUP8H12.4 | Unknown | CER461460, 461486 | CER461458&9, 461485 |
| 1,560 | F8K7.10 | Unknown | CER432984&5 | None listed |
| 2,700 | T27G7.20 | Putative oleoyl-(acyl-carrier-protein) hydrolase | CER459954 | CER445479, 459953, 459955 |
| 2,889 | F24B9.2 | l-ascorbate peroxidase | CER475657 | None listed |
| chr 5 | ||||
| 4,100 | T24H18.200 | Unknown | CER477942 | None listed |
| 4,649 | MLN1.10 | hox7 | CER438449-454 | None listed |
| 4,789 | MFC16.10 | Berberine bridge enzyme | None listed | CER455619-623 |
| 5,685 | MPL12.21 | Unknown, but contains phosphatase signature | None listed | None listed |
| 5,950 | MRG7.9 | Unknown | CER439753-7, CER456926-8 | None listed |
| 5,995 | MQD22.19 | Unknown | CER456796 | CER439412, 456797 |
| 6,129 | K10A8.120 | Unknown, but contains P loop signature | None listed | None listed |
| 6,220 | MNJ7.15 | Sodium/dicarboxylate cotransporter | CER438799, 456441 | CER438796-8 |
Position refers to the physical map location for the BAC/P1 clone corresponding to the gene of interest. Polymorphisms between ecotypes Col and Ler were identified using the Cereon/Monsanto collection available to public-sector investigators, and are designated using the CER numbering scheme. Polymorphisms within the predicted coding sequence (within CDS) are distinguished from those differences found with 500 nucleotides of the transcriptional start and stop sites (near CDS). If no polymorphisms were identified found in a particular interval, “none listed” is given
DISCUSSION
This study is based on an integrated investigation of the genetics and physiology of Al tolerance in Arabidopsis, using a set of RILs derived from the cross between Al-sensitive Ler and Al-tolerant Col. Our findings indicate that Al tolerance in this cross between Arabidopsis ecotypes is mediated by a genetically complex but physiologically simple mechanism. Composite interval analysis identified two major loci for Al tolerance that interact to explain approximately 40% of the variance observed. Thus, other loci of smaller effect appear to be segregating in this population. Whereas other QTL were resolved using MRA, we have focused our attention on those two loci identified by the more conservative methodology. However, our findings also indicate that the difference in Al tolerance is physiologically simple when considered at the whole-plant level, as only one mechanism—root malate release—is required to explain 95% of the variance in root length among a select sample of the RI lines.
The two Al tolerance QTL identified in this study appear to affect constitutive and Al-activated root exudation of malate, involving an Al tolerance mechanism that appears to be similar in general to that suggested for wheat (Delhaize et al., 1993b; Pellet et al., 1995). Furthermore, the two QTL have a significant statistical interaction, which suggests that they may affect malate release through a shared mechanistic pathway. The close correspondence between root malate release and Al tolerance in the selected RIL and the parental ecotypes (Fig. 6) has lead us to propose a model in which the extent of malate release, controlled by multiple interacting genetic factors, underlies nearly all of the difference in tolerance between these two ecotypes. It should be noted that linkage or pleiotropy could also be at work, but the fact that the two detected QTL and the collective action of the residual QTL influence root malate release in the same direction and with a magnitude proportional to the observed differences in tolerance is strong evidence that the correlation is a causative one. There is a high degree of agreement between the two measures of tolerance—Al-dependent inhibition of root growth in a solid medium and Al-dependent release of malate in a liquid medium—despite the dramatic differences in growth environment, suggesting that Al tolerance is a highly penetrant phenotype in Arabidopsis.
A recent report from Kobayashi and Koyama (2002) underscores the reproducibility of the Al tolerance phenotype, because these investigators also used QTL mapping to identify genes important for Al tolerance in the same Ler × Col RIL set. The study by Kobayashi and Koyama focused solely on QTL analysis of root growth in Al-containing hydroponic media and did not attempt to elucidate the mechanisms underlying the QTLs. In that study, it was reassuring to find that the factor on chromosome 1 (QTL 1) was also identified as the major tolerance locus. However, the remainder of the QTLs identified did not agree with the results presented in our investigation. This is perhaps not surprising given the differences in growth conditions used in each study, which differed in pH (5.0 from Kobayashi and Koyama [2002] versus 4.2 in this study), Al level (as estimated by GEOCHEM: 0.12 μm Al3+ activity from Kobayashi and Koyama [2002] versus 12 μm Al3+ activity in this study), and ionic strength of the medium (3.1 × 10-4 from Kobayashi and Koyama [2002] versus 2.4 × 10-2 in this study; Fujiwara et al., 1992; Parker et al., 1995). Statistical analyses and sampling approaches differed between studies, which may have also contributed to the differences in conclusions. The robustness of QTL 1 identified in the two different studies has given us further impetus to focus on isolating the molecular determinant underlying this QTL in ongoing research.
