Abstract
NRT1.1 and NIA1 genes, which encode a nitrate (NO3–) transporter and the minor isoform of NO3– reductase (NR), respectively, are overexpressed in roots of NR-deficient mutants of Arabidopsis grown on nutrient solution containing NO3– and reduced N. The overexpression is found only in mutants with reduced NIA2 activity, and disruption of the NIA1 gene alone has no effect on NRT1.1 expression. Because the up-regulation of NRT1.1 and NIA1 is observed in N-sufficient NR mutant plants, it cannot be related to a release of the general feedback repression exerted by the N status of the plant. Our data do not support the hypothesis of overinduction of these genes by an increased concentration of NO3– in tissues. Furthermore, although a control by external pH might contribute to the regulation of NRT1.1, changes in external pH due to lack of NR activity cannot alone explain the up-regulation of both genes. The stimulation of NRT1.1 and NIA1 in NR mutants in these conditions suggests that NR activity is able to repress directly the expression of both genes independently of the availability of reduced N metabolites in wild-type plants. Accordingly, nitrite (NO2–) strongly represses NRT1.1 and NIA1 transcript accumulation in the roots. This effect is rapid, specific, and reversible. Furthermore, transport studies on plants exposed to NO2– show that down-regulation of the NRT1.1 gene is associated with a decrease in NO3– influx. These results indicate that feedback regulation of genes of NO3– assimilation relies not only on the repression exerted by reduced N metabolites, such as NH4+ or amino acids, but may also involve the action of NO2– as a regulatory signal.
Nitrate (NO3–), which is the most important source of mineral nitrogen for most crop species, is acquired by higher plants from the soil through the combined activities of high- and low-affinity uptake systems. Subsequently, NO3– may be accumulated or reduced in root cells, transported via the xylem vessels to be assimilated or stored in the shoot, or released outside of the root via efflux systems. The reduction of NO3– involves two enzymatic steps, reduction of NO3– to nitrite (NO2–) by NO3– reductase (NR), and reduction of NO2– to NH4+ by NO2– reductase (NiR).
Several structural genes encoding transporters of the uptake systems and assimilatory enzymes have been identified in Arabidopsis. To date, the genes encoding NO3– transporters belong to two different families (NRT1 and NRT2). Each family is represented by multiple genes that are differentially regulated and may encode transporters with different regulatory or kinetic properties (Forde, 2000; Orsel et al., 2002; Glass et al., 2002). In Arabidopsis, NRT2.1, the major member of the NRT2 family (seven members) has been characterized as a component of the high-affinity and low-capacity NO3– transport system (Filleur and Daniel-Vedele, 1999; Filleur et al., 2001; Orsel et al., 2002). The NRT1 genes belong to a large family (52 genes)—the so-called peptide transporter (PTR) family—that also contains two transporters able to mediate oligopeptide and His uptake in yeast (Frommer et al., 1994; Steiner et al., 1994). Two members of the NRT1 family (NRT1.1 and NRT1.2) were shown to be involved in the low-affinity and high-capacity transport of NO3– (Tsay et al., 1993; Huang et al., 1996; Touraine and Glass, 1997). Subsequent studies revealed that NRT1.1 is also involved in high-affinity NO3– uptake, thus behaving as a dual affinity transporter (Wang et al., 1998; Liu et al., 1999). The gene is expressed in nascent organs, preferentially in root tips, and is up-regulated in response to exogenous addition of auxin (Tsay et al., 1993; Huang et al., 1996; Guo et al., 2001, 2002). In Arabidopsis, two genes, NIA1 and NIA2, encode the two isoforms of the NR apoprotein with divergent sequences but similar structure (Wilkinson and Crawford, 1993), whereas a single gene encodes the NiR apoprotein (Tanaka et al., 1994). The NIA isoforms do not contribute equally to the NR activity (NRA) of the plant. The G5 mutant, which lacks the NIA2 gene, retains only 10% of the wild-type (WT) NRA in the leaves (Wilkinson and Crawford, 1991). This activity is reduced to 0.5% in the G′4-3 double mutant, which carries an additional point mutation in NIA1 (Wilkinson and Crawford, 1993). Despite a common catalytic function, the expression patterns of NIA1 and NIA2 are not similar in response to NO3– induction, light, and cytokinins (Cheng et al., 1992, 1991; Lin and Cheng, 1997; Yu et al., 1998).
