Abstract
Phosphoenolpyruvate carboxylase (PEPC) is distributed in plants and bacteria but is not found in fungi and animal cells. Important motifs for enzyme activity and structure are conserved in plant and bacterial PEPCs, with the exception of a phosphorylation domain present at the N terminus of all plant PEPCs reported so far, which is absent in the bacterial enzymes. Here, we describe a gene from Arabidopsis, stated as Atppc4, encoding a PEPC, which shows more similarity to Escherichia coli than to plant PEPCs. Interestingly, this enzyme lacks the phosphorylation domain, hence indicating that it is a bacterial-type PEPC. Three additional PEPC genes are present in Arabidopsis, stated as Atppc1, Atppc2, and Atppc3, encoding typical plant-type enzymes. As most plant PEPC genes, Atppc1, Atppc2, and Atppc3 are formed by 10 exons interrupted by nine introns. In contrast, Atppc4 gene has an unusual structure formed by 20 exons. A bacterial-type PEPC gene was also identified in rice (Oryza sativa), stated as Osppc-b, therefore showing the presence of this type of PEPC in monocots. The phylogenetic analysis suggests that both plant-type and bacterial-type PEPCs diverged early during the evolution of plants from a common ancestor, probably the PEPC from γ-proteobacteria. The diversity of plant-type PEPCs in C3, C4, and Crassulacean acid metabolism plants is indicative of the evolutionary success of the regulation by phosphorylation of this enzyme. Although at a low level, the bacterial-type PEPC genes are expressed in Arabidopsis and rice.
Phosphoenolpyruvate carboxylase (PEPC, EC 4.1.1.31) catalyzes the β-carboxylation of phosphoenolpyruvate in the presence of Mg2+ to yield oxaloacetate and inorganic phosphate (Chollet et al., 1996). In vivo, the active enzyme acts as a homotetramer composed by subunits of 100 to 110 kD (O'Leary, 1982). This enzyme is widely distributed in plants, algae, and bacteria, but is absent from animals, fungi, and yeast (Lepiniec et al., 1994; Toh et al., 1994; Gehrig et al., 1998a). The crystal structure of PEPC from Escherichia coli has been determined (Kai et al., 1999); therefore, the three-dimensional structure and the location of residues important for catalysis and regulation of enzyme activity are known for the bacterial enzyme. All motifs important for enzyme structure and the residues forming the active site are highly conserved in PEPCs from plants and bacteria.
The most significant feature distinguishing plant and bacterial PEPCs is the presence of a Ser residue at the N terminus of plant enzymes forming part of the invariant Acid-Base-XX-SIDAQLR motif, which is absent in PEPCs from bacteria (Vidal and Chollet, 1997). This is a very important difference because the plant enzyme, but not the bacterial enzyme, is reversibly phosphorylated at this Ser residue by PEPC kinases, which have been recently cloned from Kalanchoe fedtschenkoi and Arabidopsis (Hartwell et al., 1999). The reversible phosphorylation of plant PEPCs is a critical regulatory mechanism of primary carbon fixation in C4 and Crassulacean acid metabolism (CAM) photosynthesis (Vidal and Chollet, 1997; Nimmo, 2000). This regulatory mechanism is also operative in PEPCs from non-photosynthetic tissues as has been shown in developing and germinating cereal grains (Osuna et al., 1996, 1999).
In bacteria, PEPC plays an anaplerotic function replenishing intermediates of the citric acid cycle because it is able to recapture CO2 produced in respiration to generate oxaloacetate, which is readily converted to malate by the action of malate dehydrogenase. In plants, PEPCs are encoded by small gene families and show differential expression in plant organs that probably reflect specific functions for each gene as has been shown in maize (Zea mays; Kawamura et al., 1990; Dong et al., 1998), Sorghum bicolor (Lepiniec et al., 1993), Flaveria trinervia (Ernst and Westhoff, 1997), or Vanilla planifolia (Gehrig et al., 1998b). Therefore, it may be distinguished the so-called housekeeping PEPC genes, expressed in tissues with a high metabolic activity, likely with an anaplerotic function, and CAM- or C4-specific genes, related to photosynthetic CO2 fixation, are preferentially expressed in green tissues.
Genes encoding PEPC in plants show a highly conserved structure. Most plant PEPC genes (from C4, CAM, or C3 plants) are formed by 10 exons interrupted by nine introns located at conserved positions with respect to the coding sequence (Chollet at al., 1996). This high degree of conservation, and the fact that this enzyme is involved in primary carbon fixation in C4 and CAM photosynthesis, has made PEPC genes very useful molecular markers for the phylogenetic analysis of the different ways of carbon fixation in plants (Gehrig et al., 1998a, 2001; Engelmann et al., 2002). These analyses have established that bacterial and plant PEPCs, which have a similar primary structure except for the phosphorylation motif, form two different subgroups (Toh et al., 1994; Gehrig et al., 1998a, 2001). Among plant PEPC genes, it has been proposed that C4 PEPCs have evolved from ancestral C3-type PEPCs (Bläsing et al., 2002; Engelmann et al., 2002) and proceed faster in monocots than in dicots. In addition, these phylogenetic analysis show the polyphyletic origin of CAM PEPCs (Cushman and Bohnert, 1999; Gehrig et al., 1998a, 2001).