When an Al tolerance mechanism based on Al-activated root malate exudation is studied in detail, it is possible to see why such a trait could be expected to be physiologically simple (employing a single mechanism at the whole-plant level) but could be conditioned by a number of different loci. Al-activated malate release requires the participation of at least three separate cellular processes: (a) perception of toxic Al; (b) synthesis and possibly compartmentation of malate in the cytosol; and, (c) malate transport from the cytosol to the root cell apoplast. Each of the steps could also involve multiple components, including different enzymes, transporters, and membrane-associated receptors and other possible signal transduction molecules. Hence, it is reasonable to speculate that the variation in Al tolerance seen in these Arabidopsis genotypes involves a number of genes associated with different aspects of organic acid synthesis, transport, and Al perception, all of which act on the same overall physiological mechanism.
It also is interesting to note that there is a strong correlation for constitutive and Al-activated root malate exudation with Al tolerance (see Fig. 5). For example, the Col ecotype, which is considerably more tolerant than the next most Al-tolerant genotype (CC RIL class), exhibits a 2-fold higher constitutive and Al-activated malate exudation compared with the CC RIL class, and a 7-fold higher constitutive and Al-activated malate release compared with the most Al-sensitive genotype, the Ler ecotype. Hence, Al-activated malate exudation may not be as important as overall root malate release for Arabidopsis Al tolerance. One possible scenario to explain these results would be that both Al-tolerant and -sensitive Arabidopsis genotypes contain the same Al-sensing and malate transport machinery in their root-cell plasma membranes. On the basis of the physiological findings presented here, this transporter would have some capacity to release malate in the absence of Al, and this transport capacity would be stimulated by exposure to Al. The difference in Al tolerance would then be associated with a greater expression of these components in genotypes exhibiting more Al tolerance. That is, the more Al-tolerant genotypes would have a greater density of these key components in their root cells, resulting in greater constitutive and Al-activated malate fluxes.
Support for some aspects of this speculative model comes from an electrophysiological study of the Al-activated malate transporter in wheat roots (Zhang et al., 2001). In that patch clamp study of the Al-activated malate transporter in protoplasts isolated from root tips of Al-tolerant and -sensitive wheat, the authors found that root tip cells from both the tolerant and sensitive genotypes exhibited Al-activated malate transporter activity. These findings suggest that in wheat as in Arabidopsis, the Al-sensitive genotypes also contain all or most of the molecular machinery involved in Al tolerance.
How will we identify the different pieces of this molecular machinery? On the basis of the QTL analysis, we can locate two important factors (QTL 1 and 2) to genomic regions containing hundreds of genes. The gene expression profiling experiments indicate that only a small fraction of those genes increase mRNA levels during Al stress, and a majority of those genes do contain nucleotide polymorphisms between Col and Ler (Table IV). Fine-scale genetic mapping should eliminate most of these candidate tolerance genes, leaving a small number that will require verification using additional alleles (e.g. T-DNA knockouts, allele replacement, or overexpression) and physiological assessment using both root growth and malate release. Given the incomplete genome coverage of the AFGC array, it is very possible that we have not yet identified the correct candidate gene. However, this type of genomics-based approach is being integrated with ongoing positional mapping to focus our attention on the portion of the Arabidopsis genome that harbors the genes responsible for the Al tolerance QTL. Our understanding of the physiological basis for Al tolerance gained from this study facilitates this effort and should permit us to make better decisions in selecting candidate genes for future analysis.