The recent advances in the understanding of the regulation of NO3– uptake and assimilation (for review, see Crawford and Glass, 1998; Stitt, 1999; Forde, 2002) have shown that these functions are highly integrated at the whole-plant level, through a complex control system involving the action of signals related to metabolism and nutritional status (probably many of them remain unknown). At least three major types of regulation have been identified. The first one is the induction by NO3– itself. The second one is the coordination with photosynthesis through the stimulation of various steps of NO3– acquisition and metabolism by sugars. The third one is the control of the N status of the whole plant through feedback repression exerted by downstream N metabolites such as NH4+ or amino acids. A similar complexity is found at the molecular level. NRT1.1 and NRT2.1, as NIA1 and NIA2, are inducible by NO3– and up-regulated by sugars (Cheng et al., 1991; Filleur and Daniel-Vedele, 1999; Lejay et al., 1999; Zhuo et al., 1999). These two characteristics are common features of the expression of most of the Arabidopsis genes involved in NO3– uptake and assimilation (Stitt, 1999; Coruzzi and Bush, 2001; Forde, 2002; Glass et al., 2002), with the exception of the expression of NRT1.2, that does not depend on the presence of NO3– (Huang et al., 1999). Expression of NRT1.1 and NRT2.1 has been studied in detail. So far, NRT2.1 is the only NO3– transporter gene known to be under feedback repression by N metabolites (Filleur and Daniel-Vedele, 1999; Lejay et al., 1999; Zhuo et al., 1999). Its expression is strongly increased by N deficiency (Lejay et al., 1999; Gansel et al., 2001), and the gene has been characterized functionally as a major component mediating the response of NO3– uptake to variation of the N status of the whole plant (Cerezo et al., 2001). In contrast to NRT2.1, the NRT1.1 gene is neither repressed by reduced N metabolites nor induced by N limitation (Lejay et al., 1999).
The use of NR-deficient mutants has been a powerful tool to unravel the specific regulatory effects of NO3– and of products of its assimilation (Scheible et al., 1997; Krapp et al., 1998; Filleur and Daniel-Vedele, 1999; Zhang et al., 1999). When NRA is strongly reduced compared with WT, mutant plants are under N limitation when supplied with NO3– as sole nitrogen source but are N sufficient if a reduced nitrogen form is added together with NO3– in the nutrient solution. This allows investigation of the response of genes to feedback repression by N metabolites without modifying the supply of NO3–. Using this strategy, we have previously shown that NRT2.1 is strongly up-regulated in the G′4-3 Arabidopsis NR-deficient mutant compared with WT when both genotypes are grown on NO3– as sole nitrogen source (Lejay et al., 1999). However, this up-regulation of NRT2.1 in the NR-deficient mutant is suppressed as soon as NH4+ is added to the nutrient solution. This indicates that NRT2.1 expression is not directly controlled by NRA, but rather by the availability of downstream N metabolites such as NH4+ or amino acids. Accordingly, exogenous supply of amino acid or NH4+ results in a strong decrease in NRT2.1 transcript accumulation (Zhuo et al., 1999). An unexpected outcome of our study was to find the NRT1.1 transcript level markedly increased in the roots of G′4-3 NR-deficient mutant compared with WT (Lejay et al., 1999). However, in contrast to NRT2.1, the up-regulation of NRT1.1 in the G′4-3 mutant is also observed in N-sufficient plants cultivated on NH4NO3 and cannot be attributed to the release of feedback repression exerted by NH4+ or amino acids. To explain these results, we have proposed the existence of an as yet unknown regulatory mechanism, corresponding to a direct repression of NRT1.1 expression by NR independently of the N status of the plant.