We are interested in the analysis of PEPC genes and function in C3 plants. Taking advantage of the sequence of the Arabidopsis genome, we have analyzed the complexity of the PEPC gene family in this plant as a model of dicot, C3 plants. The Arabidopsis PEPC gene family is composed of four members. Three of these genes share a high similarity among them and encode typical plant PEPCs. Surprisingly, a PEPC gene was identified, which shows higher similarity to bacterial than to plant PEPCs. Most notably, it encodes an enzyme without the phosphorylation motif at the N terminus; that is, a bacterial-type PEPC. A gene encoding a bacterial-type PEPC was also identified in rice (Oryza sativa). Both PEPC genes, which are not silent, have an unusually high number of introns, suggesting a different evolutionary origin from other known plant PEPC genes.
RESULTS
The Arabidopsis PEPC Gene Family Is Composed of Four Genes, One of Them Encoding a Bacterial-Type Enzyme
A search of the Arabidopsis genome database identified four genes putatively encoding PEPC, here stated as Atppc1 (on chromosome I), Atppc2 (on chromosome II), Atppc3 (on chromosome III), and Atppc4 (on chromosome I). Only the cDNA corresponding to Atppc3 gene had previously been isolated (EMBL Nucleotide Sequence Database accession no. AF071788, deposited by K.M. Paterson and H.G. Nimmo, Glasgow). To isolate the full-length cDNAs corresponding to the other PEPC genes from Arabidopsis, we used oligo(dT)-primed cDNA synthesized with mRNA isolated from rosette leaves and PCR amplification with gene-specific oligonucleotides encompassing the initial ATG codon and 3′-untranslated region sequences according to the Arabidopsis database. This strategy was successful to isolate the full-length cDNAs corresponding to Atppc1 and Atppc2 genes (deposited at the EMBL Nucleotide Sequence Database with accession nos. AJ532901 and AJ532902, respectively). However, we were unable to isolate the full-length cDNA of Atppc4 gene, probably due to the low representation of this cDNA as a consequence of the low abundance of Atppc4 mRNA (see below). To obtain the full-length sequence of this gene, a cDNA library was constructed with random primers and mRNA isolated from flowers, the organ showing higher expression of Atppc4 gene (see below). Partial, overlapping cDNA fragments of Atppc4 gene were then obtained by PCR. Therefore, the full-length cDNA sequence corresponding to the Atppc4 gene was deduced from the partial cDNA fragments (EMBL Nucleotide Sequence Database accession no. AJ532903). It should be noted that Atppc1 and Atppc3 genes are correctly annotated in the Arabidopsis database; however, important errors were detected in Atppc2 and Atppc4 genes due to incorrect assignation of intron/exon sequences. AtPPC2 appears annotated with a deletion of 41 residues due to the mis-annotation of sequence that corresponds to exon 9, according to our cDNA data, as intron sequence. Two errors were detected in the annotation of the Atppc4 gene: Exon 3 is actually longer, and the cDNA sequence revealed a new exon (exon 10, see below), which is annotated as intron sequence.
The amino acid sequence of the polypeptides deduced from the cDNAs of the Arabidopsis PEPC genes were aligned and are shown in Figure 1. Atppc1 and Atppc3 code for polypeptides with expected molecular masses of 110.3 and 110.2 kD, respectively, and share a high degree of identity (91% at the amino acid level). Atppc2 codes for a slightly lower molecular mass polypeptide (109.7 kD) and shares lower level of identity (85% with AtPPC1 and 84% with AtPPC3). However, the Atppc4 gene codes for a polypeptide with an expected molecular mass of 116.6 kD, and shares a low level of identity with the other PEPC genes of Arabidopsis (39% with AtPPC1 and 40% with AtPPC2 and AtPPC3). Interestingly, the polypeptide deduced from Atppc4 shares slightly more identity with the PEPC from E. coli (42%). The molecular mass of AtPPC4 is larger than that of E. coli and even larger than any other PEPC from higher plants. Because the largest PEPCs so far reported are from cyanobacteria, we included in the comparison shown in Figure 1 the amino acid sequence of PEPC from Synechocystis sp. PCC 6803. It may be observed that the insertion responsible of the large size of AtPPC4 (residues 328–423) do not correspond with the insertions found in the cyanobacterial enzyme (Fig. 1). In addition, the level of identity of AtPPC4 with PEPC from Synechocystis sp. PCC 6803 (30%) is lower than with the E. coli enzyme.
Figure 1.
Sequence alignment of PEPC from different sources. The protein sequences deduced from Atppc1, Atppc2, Atppc3, Atppc4, and Osppc-b genes were compared with the E. coli and Synechocystis sp. PCC 6803 enzymes using the program BioEdit sequence alignment editor (Hall, 1999). The accession numbers of these sequences are described in ”Materials and Methods.” The asterisk marks the phosphorylable Ser residue at the phosphorylation domain (underlined). Boxes I to V mark domains important for enzyme catalysis.