In summary, we have identified and characterized two major loci that contribute to the Al tolerance differences observed between Col and Ler. Root malate exudation closely correlated with Al tolerance in this cross between Arabidopsis ecotypes; a similar correlation between Al tolerance and root malate release was previously observed in a survey of Al-tolerant and -sensitive wheat cultivars (Ryan et al., 1995). This suggests that information gained from cloning and characterizing the genes that underlie malate release and Al tolerance in Arabidopsis will be of immediate utility to crop improvement. As mentioned previously, QTL mapping is the first step toward the positional cloning of genes underlying complex phenotypes (Frary et al., 2000; Yano et al., 2000; Jander et al., 2002). Positional cloning of the genes responsible for the differences in Al tolerance and malate release is the obvious extension of the present research. Tight integration of physiological, genetic, and genomic approaches has been an efficient and effective strategy to investigate the complex problem of Al tolerance and should ultimately allow us to identify the molecular mechanisms responsible for this important trait.
MATERIALS AND METHODS
Plant Growth Experiments: Identification of QTL
The Ler × Col RIL collection was obtained from the Arabidopsis Biological Resource Center (Columbus, OH) as stock number CS1899. The core collection of 100 RIL was used for our root growth experiment. However line CL35 failed to germinate in sufficient numbers to include in this analysis. Plants were grown on solid (gellan gum), acidic pH media with and without Al for 8 d. Tolerance to Al stress was estimated by measuring primary root length after 6 and 8 d of growth.
The solid media plates used a nutrient solution adapted from Larsen et al., 1996, which contained 0.2 mm KH2PO4, 3 mm MgSO4, 0.25 mm (NH4)2SO4, 3 mm Ca(NO3)2, 2 mm K2SO4, 1 μm MnSO4, 5 μm H3BO3, 0.05 μm CuSO4, 0.2 μm ZnSO4, 0.02 NaMoO4, 0.1 μm CaCl2, 0.001 μm CoCl2, 1% (w/v) Suc, and 0.125% (w/v) gellan gum. The pH of the medium was adjusted to 4.2 with the addition of 0.1 m HCl. The medium was autoclaved to sterilize the solution; 85 mL of media was dispensed per 25- × 100-mm petri dish under sterile conditions. Al was added through the use of a soaking solution identical to the nutrient solution, with the following modifications: 0.1 mm KH2PO4, 1.1 mm K2SO4, and 1 mm AlCl3. Soak solution was applied to the plates for 2 d to allow equilibration (20 mL soak 85 mL-1 media). If equilibrium was reached between the soak solution and the medium, we estimate that the Al activity was on the order of 12 to 15 μm Al3+ using GEOCHEM to model the speciation chemistry (Parker et al., 1995). Plates were rinsed briefly with sterile Milli-Q grade water. Seeds were surface sterilized (30% [v/v] bleach and 1% [v/v] Triton X-100; 15 min), resuspended in 0.1% (w/v) agar, and planted to the solid media plates. Plates were then placed at 4°C for 5 d to stratify the seeds to ensure robust, synchronous germination. Ten seeds were planted for each RIL per treatment, in triplicate. Plants were grown for 8 d at 20°C in continuous light (50 μE m-2s-1) in a growth chamber.
Root length measurements were collected using an electronic digital caliper (Fowler ProMax, Kelley and Kelly Industrial Supply, Syracuse, NY) connected to a computer via an RS-232 cable using the OPTO-RS computer program (Fred Fowler Co., Newton, MA).
Statistical Methods
We performed QTL analysis using Qgene (Nelson, 1997) and QTL Cartographer (Basten et al., 1999). The traits examined included the four raw variables RLAl6, RLAl8, RLC6, and RLC8, where subscript Al indicates that Al was added, C that it was not, and the subscripts 6 and 8 indicate the day upon which the measurement was taken. In addition, we examined several different composite variables, including (a) the difference in root length between control and Al-treated plants at the two different time periods, D6 = RLC6 - RLAl6 and D8 = RLC8 - RLAl8; (b) the difference in root length between the two time periods for each Al treatment, DA = RLAl8 - RLAl6 and DC = RLC8 - RLC6; (c) the ratios describing the proportional difference in growth between treatments over the first 6 d, R1 = (RLC6 - RLAl6)/RLC6 and between the 6th and 8th d, R2 = [(RLC8 - RLAl8) - (RLC6 - RLAl6)]/(RLC8 - RLC6); and (d) percent RRG, which described the ratio between root growth for Al-treated and control plants, on either d 6 or 8, RRG = RLAl/RLC × 100.
We chose markers from among the available “framework” markers so as to provide complete coverage of all five chromosomes at intervals of less than 5 centiMorgans (cM), on average. We obtained genotypic data for the 99 lines, and map location data for 113 chosen markers, from the Nottingham Arabidopsis Stock Center Web site (http://nasc.nott.ac.uk/new_ri_map.html).