The aim of the present work was to investigate this hypothesis. To demonstrate the generality of the observations made on the G′4-3 NR mutant, expression of NRT1.1 has been analyzed in various other mutants impaired either in the NR apoprotein isoform or in the NR molybdenum cofactor (MoCo) biosynthesis. Investigations concerning the effect of the nitrogen source (reduced nitrogen and NO3–) and the external pH on the expression of NRT1.1 in WT and NR-deficient plants are described. Finally, the action of NO2–, the direct product of the reaction catalyzed by NR, has been investigated on both NRT1.1 expression and root NO3– influx.
RESULTS
NRT1.1 and NIA1 Are Up-Regulated in Various NR-Deficient Mutants
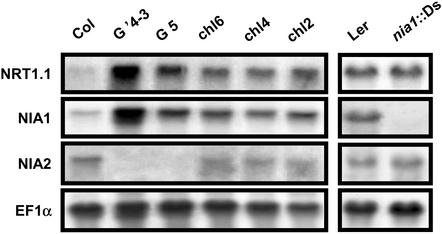
Several NR-deficient mutants were investigated to determine whether one specific component of NRA (NIA1 or NIA2 apoenzymes, MoCo) is responsible for the repression of NRT1.1 expression in roots. The G5 mutant has a deletion in the NIA2 gene encoding the major isoform of Arabidopsis NR apoenzyme (Wilkinson and Crawford, 1991). The G′4-3 double mutant, derived from G5, has an additional point mutation in the NIA1 gene that encodes the minor isoform of the NR apoenzyme (Wilkinson and Crawford, 1993). This mutation reduces NRA but does not abolish NIA1 transcript accumulation. The nia1::Ds mutant carries a Ds nonautonomous transposable element inserted in the coding sequence of the NIA1 gene (Parinov et al., 1999). The chl6, chl4, and chl2 mutants possess intact WT NIA1 and NIA2 apoproteins, but are deficient in MoCo biosynthesis, which leads to altered activities of both enzymes (LaBrie et al., 1992).
In plants grown hydroponically on 1 mm NH4NO3 as the sole nitrogen source, the amount of NRT1.1 transcript in the roots was higher in most NR-deficient mutants than in WT plants (Fig. 1). The only exception was the nia1::Ds mutant, in which only the NIA1 gene is disrupted and which displayed unaltered NRT1.1 transcript accumulation compared with Landsberg erecta (Ler) plants. Thus, comparison between mutants indicates that NRT1.1 is overexpressed only when NIA2 activity is altered, due to either the absence of the NIA2 gene (G5 and G′4-3 mutants) or the mutation of MoCo biosynthesis (chl2, chl4, and chl6 mutants). This indicates that NIA2 plays a predominant role in the regulation of NRT1.1 expression. An unexpected result of these studies was that NIA1 transcript was accumulated in parallel to NRT1.1 transcript in roots, indicating that expression of NIA1 also is probably under the same control as NRT1.1. Interestingly, transcripts of NIA1 and NIA2 do not display the same behavior in the various NR mutants. Neither the loss of NIA1 isoform (nia1::Ds mutant) nor the altered NRA resulting from mutations on MoCo biosynthesis (chl2, chl4, and chl6 mutants) significantly alters NIA2 transcript accumulation.
Figure 1.
Transcript accumulation of NRT1.1, NIA1, and NIA2 in the roots of NR-deficient mutants compared with WT (Col and Ler). Plants (8 weeks old) were grown hydroponically on nutrient solution containing 1 mm NH4NO3 as the nitrogen source. Total RNAs were analyzed by northern blot.