An important and invariable property of plant PEPCs is its regulation by reversible phosphorylation of a Ser residue forming part of the sequence Acid-Base-XX-SIDAQLR in an extended stretch at the N terminus of these enzymes (Chollet et al., 1996; Vidal and Chollet, 1997; Nimmo, 2000). The phosphorylation domain was found in the polypeptides AtPPC1, AtPPC2, and AtPPC3, with the only variation of residues VH instead of AQ in AtPPC1 (underlined in Fig. 1). Surprisingly, no phosphorylation domain was found in the polypeptide deduced from the Atppc4 gene (Fig. 1). This finding, together with the higher level of identity with the E. coli enzyme, suggests that this plant gene encodes a bacterial-type PEPC. Additional residue variations support this proposal, this is the case of the highly conserved QNTG residues at the C terminus of plant PEPCs, found in AtPPC1, AtPPC2, and AtPPC3, whereas in AtPPC4 the C terminus is formed by the residues RNTG as in E. coli and other bacterial PEPCs (Fig. 1).
To our knowledge, this is the first gene encoding a bacterial-type PEPC described in plants; therefore, we tested the presence of this unusual type of PEPC gene in other plants. The recent release of the rice genome sequence allowed the identification of a similar gene in rice, called Osppc-b here. The polypeptide deduced from Osppc-b (included in the comparison of Fig. 1) shares a high degree of identity with AtPPC4 (70%). It contains all the motifs important for PEPC activity and structure, lacks the phosphorylation domain at the N terminus, and contains the sequence RNTG at the C terminus (Fig. 1), thus showing the presence of this unusual type of PEPC gene also in monocots.
The four polypeptides deduced from the Arabidopsis PEPC genes and from Osppc-b contain the characteristic domains and residues important for PEPC structure and activity as reported for the E. coli enzyme (Kai et al., 1999). Essential residues contributing to the active site of PEPC are summarized in Figure 1. These are His-138 (E. coli numbering), forming part of the sequence VLTAHPT (Box I) and His-579, Arg-581, and Arg-587 in the sequence FHGRGGXXGRGG (Box IV). It should be noted that the variable residues of this sequence are TV in the polypeptides deduced from Atppc1, Atppc2, and Atppc3, but SI in the polypeptides deduced from Atppc4 and Osppc-b as in the E. coli enzyme (Fig. 1, Box IV). Additional residues important for PEPC function are Arg-396, forming part of the sequence DXRQE (Box II), and Lys-546 in the sequence GYSDSG/AKDAG (Box III). This residue is probably involved in bicarbonate binding together with Arg-581, conserved in bacterial and plant-type PEPCs (Box IV), and Arg-699, which forms part of the sequence GSRPXXR (Box V), conserved in E. coli and plant PEPCs.
Gene Structure and Phylogenetic Relationship among PEPC Genes from Different Sources
Almost all plant PEPC genes have a very conserved structure composed of 10 exons interrupted by nine introns, regardless of whether they are from C3, C4, or CAM plants (Chollet et al., 1996). Atppc1, Atppc2, and Atppc3 genes, encoding plant-type PEPCs from Arabidopsis, also show this conserved gene structure (Fig. 2). However, the genes encoding bacterial-type PEPCs, Atppc4 and Osppc-b, show a different, non-conserved structure: Atppc4 is composed of 20 exons, and Osppc-b is composed of 16 exons (Fig. 2).
Figure 2.
Structure of the Arabidopsis PEPC gene family and Osppc-b gene from rice. Black rectangles, exons; lines, introns. The numbers indicate intron size in nucleotides. Exons and introns in the Arabidopsis genes were deduced by comparing the corresponding cDNA with the gene sequence obtained from the Arabidopsis database (http://www.Arabidopsis.org). The structure of Osppc-b was deduced with the GENSCAN program (http://genes.mit.edu/GENSCAN.html).
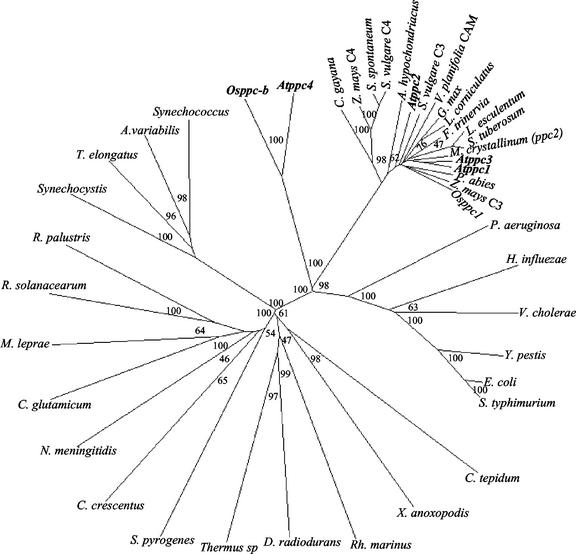
To establish the phylogenetic relationship of these new plant genes with PEPC genes from different sources, an unrooted phylogenetic tree was constructed using the neighbor-joining method with full-length PEPC amino acid sequences from different bacteria and plants found in accessible databases (Fig. 3). The phylogenetic tree displays two major groups: One of them is formed exclusively by bacterial PEPCs, whereas the other includes plant and bacterial PEPCs from enterobacteria, Pseudomonas aeruginosa, Vibrio cholerae, and Haemophilus influenzae. In this group, plant-type PEPCs, including representatives of C3, C4, and CAM plants, form a homogeneous cluster clearly distinguished from the plant genes encoding bacterial-type PEPCs, Atppc4, and Osppc-b (Fig. 3). All these branches were supported by high bootstrap values. The phylogenetic tree shows that both types of plant PEPCs diverged early during the evolution of plants from a common ancestor gene, related to PEPC genes from bacteria such as P. aeruginosa, V. cholerae, H. influenzae, and enterobacteria. The divergence of Osppc-b and Atppc4 probably reflects the monocot-dicot divergence. The fact that the bacterial-type PEPC genes found in plants show a different gene structure than the plant-type genes further supports the independent evolution of both types of PEPC genes in plants.