To map the QTL, we performed composite interval analysis (Zeng, 1994), which controls for the effects of unlinked QTL on the chromosomal region being tested. This is important in an RIL population, in which substantial deviations from Mendelian segregation have the potential to create spurious associations. We first performed regression analysis for each trait on each marker singly. Then, all markers from the chromosome under consideration greater than 10 cM from the test position as well as the two markers from each chromosome with the highest single-marker regression F scores were included as cofactors for composite interval analysis. A LR test statistic measuring the strength of support for a QTL at a given position was calculated at 1- to 2-cM intervals along the length of each chromosome.
We used permutation tests to calculate appropriate experiment-wise significance levels for the LR statistic that would be robust to the non-normality in the data and correct for multiple comparisons (Churchill and Doerge, 1994). The trait values were shuffled among the genotype scores N times, and interval and composite interval analysis was performed on each of the shuffled datasets. The critical value of the LR statistic at an experiment-wise significance level of α is the minimum value of the LR statistic greater than N-αN of those observed among the permuted replicates. In our experiments, N = 1,000 and α = 0.05. For each QTL that surpasses the critical value, we have reported α, the estimated additive effect, and r2, the proportion of the phenotypic variation explained, conditioned on the selected background markers.
Pair wise epistatic interactions between QTL were tested by an ANOVA. The markers closest to each QTL LR peak were selected as factors for all possible pair wise ANOVAs. A significant interaction effect between factors was considered to be evidence for a statistical interaction between the QTL associated with each marker. ANOVA was also used to test the correlation between QTL genotypes and patterns of organic acid release.
Broad sense heritability (hb2) was estimated from one way ANOVA of the root length data at 6 d using the formula, hb2 = σg2/((σe2/r) + σg2) where σg2 is the variance between RIL, σe2 is the variance within RIL, and r is the average number of measurements per RIL.
Plant Growth Experiments: Assessment of QTL
Al-inducible release of Al-chelating ligands is the most common and best understood protective mechanism in plants. Experiments were conducted to test whether the QTL cosegregated with patterns of ligand release that would correlate with tolerance. Seeds were weighed to estimate number (10 mg = 500 seeds), surface sterilized, and stratified as described above. Magenta GA-7 culture vessels (Sigma-Aldrich, St. Louis) were adapted for sterile hydroponic growth using 250-μm polypropylene mesh as substrate for plant growth (Small Parts, Miami Lakes, FL) and a support stand constructed from two notched rectangles of polycarbonate (Laird Plastics, Syracuse, NY). The hydroponic growth solution contained 0.2 mm KH2PO4, 2 mm MgSO4, 0.25 mm (NH4)2SO4, 2 mm Ca(NO3)2, 2 mm K2SO4, 1 μm MnSO4, 5 μm H3BO3, 0.05 μm CuSO4, 0.2 μm ZnSO4, 0.02 NaMoO4, 0.1 μm CaCl2, 0.001 μm CoCl2, and 1% (w/v) Suc. Culture vessels were assembled, filled with 150 mL of media, and sterilized by autoclaving. Duplicate sterile vessels were used for each RIL for each condition (control and Al treated). Stratified seeds were planted to the polypropylene mesh cloth under sterile conditions; KH2PO4 was added at the time of planting to avoid precipitation during autoclaving. Plants were grown for 6 d at 20°C in continuous light (50 μE m-2 s-1) in a growth chamber. After 6 d, a second, low-strength nutrient solution was prepared for ligand exudation. This nutrient solution contained 275 μm MgCl2, 275 μm CaCl2, 275 μm KCl, 33.4 μm Ca(NO3)2, 33.4 μm MgSO4, 16.7 μm K2SO4, 8.35 μm (NH4)2SO4, 3.0 mm Homopipes buffer, 1% (w/v) Suc, and micronutrients identical to the prestress medium. AlCl3 was added to the Al-treatment media (50 μm concentration and 25 μm activity). Homopipes was used to buffer the pH of the nutrient solution at 4.2. The +Al nutrient solution was filter sterilized and dispensed into 25- × 100-mm petri dishes (20 mL). Plants were transferred into the +Al media by transferring the polypropylene mesh and seedlings under sterile conditions. After 48 h, the +Al media was sampled and was subsequently analyzed for organic ligands (organic acids and phosphate anions) by capillary electrophoresis and spectrophotometry as described by Piñeros et al. (2002). Organic acid fluxes were standardized by the number of plant grown in each Magenta box (picomoles per plant per 2 d). Due to the low ionic strength of the +Al medium, samples did not require treatment by column chromatography to remove inorganic anions (primarily Cl-) that could interfere with ligand determination via capillary electrophoresis.