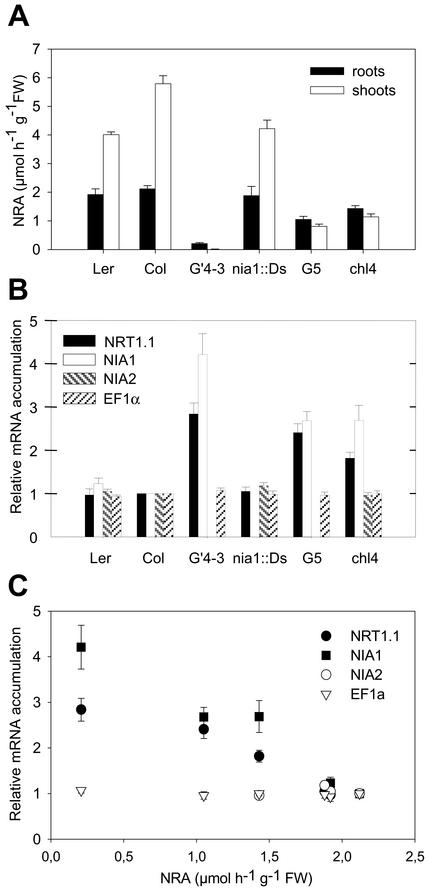
The ability of mutations affecting the MoCo biosynthesis pathway (chl2, chl4, and chl6 mutants) to stimulate expression of both NRT1.1 and NIA1 in roots indicates that the repression exerted by NR is not related only to the expression of the NR apoprotein, but requires the activity of the enzyme. Additional experiments were performed to quantify more precisely the correlation between expression of both NRT1.1 and NIA1 in the roots of the various mutants and total NRA in these organs (Fig. 2). Mutants display various levels of NR deficiencies in both roots and shoots (Fig. 2A). An inverse correlation was found between NRT1.1 and NIA1 transcript levels and total NRA in the roots in most of the mutants (Fig. 2, B and C). Similar inverse correlation was also found with total shoot NRA (data not shown). Together these data suggest that the repression of the two genes depends on the plant capacity to reduce NO3–. According to the hypothesis of a direct control exerted by root NRA, loss of NIA2 is expected to have a major effect on the repression of NRT1.1 and NIA1, because NIA2 encodes the main isoform of NR responsible for most of the catalytic activity of the plant. However, not all of the data agree with this hypothesis. In particular, G′4-3 and G5 plants (Fig. 2A) have markedly different root NRA (9% and 49% of the WT root NRA, respectively; Fig. 2A), whereas NRT1.1 is expressed at similar levels in both genotypes (Fig. 2B). Moreover, the root NRA found in the G5 mutant (deleted for NIA2) is fully attributable to NIA1. This activity is especially high in the mutant because of the overexpression of NIA1 (Figs. 1 and 2B). This indicates that NIA1-related NRA alone is unable to repress NRT1.1 and suggests that both NR isoforms are not equivalent in the regulation of this gene.
Figure 2.
NRA (A) and root transcript accumulation of NRT1.1, NIA1, NIA2, and EF1α (B) in NR-deficient mutants compared with WT (Col and Ler). C, Transcript accumulation was plotted as a function of root NRA. Plants (8 weeks old) were grown hydroponically on nutrient solution containing 1 mm NH4NO3 as nitrogen source. The values of NRA are means of six replicates ± SE. The values of relative accumulation of transcript are means of four independent experiments ± SE.
Repression of NRT1.1 and NIA1 Expression by NR Depends on NO3– Supply
The initial evidence for the up-regulation of NRT1.1 in response to NR deficiency has been obtained in G′4-3 plants grown on nutrient solution containing 1 mm NH4NO3 as the sole nitrogen source. To further investigate the mechanisms involved in the repression of NRT1.1 and NIA1 expression by NR, the effects of the nitrogen source were studied in more detail.
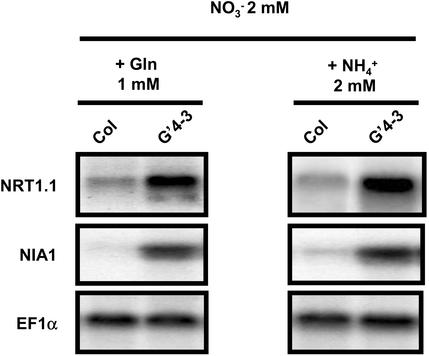
First, two sources of reduced nitrogen that can be assimilated by NR-deficient plants were compared (Fig. 3). G′4-3 plants were cultivated hydroponically with a nutrient solution containing 2 mm NO3– supplemented either with 1 mm Gln or with 2 mm NH4+. In both conditions, higher levels of NRT1.1 and NIA1 transcripts were found in the roots of the G′4-3 mutant than in those of the WT. Thus, up-regulation of NRT1.1 and NIA1 in NR-deficient mutants cannot be attributed to a specific effect of the exogenous supply of NH4+.