Figure 3.
Phylogenetic tree of PEPCs from different sources. The phylogenetic tree was constructed with full-length PEPC amino acid sequences using the neighbor-joining method of the ClustalX version 1.8 program (Thompson et al., 1997). Bootstrap analysis was computed with 1,000 replicates. Numbers in branches indicate bootstrap values (percent). The accession numbers are described in ”Materials and Methods.”
Differential Expression of PEPC Genes in Arabidopsis and Rice
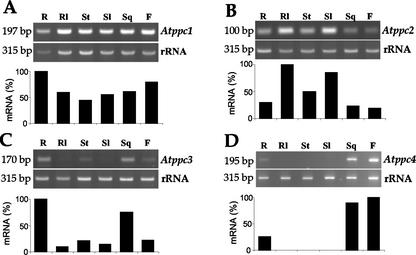
To distinguish among the different PEPC genes and to detect even low-abundant transcripts, we used the relative quantitative reverse transcriptase (RT)-PCR technique with gene-specific primers to analyze the expression of this gene family in Arabidopsis. Figure 4A shows a wide distribution of Atppc1 transcripts in all plant organs, though accumulated at higher level in roots. Atppc2 gene transcripts were also detected in all plant organs but were more abundant in green tissues, such as rosette and stem leaves (Fig. 4B). In contrast, Atppc3 was almost not expressed in green tissues; these transcripts were more abundant in roots and siliques (Fig. 4C). An important question addressed in this analysis was to establish whether or not Atppc4, the bacterial-type gene, is expressed. The relative quantitative RT-PCR analysis detected Atppc4 transcripts exclusively in siliques, flowers, and, to a lower level, in roots (Fig. 4D). It should be mentioned that Atppc4 is expressed at a lower level than the other PEPC genes, as shown by the higher number of cycles needed to obtain the linear range of amplification in the RT-PCR analysis (26 for Atppc1, Atppc2, and Atppc3 and 32 for Atppc4).
Figure 4.
Differential expression of PEPC genes in Arabidopsis. DNA-free total RNA (0.8 μg) isolated from different Arabidopsis tissues were analyzed by relative quantitative RT-PCR using gene-specific primers in the presence of 18S rRNA primers and competimers. Experiments were repeated at least three times obtaining similar results. The expected sizes of the bands corresponding to the different transcripts are: Atppc1, 197 bp; Atppc2, 100 bp; Atppc3, 170 bp; Atppc4, 195 bp and rRNA, 315 bp, as indicated on the left. Ethidium bromide-stained bands for each gene were quantified with the ScionImage software and compared with the corresponding rRNA band. The highest value in each analysis was arbitrarily considered as 100%. R, Root; Rl, rosette leaves; St, stem; Sl, stem leaves; Sq, silique; F, flower.
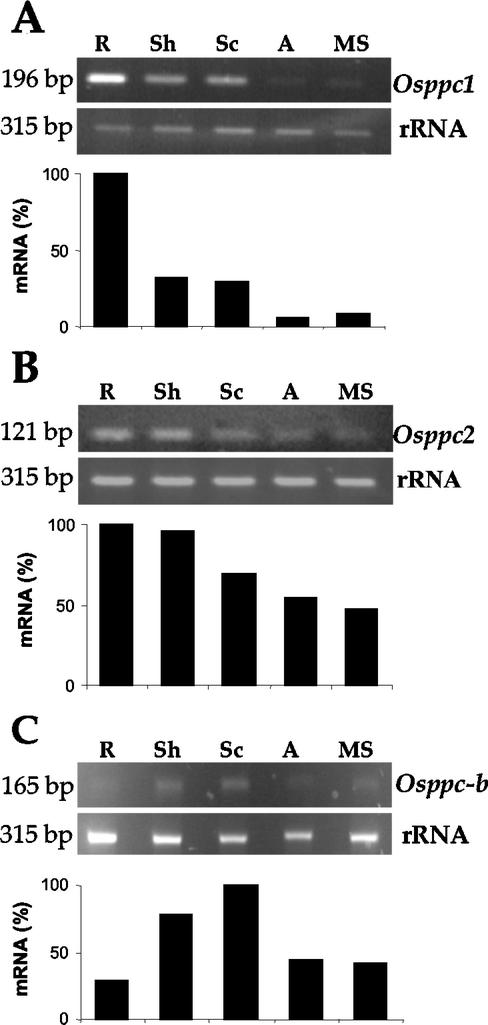
Similarly, we compared the level of expression of Osppc-b gene with other PEPC genes in rice. A search of the databases identified two additional genes in the rice cv japonica group, a root-type PEPC isoform, called Osppc1 here (accession no. AF271995) and a PEPC gene identified on chromosome 1 (accession no. AP003409, called Osppc2 here). Both genes are typical plant-type genes. Relative quantitative RT-PCR analysis with gene-specific primers confirmed the high accumulation of Osppc1 transcripts in roots (Fig. 5A). At a lower level, transcripts were also detected in shoots from 5-d-old seedlings and in the scutellum of seeds after 5 d of imbibition and were almost undetectable in the aleurone layer or mature seeds (Fig. 5A). In contrast, the Osppc2 gene shows a similar level of expression in vegetative tissues, roots, and shoots from 5-d-old seedlings and a lower level of accumulation in seed tissues (Fig. 5B). This analysis revealed the expression of the bacterial-type Osppc-b gene both in seeds (mature seeds and aleurone and scutellum from seeds after 5 d of imbibition) and in roots and shoots from 5-d-old seedlings (Fig. 5C). The linear range of amplification for the Osppc-b gene was of 34 cycles; thus, it also showed a very low level of expression of this gene, as compared with the other PEPC genes from rice, which required a lower number of cycles to obtain the linear range of amplification (26 for Osppc1 and 28 for Osppc2). Therefore, the RT-PCR analysis shows that the bacterial-type PEPC genes, Atppc4 and Osppc-b, are not silent genes, although the level of expression is rather low and, in the case of Arabidopsis, restricted exclusively to some organs.