Candidate Tolerance Gene Discovery
Microarray experiments were conducted in collaboration with the AFGC to identify genes regulated by Al stress. The AFGC generated microarrays containing 11,500 spotted cDNAs for public consumption. Ecotype Col plants were grown in hydroponic culture as described for the organic acid quantitation, with the modification that upon transfer to pH-buffered treatment media that the plants were grown in a large (150 mL) rather than small (20 mL) volume. Three sets of plants were grown for the experiment: those harvested for root tissue 3, 8, and 24 h after transfer to fresh experimental medium (± Al containing media). Total RNA was isolated using the TRIzol reagent (Invitrogen, Grand Island, NY) following standard protocols from each of the six groups of plants (three control and three Al treated). Poly(A+) RNA was isolated from total RNA using the Poly(A) Pure kit (Ambion, Austin, TX) following manufacturer's instructions and was quantitated using a spectrophotometer. Equal amounts of poly(A+) RNA were pooled from each time group to form a control pool and an Al-treated pool. Pooled poly(A+) RNA samples were sent to the AFGC (East Lansing, MI) for probe labeling, array hybridization, and scanning. Gene expression patterns were compared between ± Al exposed plants to identify genes influenced by the Al treatment. All of the raw and processed data from these experiments can be accessed from the Stanford Microarray Database (http://smd.stanford.edu) filed under Kochian, as corresponding investigator. Individual array elements can also be queried through http://www.Arabidopsis.org/tools/bulk/microarray/index.html. Results were analyzed using simple Boolean strategies in a FileMaker database (v5 for Macintosh). Spots that met threshold requirements (signal >350 in every probe set) and reproducibility guidelines (Al-treated to control signal ratio equal to or greater than 1.5 in both replicates) were considered as candidate Al-inducible genes (see Supplemental Table I for complete list of genes along with signal intensity data). The Cereon collection of nucleotide polymorphisms between ecotypes Col and Ler was downloaded from http://www.Arabidopsis.org/cereon/index.html and compiled in a FileMaker database. The candidate Al-inducible genes were located on the physical map using locus alias (e.g. yUP8H12.4) and locus name (e.g. At1g05340). Genes that fell within QTL containing regions were then inspected for polymorphisms using the Cereon database.
Acknowledgments
We acknowledge the technical assistance provided by Holly Manslank, William Shaben, Lydiah Kemunto Bosire, Elizabeth Ogilvie, and Yasin Senbabaoglu (Cornell University); by the greenhouse staff of the Boyce Thompson Institute for Plant Research; by the Arabidopsis Biological Resource Center for seed stocks; by the Arabidopsis Functional Genomics Consortium for the microarray experiments; and by the instructive discussions with Dr. Andrew Cary (Yale University) and Dr. Edward Buckler (North Carolina State University/U.S. Department of Agriculture-Agricultural Research Service).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.023085.
This work was supported by the U.S. Department of Agriculture-National Research Initiative (proposal nos. 97–35100–5050 to S.H.H. and L.V.K. and 02–35100–12058 to O.A.H. and L.V.K.).