Figure 3.
Transcript accumulation of NRT1.1 and NIA1 in roots of WT (Col) and G′4-3 plants grown on nutrient solution containing 2 mm KNO3 with either 1 mm Gln or 2 mm NH4Cl. Plants (4 weeks old) were grown hydroponically under sterile conditions. Total RNAs were analyzed by northern blot.
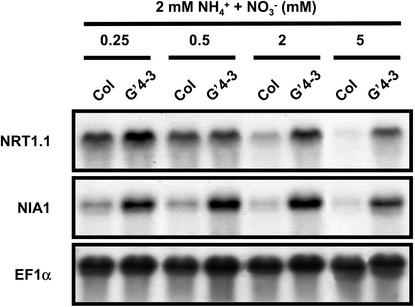
Second, the effect of the level of NO3– supply on NRT1.1 and NIA1 expression was analyzed in both Columbia (Col) and G′4-3 plants. The two genotypes were cultivated on nutrient solution containing 2 mm NH4+ (to ensure N-sufficiency), supplemented with NO3– at various concentrations (0.25, 0.5, 2, and 5 mm). The increase of NO3– concentration in the nutrient solution resulted in a strong decrease of both NRT1.1 and NIA1 transcript accumulations in the roots of WT plants, but had no effect on the expression of these genes in roots of G′4-3 plants (Fig. 4). This indicates that high levels of NO3– promote down-regulation of NRT1.1 and NIA1 through a mechanism dependent on NO3– reduction. This confirms the inverse relationship between the reduction of NO3– and the repression of NRT1.1 and NIA1 (Fig. 2C). Moreover, repression of NRT1.1 in WT plants by high availability of NO3– suggests that the rate of NO3– reduction present in roots rather than total reduction capacity is probably involved in the repression of NRT1.1 and NIA1.
Figure 4.
Transcript accumulation of NRT1.1 and NIA1 in roots of WT (Col) and G′4-3 plants grown on nutrient solution containing 2 mm NH4Cl and various concentrations of KNO3 as a nitrogen source. Plants were grown hydroponically under sterile conditions on 2 mm NH4NO3 for 3 weeks and transferred to the various conditions of NH4+ and NO3– supply 1 week before harvest. Total RNAs were analyzed by northern blot.
Expression of NRT1.1 and NIA1 Is Repressed by NO2–
The ability of NO3– reduction to trigger the repression of NRT1.1 and NIA1 in the presence of NH4+ or Gln (Figs. 3 and 4) suggests that products of NO3– reduction upstream of NH4+ are involved in this down-regulation. NR deficiency may reduce cellular levels of NO2–. Because the reduction of NO3– generates OH–, which is generally excreted by the plant, NR deficiency may also promote a decrease of both internal and external pH.