Figure 5.
Differential expression of PEPC genes in rice seeds and seedlings. Rice seeds were allowed to germinate for 5 d and aleurone layer (A) and scutellum (Sc) were dissected. Roots (R) and shoots (Sh) were dissected from 5-day-old seedlings. Total RNA was extracted from these tissues and from mature seeds (MS) and analyzed by relative quantitative RT-PCR using gene-specific primers in the presence of 18S rRNA primers and competimers. Experiments were repeated at least three times obtaining similar results. The expected sizes of the bands corresponding to the different transcripts are: Osppc1, 196 bp; Osppc2, 121 bp; Osppc-b, 165 bp; and rRNA, 315 bp, as indicated on the left. Ethidium bromide-stained bands were quantified with the ScionImage software and compared with the corresponding rRNA band. The highest value in each analysis was arbitrarily considered as 100%.
DISCUSSION
In this report, we describe new and unusual PEPC genes, stated Atppc4 and Osppc-b, from Arabidopsis and rice, respectively. Despite the low level of similarity of Atppc4 and Osppc-b genes with other PEPC genes from plants, these genes undoubtedly encode for PEPC as shown by the high conservation of all the residues important for PEPC activity and structure (Fig. 1). Based on the fact that the polypeptides deduced from these genes show slightly more identity with PEPC from E. coli than with plant PEPCs and, most important, the lack of the phosphorylation domain at the protein N terminus, a hallmark used to differentiate plant and bacterial PEPCs (Toh et al., 1994), we conclude that Atppc4 and Osppc-b genes encode bacterial-type PEPCs. In addition, these genes have a peculiar gene structure. Unlike most plant PEPC genes, which are composed of 10 exons (Chollet et al., 1996), Atppc4 and Osppc-b genes have an unusually high number of exons (Fig. 2). The average size of introns is larger in Osppc-b than in Atppc4. This finding is in agreement with the comparison of the intron size in Arabidopsis and rice, based on the sequence and structure of rice chromosome 1, which revealed an average intron size about 3.6 times larger in rice (Sasaki et al., 2002).
PEPC is a useful molecular marker in phylogenetic studies of plant photosynthetic metabolism due to its wide distribution in C4, CAM, and C3 plants. The proposal of this analysis is that the C4 and CAM forms of PEPC have evolved from an ancestral C3 gene (Gehrig et al., 1998a, 2001; Bläsing et al., 2000, 2002). The presence of PEPC in cyanobacteria led to speculate that the plant enzyme might have arisen from an endosymbiotic origin (Lepiniec et al., 1994). The identification of bacterial-type PEPC both in mono- and dicotyledoneous plants supports the prokaryotic origin of plant PEPCs. AtPPC4 along with the cyanobacterial PEPCs are the largest PEPCs reported to date. This finding might suggest a cyanobacterial origin for plant PEPC genes; however, the bacterial-type PEPCs from plants are more closely related to PEPC from bacteria such as E. coli or P. aeruginosa than to cyanobacteria. Therefore, based on the phylogenetic analysis and the lower level of similarity with the cyanobacterial PEPCs, a chloroplastidic origin for plant PEPCs is unlikely. Rather, the finding of these new plant genes encoding bacterial-type PEPCs lends a strong support to a previous proposal of the γ-proteobacterial PEPC as the origin of plant PEPCs (Cushman and Bohnert, 1999). The phylogenetic analysis reported here also suggests that the two types of plant PEPC genes originated very early during the evolution of plants from the common ancestor gene. The different number of introns in both types of genes clearly suggests that they have evolved independently since then. The divergence of Osppc-b and Atppc4 (Fig. 3) is indicative of the divergence of monocots and dicots, which was estimated to occur at 200 million years ago (Wolfe et al., 1989).