References
- Alonso-Blanco C, Koornneef M (2000) Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci 5: 22-29 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815 [DOI] [PubMed] [Google Scholar]
- Basten CJ, Weir BS, Zeng ZB (1999) QTL Cartographer. Computer Program Version 1.13. North Carolina State University, Raleigh, NC
- Baykov AA, Evtushenko OA, Avaeva SM (1988) A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal Biochem 171: 266-270 [DOI] [PubMed] [Google Scholar]
- Berzonsky WA (1992) The genomic inheritance of Al tolerance in ′Atlas 66′ wheat. Genome 35: 689-693 [Google Scholar]
- Burr B, Burr FA (1991) Recombinant inbreds for molecular mapping in maize: theoretical and practical considerations. Trends Genet 7: 55-60 [DOI] [PubMed] [Google Scholar]
- Camargo CEO (1981) Melhoramento do trigo: I. Hereditariedade do tolerancia a toxicidade do aluminio. Bragantia 40: 33-45 [Google Scholar]
- Camargo CEO (1984) Melhoramento do trigo: VI. Hereditariedade de tolerancia a tres concentracoes de aluminio em solucao nutritiva. Bragantia 43: 279-291 [Google Scholar]
- Churchill G, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138: 963-971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Craig S, Beaton CD, Bennet RJ, Jagadish VC, Randall PJ (1993a) Al tolerance in wheat (Triticum aestivum L.): I. Uptake and distribution of Al in root apices. Plant Physiol 103: 685-693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR, Randall PJ (1993b) Al tolerance in wheat (Triticum aestivum L.): II. Al-stimulated excretion of malic acid from root apices. Plant Physiol 103: 695-702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerge RW (2002) Mapping and analysis of quantitative trait loci in experimental populations. Nat Rev Genet 3: 43-52 [DOI] [PubMed] [Google Scholar]
- Foy CD (1988) Plant adaptation to acid, Al-toxic soils. Commun Soil Sci Plant Anal 19: 959-987 [Google Scholar]
- Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD (2000) fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85-88 [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Hirai YM, Chino M, Komeda Y, Naito S (1992) Effect of sulfur nutrition on expression of soybean seed storage protein genes in transgenic petunia. Plant Physiol 99: 263-268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson T, Reisenauer H (1984) Crop response to lime in western United States. In F Adams, ed, Soil Acidity and Liming. American Society of Agronomy, Crop Science Society of America, Soil Science Society of America, Madison, WI, pp 333-347
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129: 440-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juenger T, Purugganan M, Mackay TF (2000) Quantitative trait loci for floral morphology in Arabidopsis thaliana. Genetics 156: 1379-1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerridge PC, Kronstad WE (1968) Evidence of genetic resistance to Al toxicity in wheat (Triticum aestivum Vill., Host). Crop Sci 60: 710-711 [Google Scholar]
- Kliebenstein D, Pedersen D, Barker B, Mitchell-Olds T (2002) Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidopsis thaliana. Genetics 161: 325-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Koyama H (2002) QTL analysis of Al tolerance in recombinant inbred lines of Arabidopsis thaliana. Plant Cell Physiol 43: 1526-1533 [DOI] [PubMed] [Google Scholar]
- Kochian LV (1995) Cellular mechanisms of Al toxicity and resistance in plants. Annu Rev Plant Physiol Plant Mol Biol 46: 237-260 [Google Scholar]
- Lambrix V, Reichelt M, Mitchell-Olds T, Kliebenstein DJ, Gershenzon J (2001) The Arabidopsis epithiospecifier protein promotes the hydrolysis of glucosinolates to nitriles and influences Trichoplusia in herbivory. Plant Cell 13: 2793-2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PB, Degenhardt J, Tai CY, Stenzler LM, Howell SH, Kochian LV (1998) Al-resistant Arabidopsis mutants that exhibit altered patterns of Al accumulation and organic acid release from roots. Plant Physiol 117: 9-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PB, Tai CY, Kochian LV, Howell SH (1996) Arabidopsis mutants with increased sensitivity to Al. Plant Physiol 110: 743-751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister C, Dean C (1993) Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J 4: 745-750 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 123: 795-805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnavaca R, Gardner CO, Clark RB (1987) Inheritance of Al tolerance in maize. In HW Gabelman, BC Loughman, eds, Genetic Aspects of Plant Mineral Nutrition. Martinus Nijhoff, Dordrecht, The Netherlands, pp 201-212
- Miftahudin, Scoles G, Gustafson JP (2002) AFLP markers tightly linked to the Al-tolerance gene Alt3 in rye (Secale cereale L.). Theor Appl Genet 104: 626-631 [Google Scholar]