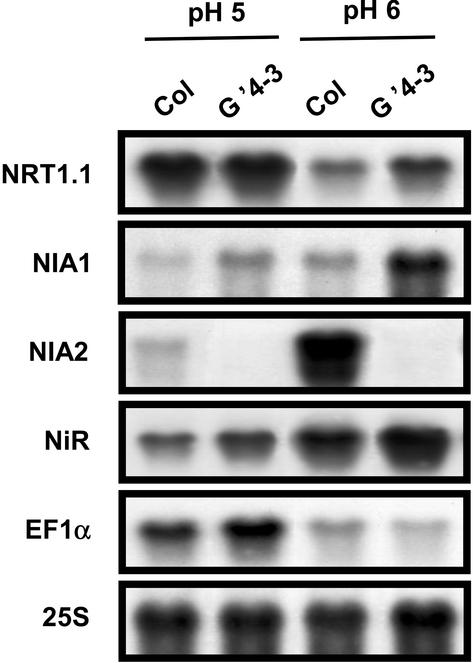
The pH hypothesis was examined in detail because expression of NRT1.1 is known to be up-regulated by acidification of the nutrient medium (Tsay et al., 1993). In our standard hydroponic conditions, no significant differences of acidification of the external medium were observed between G′4-3 and WT plants. However, it cannot be ruled out that subtle variations might trigger the up-regulation observed in NR-deficient mutants. To investigate the role of the external pH, WT and G′4-3 plants were cultivated in the same container, and the pH of the nutrient solution was maintained at pH 5 or 6 in the presence of organic buffers. In both pH conditions, NIA1 and NRT1.1 were up-regulated in the mutant when compared with the WT (Fig. 5). Acidification of the external solution from pH 6 (initial pH of the standard nutrient solution) to pH 5 is correlated with large variations in all genes studied including NRT1.1 (increased expression) and NIA1 (reduced expression). These divergent responses of NIA1 and NRT1.1 upon pH changes make it highly unlikely that a common up-regulation of both genes in the G′4-3 mutant results from acidification related to NR deficiency. However, because the difference of NRT1.1 expression between the G′4-3 mutant and the wild type is reduced when pH is maintained by organic buffer particularly at pH5 when NRT1.1 expression is high, a possible involvement of external pH in the up-regulation of NRT1.1 in G′4-3 cannot be totally ruled out.
Figure 5.
Effect of the external pH on the transcript accumulation of NRT1.1, NIA1, NIA2, and NiR in roots of WT (Col) and G′4-3 plants. Plants were grown hydroponically on standard nutrient solution containing 1 mm NH4NO3 as nitrogen source for 7 weeks and transferred for 1 week to solutions buffered with 4.4 mm MES (pH 6 or pH 5 with Tris). Total RNAs were analyzed by northern blot.
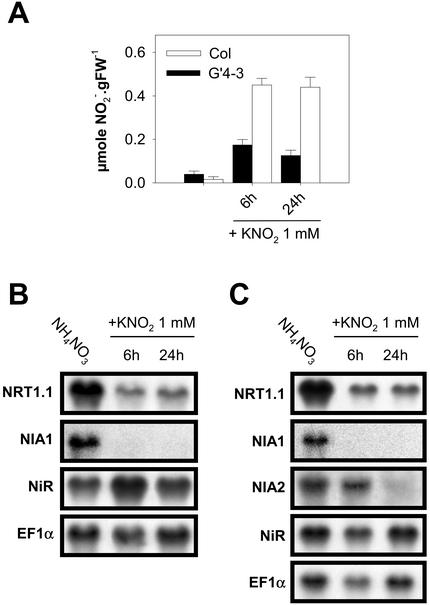
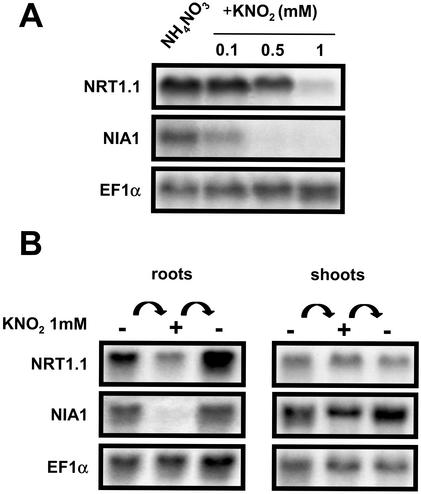
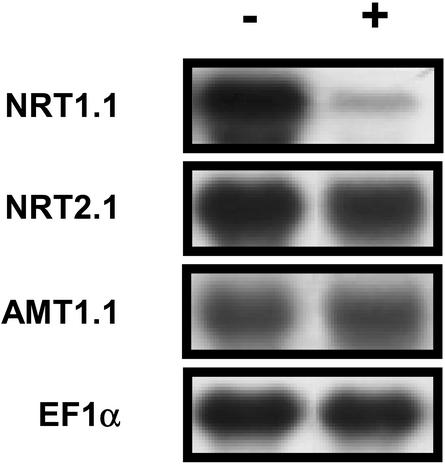
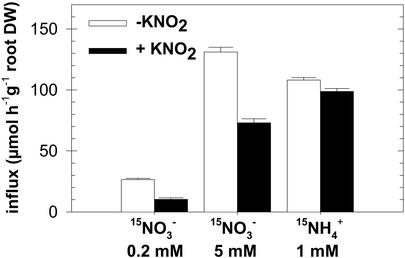
To examine the hypothesis of a NO2–-mediated regulation, the effect of exogenous supply of NO2– was investigated. In these experiments, NO2– was added as 1 mm KNO2 to the nutrient solution containing 1 mm NH4NO3 as the sole nitrogen source. Net NO2– uptake rate was measured at 24.0 ± 6.5 μmol g-1 root dry weight h-1 (±se, n = 7) on G′4-3 plants exposed for 5 h to 1 mm K15NO2. No visible symptoms of toxicity were noticed in response to the exogenous supply of NO2–, at least during the first 48 h. This treatment had no effect on the pH of the bulk solution and did not modify NO2– accumulation in the shoots (data not shown). However, root NO2– content increased in both WT and G′4-3 plants during the first 6 h after addition of NO2–, and remained almost stable thereafter until 24 h (Fig. 6A). After 6 h of treatment, NRT1.1 and NIA1 transcript levels were markedly reduced in