We have not found PEPC genes after searching the archaeobacteria genomes accessible in public databases. However, a PEPC enzyme has been purified and characterized from the archaeon Methanothermus sociabilis, which is different from plant and bacterial PEPCs (Sako et al., 1996). This finding suggests that the PEPC genes are present in archaeobacteria; however, their sequence must have diverged significantly from PEPC genes characterized thus far.
The plant-type PEPC genes, which possess the regulatory mechanism of phosphorylation, form a homogeneous cluster in the phylogenetic tree (Fig. 3). This PEPC is the form that has diverged in different types, allowing the appearance of CAM and C4 photosynthetic metabolisms, which is indicative of the evolutionary success of this mechanism of regulation of PEPC activity. Despite the low level of expression, the bacterial-type genes are not silent and, based on our sequence analysis of Atppc4 cDNA, the introns are correctly processed. These data suggest that the bacterial-type PEPC still plays a role in plant metabolism, but further analysis is required to understand the function of this new type of PEPC in plants.
It is surprising that genes encoding bacterial-type PEPCs in plants have escaped previous analysis of PEPC genes carried out over the years in different plants. A possibility, which we think is unlikely, is that these genes are not present in C4 and CAM plants, in which PEPC genes have been more intensively studied. More likely is that, due to the low level of expression of these genes, they are poorly represented in expression libraries and have not been detected. The complete genome sequences of Arabidopsis and rice were necessary for these unexpected PEPC genes to be identified.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (ecotype Columbia) was grown on moist vermiculite supplemented with Hoagland medium (Jones, 1982) in culture chambers set at 20°C in the light and 18°C in the dark, under a 16-h-white-light/8-h-dark photoperiod. After 30 d of growth, roots, siliques, stems, flowers, and leaves from stems and rosette were dissected, frozen, and kept at -80°C until needed. Rice (Oryza sativa cv Nipponbare) seeds were sterilized in 2% (v/v) NaOCl for 20 min, washed twice with sterile water, once with 0.01 m HCl, and then thoroughly with sterile distilled water. Sterilized seeds were allowed to germinate at 30°C on sterile filter paper soaked with water. Roots and shoots were dissected from 5-day-old seedlings, and scutellum and aleurone layers were dissected from seeds after 5 d of imbibition. This material was frozen in liquid nitrogen and kept at -80°C until needed.
Cloning of the cDNAs Corresponding to the Arabidopsis PEPC Genes
To isolate the cDNAs of the Arabidopsis PEPC gene family, an oligo(dT)-primed cDNA was synthesized from poly(A+) RNA isolated from rosette leaves using the Marathon cDNA Amplification Kit (CLONTECH Laboratories, Palo Alto, CA) according to the manufacturer's instructions. cDNAs of the different genes were then isolated by PCR using the following gene-specific primers: Atppc1 (5′-ATGGCGAATCGGAAGTTA-3′ and 5′-AGCATGTGTTCAATGATC-3′) and Atppc2 (5′-ATGGCTGCGAGAAATTTG-3′ and 5′-CATTAAAAAGCGTACGGG-3′). The cDNA of the Atppc4 gene was deduced from the sequence of partial, overlapping cDNA fragments isolated by PCR from a random-primed cDNA, which was synthesized from poly(A+) RNA from flowers as indicated above. Five cDNA fragments (At4-1 to At4-5) were obtained ranging in size from 400 to 1,500 bp with the following primers:At4-1(5′-ATGACGGACACAACAGACGAT-3′and5′-ATTCTAATATGCTTGTACTGC-3′), At4-2 (5′-ATTGTTCTTACTGCTCATCC-3′ and 5′-CTTTGCCGTGACATTTGG-3′), At4-3 (5′-TTTACAGGGAAGCCACTT-3′ and 5′-CTTTTCTTTTACTTCTCCAAG-3′), At4-4 (5′-AAGCAGAATGAACAAGACTTT-3′ and 5′-TACATTGAGGAACAAGAGGTC-3′), and At4-5 (5′-AACTCGACTTACGCCAGGAA-3′ and 5′-TCCTGCAGCAATACCATTGA-3′). All cDNAs were cloned in pGEMt vectors and automatically sequenced by NewBiotechnic (Sevilla, Spain).
RNA Isolation and Relative Quantitative RT-PCR Analysis
Total RNA was extracted from liquid nitrogen-frozen tissue and precipitated in 3 m LiCl. RNA samples (100 μg) were treated with 2 units of DNase I for 1 h at 37°C, then DNase I was inactivated by treatment with DNase I Inactivating Reagent (Ambion, Austin, TX). This DNA-free RNA (0.8 μg) was retrotranscribed with a random primers and oligo(dT) mix (1:1 [v/v] ratio) and SuperScript RT (Life Technologies/Gibco-BRL, Cleveland) at 42°C for 1 h. Samples of the first strand cDNA were then used in PCR reactions with gene-specific primers and the Quantum RNA 18S Internal Standards (Ambion). For each gene (Atppc1, Atppc2, Atppc3, and Atppc4 from Arabidopsis and Osppc1, Osppc2, and Osppc-b from rice), the linear range of amplification was determined to establish the number of cycles of the PCR reactions.