- Miyasaka SC, Buta JG, Howell RK, Foy CD (1991) Mechanisms of Al tolerance in snapbeans: root exudation of citric acid. Plant Physiol 96: 737-743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J (1997) QGene: software for marker-based genomic analysis and breeding. Mol Breed 3: 229-235 [Google Scholar]
- Nguyen V, Burow M, Nguyen HT, Le B, Le T, Paterson A (2001) Molecular mapping of genes conferring Al tolerance in rice (Oryza sativa L.). Theor Appl Genet 102: 1002-1010 [Google Scholar]
- Nguyen VT, Nguyen BD, Sarkarung S, Martinez C, Paterson AH, Nguyen HT (2002) Mapping of genes controlling Al tolerance in rice: comparison of different genetic backgrounds. Mol Genet Genomics 267: 772-780 [DOI] [PubMed] [Google Scholar]
- Parker D, Norvell W, Chaney R (1995) GEOCHEM-PC: a chemical speciation program for IBM and compatible personal computers. In R Loepper, ed, Chemical Equilibrium and Reaction Models, Vol 2. Soil Science Society of America, Madison, WI, pp 253-269 [Google Scholar]
- Pellet DM, Grunes DL, Kochian LV (1995) Organic acid exudation as an Al-tolerance mechanism in maize (Zea mays L.). Planta 196: 788-795 [Google Scholar]
- Perez-Perez JM, Serrano-Cartagena J, Micol JL (2002) Genetic analysis of natural variations in the architecture of Arabidopsis thaliana vegetative leaves. Genetics 162: 893-915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeros MA, Magalhaes JV, Carvalho Alves VM, Kochian LV (2002) The physiology and biophysics of an Al tolerance mechanism based on root citrate exudation in maize. Plant Physiol 129: 1194-1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioli AJ (1987) Analise genetica da tolerancia a toxidez do alumino em milho (Zea mays L). PhD. Universidade Estadual de Campinas, Campinas, SP, Brazil
- Quesada V, Garcia-Martinez S, Piqueras P, Ponce MR, Micol JL (2002) Genetic architecture of NaCl tolerance in Arabidopsis. Plant Physiol 130: 951-963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao IM, Zeigler RS, Vera R, Sarkarung S (1993) Selection and breeding for acid-soil tolerance in crops. Bioscience 43: 454-465 [Google Scholar]
- Ryan PR, Delhaize E, Randall PJ (1995) Malate efflux from root apices and tolerance to Al are highly correlated in wheat. Aust J Plant Physiol 22: 531-536 [Google Scholar]
- Sawazaki E, Furlani ER (1986) Genetica da tolerancia ao alumino em linhagens de milho cateto. In XVI Congresso Nacional de Milho e Sorgo. Belo Horizonte, Brazil, pp 382-392
- Silva IR, Smyth TJ, Raper CD, Carter TE, Rufty TW (2001) Differential Al tolerance in soybean: an evaluation of the role of organic acids. Physiol Plant 112: 200-210 [DOI] [PubMed] [Google Scholar]
- Sokal RR (1995) Data in Biology. In RR Sokal, FJ Rohlf, eds, Biometry: The Principles and Practice of Statistics in Biological Research. Freeman Publishing, New York, pp 17-19
- Tang Y, Sorrells ME, Kochian LV, Garvin DG (2000) Identification of RFLP markers linked to barley Al tolerance gene Alp. Crop Sci 40: 778-782 [Google Scholar]
- von Uexküll HR, Mutert E (1995) Global extent, development and economic impact of acid soils. In RA Date, NJ Grundon, GE Raymet, ME Probert, eds, Plant-Soil Interactions at Low pH: Principles and Management. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 5-19
- Weller JI (1986) Maximum likelihood techniques for the mapping and analysis of quantitative trait loci with the aid of genetic markers. Biometrics 42: 627-640 [PubMed] [Google Scholar]
- Wisman E, Ohlrogge J (2000) Arabidopsis microarray service facilities. Plant Physiol 124: 1468-1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood S, Sebastian K, Scherr S (2000) Soil Resource Condition. In Pilot Analysis of Global Ecosystems: Agroecosystems. International Food Policy Research Institute and the World Resources Institute, Washington, DC, pp 45-54
- Wu P, Liao C, Hu B, Yi K, Jin W, Ni J, He C (2000) QTLs and epistasis for Al tolerance in rice (Oryza sativa L.) at different seedling stages. Theor Appl Genet 100: 1295-1303 [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473-2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky JM, Casal JJ, Luppi JP (1997) The VLF loci, polymorphic between ecotypes Landsberg erecta and Columbia, dissect two branches of phytochrome A signal transduction that correspond to very-low-fluence and high-irradiance responses. Plant J 12: 569-667 [DOI] [PubMed] [Google Scholar]
- Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136: 1457-1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WH, Ryan PR, Tyerman SD (2001) Malate-permeable channels and cation channels activated by Al in the apical cells of wheat roots. Plant Physiol 125: 1459-1472 [DOI] [PMC free article] [PubMed] [Google Scholar]