both genotypes as compared with control plants left on 1 mm NH4NO3 without KNO2 (Fig. 6, B and C). A similar decrease could be observed already after 3 h of exposure to NO2– (data not shown). Repression was dependent on the concentration of NO2– present in the nutrient solution (Fig. 7A). Addition of 0.1 mm or 0.5 mm KNO2 was able to trigger a significant reduction of the expression of NIA1 and NRT1.1, respectively. The rapid and strong inhibition of NRT1.1 and NIA1 expression by exogenous NO2– supply was not part of a general response. First, transcript levels of EFIα, NiR, AMT1.1 (encoding an ammonium transporter; Ninnemann et al., 1994), or NRT2.1 were not decreased by the treatment in either genotype (Fig. 6, B and C; Fig. 8). The expression level of NIA2 was also inhibited by NO2– in WT, but after a significant delay and to a lesser extent (Fig. 6C). Second, repression of NRT1.1 and NIA1 expression by exogenous NO2– supply was found only in the roots and was fully reversible (Fig. 7B). The addition of NO2– in the nutrient solution also resulted in a repression of NO3– uptake in G′4-3 plants (Fig. 9). Root influx was measured at 0.2 and 5 mm external 15NO3– concentrations to differentiate between the activities of high- and low-affinity transport systems. Supply of 1 mm NO2– for 6 h resulted in a 61% inhibition of 15NO3– influx at 0.2 mm and a 44% inhibition at 5 mm. Interestingly, NO2– exposure did not affect 15NH4+ influx.
Figure 6.
Effect of exogenous supply of 1 mm KNO2 on NO2– accumulation (A) and NRT1.1, NIA1, and NiR transcript accumulation in roots of G′4-3 (B) and WT (Col; C) plants. Plants (8 weeks old) were grown hydroponically on a nutrient solution containing 1 mm NH4NO3. KNO2 was added to the medium for 6 or 24 h before harvest (middle of the light period). The values of NO2– content are the mean of 10 replicates ± SE. Total RNAs were analyzed by northern blot.
Figure 7.
Dose-response and reversibility of the repression of NRT1.1 and NIA1 by exogenous supply of KNO2 in G′4-3 mutants. Plants were grown hydroponically for 8 weeks on a nutrient solution containing 1 mm NH4NO3. Total RNAs were analyzed by northern blot. A, Plants were grown during 24 h on nutrient solution containing 0.1, 0.5, or 1 mm KNO2 before harvest. Plants grown without exogenous supply of KNO2 were used as a control. B, Plants were grown during 24 h on a nutrient solution containing 1 mm KNO2, and roots were washed 5 min in CaSO4 0.1 mm and returned for 24 h to standard nutrient solution before harvest.
Figure 8.
Effect of exogenous supply of KNO2 on NRT1.1, NRT2.1, and AMT1.1 on transcript accumulation in roots of G′4–3 plants. Plants (8 weeks old) were grown hydroponically on a nutrient solution containing 2mm NH4NO3; (+) 2 mm KNO2 was added to the medium for 24 h before harvest (middle of the light period); (-) control plants were left on the same nutrient solution. Total RNAs were analyzed by Northern blot.