Gene-specific primers were designed in the region encompassing the last exon and 3′-untranslated region for each gene to amplify a cDNA fragment of 100 to 200 bp. The pair of primers were: Atppc1 (5′-GCTGCTGGTCTACAAAACAC-3′ and 5′-AGCATGTGTTCAATGATCTCG-3′), Atppc2 (5′-AAACGTTCTTGTACCATTCC-3′ and 5′CATTAAAAAGCGTACGGGA-3′), Atppc3 (5′-GTGAAAGAAAACAAAACTTCG-3′ and 5′-CTTCCAGAAACAGAGTCAGTG-3′), Atppc4 (5′-CTGCAGGAATGAGAAATACCG-3′ and 5′-TTTGTAAGAACGCCGTGTTG-3′), Osppc1 (5′-TGTCCACAAGAATGCTTCCA-3′ and 5′-CTGCAAAAGCCAAATAAGTC-3′), Osppc2 (5′-TGGTTCGTTCTGATGGTGTG-3′ and 5′-CGTTGCTTAGCATCCCCTTA-3′), and Osppc-b (5′-CTCACGTACCTCAACCCCAT-3′ and 5′-GAGGAGAGGAGGGAAGAGGA-3′). Each pair was used with a mix (1:9 [v/v] ratio) of 18S rRNA primers and competimers (Quantum RNA 18S Internal Standards, Ambion) as internal standards of the amount of RNA. Control reactions were performed using as template non-reverse transcribed RNA to rule out possible amplification from contaminating genomic DNA. Ethidium bromide-stained bands were quantified with the ScionImage software, and the amount of mRNA is represented as relative units.
Sequence Comparison and Phylogenetic Analysis
Multiple alignment of complete amino acid sequences of PEPC from different plant and bacterial sources accessible from public databases and genome projects was performed with the ClustalX version 1.8 program (Thompson et al., 1997), removing gap-only columns. This alignment was used to construct the phylogenetic tree with the neighbor-joining method using the ClustalX version 1.8 program (Thompson et al., 1997). Bootstrap analysis was computed with 1,000 replicates and excluding positions with gaps. The Accession numbers of the sequences used to construct the phylogenetic tree were: Chlorobium tepidum (NC_002932), Xanthomonas axonopodis (NC_003919), Streptococcus pyrogenes (NC_0034485), Escherichia coli (NC_002655), Haemophilus influenzae (NC_000907), Salmonella typhimurium (NC_003197), Thermosynechococcus elongatus (AP005375), Corynebacterium glutamicum (X14234), Yersinia pestis (NC_004080), Caulobacter crescentus (NC_002696), Ralstonia solanacearum (NC_003295), Synechocystis sp. PCC 6803 (NC_000911), Mycobacterium leprae (NC_001263), Deinococcus radiodurans (NC_001263), Neisseria meningitidis (NC_003116), Vibrio cholerae (NC_002505), Pseudomonas aeruginosa (NC_002516), Thermus sp. (D42166), Rhodothermus marinus (X99379), Rhodopseudomonas palustris (D89668), Anabaena variabilis (M80541), Synechococcus sp. PC 6301 (M11198), Arabidopsis Atppc1 (AJ532901), Arabidopsis Atppc2 (AJ532902), Arabidopsis Atppc3 (AF071788), Arabidopsis Atppc4 (AJ532903), rice Osppc-b (AP002882), rice Osppc1 (AF271995), Lycopersicum esculentum (ppc2; AJ313434), Chloris gayana (AF2680901), Flaveria trinervia (ppcC; AF248080), Picea abies (AF159051), Lotus corniculatus (AF135371), Amaranthus hypochondriacus (L49175), Solanum tuberosum (AJ011844), Mesembryanthemum crystallinum (ppc2; X14588), Vanilla planifolia (X87148), maize (Zea mays) C3 (X61489), maize C4 (X15238), Sorghum vulgare C3 (X65137), S. vulgare C4 (X63756), Glycine max (AB008540), and Sacharum spontaneum (AJ318338).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.019653.
This work was supported by the Ministerio de Ciencia y Tecnología (grant no. BMC2001–2366 and predoctoral fellowship to R.S.) and by Junta de Andalucía (Spain; grant no. CVI–182).
References
- Bläsing OE, Ernst K, Streubel M, Westhoff P, Svensson P (2002) The non-photosynthetic phosphoenolpyruvate carboxylases of the C4 dicot Flaveria trinervia: implications for the evolution of C4 photosynthesis. Planta 215: 448-456 [DOI] [PubMed] [Google Scholar]
- Bläsing OE, Westhoff P, Svensson P (2000) Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria, a conserved serine residue in the carboxyl-terminal part of the enzyme is a major determinant for C4-specific characteristics. J Biol Chem 275: 27917-27923 [DOI] [PubMed] [Google Scholar]
- Chollet R, Vidal J, O'Leary M (1996) Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 273-298 [DOI] [PubMed] [Google Scholar]
- Cushman JC, Bohnert HJ (1999) Crassulacean acid metabolism: molecular genetics. Annu Rev Plant Physiol Plant Mol Biol 50: 305-332 [DOI] [PubMed] [Google Scholar]
- Dong LY, Masuda T, Kawamura T, Hata S, Izui K (1998) Cloning, expression, and characterization of a root-form phosphoenolpyruvate carboxylase from Zea mays: comparison with the C4-form enzyme. Plant Cell Physiol 39: 865-873 [DOI] [PubMed] [Google Scholar]
- Engelmann S, Bläsing OE, Westhoff P, Svensson P (2002) Serine 774 and amino acids 296 to 437 comprise the major C4 determinants of the C4 phosphoenolpyruvate carboxylase of Flaveria trinervia. FEBS Lett 524: 11-14 [DOI] [PubMed] [Google Scholar]
- Ernst K, Westhoff P (1997) The phosphoenolpyruvate carboxylase (ppc) gene family of Flaveria trinervia (C4) and F. pringlei (C3): molecular characterization and expression analysis of the ppcB and ppcC genes. Plant Mol Biol 34: 427-443 [DOI] [PubMed] [Google Scholar]
- Gehrig H, Faist K, Kluge M (1998b) Identification of phosphoenolpyruvate carboxylase isoforms in leaf, stem and roots of the obligate CAM plant Vanilla planifolia Salib. (Orchidaceae): a physiological and molecular approach. Plant Mol Biol 38: 1215-1223 [DOI] [PubMed] [Google Scholar]
- Gehrig H, Heute V, Kluge M (2001) New partial sequences of phosphoenolpyruvate carboxylase as molecular phylogenetic markers. Mol Phylog Evol 20: 262-274 [DOI] [PubMed] [Google Scholar]
- Gehrig IH, Heute V, Kluge M (1998a) Toward a better knowledge of the molecular evolution of phosphoenolpyruvate carboxylase by comparison of partial cDNA sequences. J Mol Evol 46: 107-114 [DOI] [PubMed] [Google Scholar]
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41: 95-98 [Google Scholar]
- Hartwell J, Gill A, Nimmo GA, Wilkins MB, Jenkins GI, Nimmo HG (1999) Phosphoenolpyruvate carboxylase kinase is a novel protein kinase regulated at the level of expression. Plant J 20: 333-342 [DOI] [PubMed] [Google Scholar]
- Jones JB (1982) Hydroponics: its history and use in plant nutrition studies. J Plant Nutr 5: 1003-1030 [Google Scholar]
- Kai Y, Matsumara H, Inoue T, Terada K, Nagara Y, Yoshinaga T, Kihara A, Tsumura K, Izui K (1999) Three-dimensional structure of phosphoenolpyruvate carboxylase: a proposed mechanism for allosteric inhibition. Proc Natl Acad Sci USA 96: 823-828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Shigesada K, Yanagisawa S, Izui K (1990) Phosphoenolpyruvate carboxylase prevalent in maize roots: isolation of a cDNA clone and its use for analysis of the gene and gene expression. J Biochem 107: 165-168 [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Keryer E, Philippe H, Crétin C (1993) Sorghum phosphoenolpyruvate carboxylase gene family: structure, function and molecular evolution. Plant Mol Biol 21: 487-502 [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Vidal J, Chollet R, Gadal P, Crétin C (1994) Phosphoenolpyruvate carboxylase: structure, regulation and evolution. Plant Sci 99: 111-124 [Google Scholar]
- Nimmo HG (2000) The regulation of phosphoenolpyruvate carboxylase in CAM plants. Trends Plant Sci 5: 75-80 [DOI] [PubMed] [Google Scholar]
- O'Leary M (1982) Phosphoenolpyruvate carboxylase: an enzymologist's view. Annu Rev Plant Physiol 33: 297-315 [Google Scholar]
- Osuna L, González MC, Cejudo FJ, Vidal J, Echevarría C (1996) In vivo and in vitro phosphorylation of the phosphoenolpyruvate carboxylase from wheat seeds during germination. Plant Physiol 111: 551-558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna L, Pierre JN, González MC, Alvarez R, Cejudo FJ, Echevarría C, Vidal J (1999) Evidence for a slow-turnover form of the Ca2+-independent phosphoenolpyruvate carboxylase kinase in the aleurone-endosperm tissue of germinating barley seeds. Plant Physiol 119: 511-520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y, Takai K, Uchida A, Ishida Y (1996) Purification and characterization of phosphoenolpyruvate carboxylase from the hyperthermophylic archaeon Methanothermus sociabilis. FEBS Lett 392: 148-152 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Matsumoto T, Yamamoto K (2002) The genome sequence and structure of rice chromosome 1. Nature 420: 312-316 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewnisk F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignments aided by quality analysis tools. Nucleic Acids Res 25: 4876-4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H, Kawamura T, Izui K (1994) Molecular evolution of phosphoenolpyruvate carboxylase. Plant Cell Environ 17: 31-43 [Google Scholar]
- Vidal J, Chollet R (1997) Regulatory phosphorylation of C4 phosphoenolpyruvate carboxylase. Trends Plant Sci 2: 230-237 [Google Scholar]
- Wolfe KH, Gouy M, Yang Y-W, Sharp PM, Li W-H (1989) Date of the monocot-dicot divergence from chloroplast DNA sequence data. Proc Natl Acad Sci USA 86: 6201-6205 [DOI] [PMC free article] [PubMed] [Google Scholar]