Figure 9.
Effect of exogenous supply of 1 mm KNO2 on 15NO3– and 15NH4+ influxes in roots of the G′4-3 mutant. Plants (8 weeks old) were grown hydroponically on a nutrient solution containing 1 mm NH4NO3. KNO2 was added to the medium 6 h before influx measurements. Plants grown without exogenous supply of KNO2 were used as a control. Root influx was measured with complete nutrient solution containing 5 mm 15NO3– 0.2 mm 15NH4+ or 1 mm NH4+ as tracers. The values are the means of 10 replicates ± SE.
DISCUSSION
Both NRT1.1 and NIA1 Are under Feedback Repression by NO3– Reduction Independent of the N Status of the Plant
Our results confirm, with a large set of genotypes, the overexpression of NRT1.1 in the roots of NR-deficient plants as initially suggested by studies with the G′4-3 mutant (Lejay et al., 1999). The overexpression is not triggered by low N status of the plant because it is observed with N sources allowing proper growth of NR-deficient mutants (NH4NO3 or NO3– plus Gln) and because previous studies have shown that NRT1.1 is not under the control of the feedback repression by downstream N metabolites (Lejay et al., 1999). Our results suggest that NRT1.1 is under a different type of feedback regulation, exerted directly at the level of NO3– reduction. One unexpected outcome of our study is the finding that the pattern of expression of NIA1 closely parallels that of NRT1.1, indicating that NIA1 is also repressed by NO3– reduction. Thus, in addition to a likely regulation of NO3– uptake (regulation of NRT1.1 expression), this feedback repression may also correspond to an auto-regulation of NR gene expression. The compensation for the absence of NIA2 by the overexpression of the NIA1 gene in the roots of the G5 mutant (deletion of NIA2) restores a level of NRA corresponding to 49% of the WT NRA. This regulation may contribute significantly to the control of NRA in Arabidopsis roots. Overaccumulation of NIA mRNA in mutants expressing a defective NR apoenzyme and in mutants impaired in MoCo biosynthesis has already been described in leaves of various species, including wild tobacco (Nicotiana plumbaginifolia; Pouteau et al., 1989), cultivated tobacco (Nicotiana tabacum; Vaucheret et al., 1990), and Arabidopsis (LaBrie et al., 1992; Wilkinson and Crawford, 1993). Similar stimulation of NIA transcript accumulation in leaves was found in plants exposed to tungstate, an inhibitor of NRA (Deng et al., 1989). Up-regulation is not restricted to NR. The NiR gene and other genes involved in nitrogen, organic acid, and carbon metabolism are also overexpressed in NR-deficient genotypes of tobacco (Scheible et al., 1997). However, in most of these studies, it is unclear whether this up-regulation results from N limitation of the plants or not, because the N source supplied to the plants did not always include reduced N. In the present study, we show that the overexpression of NIA1 in NR-deficient plants is also found in the roots. In our case, the up-regulation can be unambiguously attributed to a mechanism directly related to NO3– reduction and not to the general control exerted by the N status of the plant.
Our results concerning the respective role of the two NR isoforms suggest that catalytic activity of NIA2 has a major role in the repression exerted by NO3– reduction. It is unclear whether NIA2 has a predominant action on NRT1.1 and NIA1 because it catalyzes the major part of total NRA or because NIA2-related activity has a specific role in the regulation of these two genes. In the absence of investigations describing NIA1- and NIA2-specific activities and distributions across the plant, we can only speculate about the possible mode of action of NIA2. It has been shown recently that expression of NRT1.1 is restricted to nascent organs, mainly in root tips (Guo et al., 2001). One hypothesis might be that NIA1, NIA2, and NRT1.1 are not expressed in the same root tissues. For instance, if NIA1 and NRT1.1 are present in two distinct cell types, whereas NIA2 is present in both, this may explain why NIA1 has apparently little action on NRT1.1 expression, while NIA2 governs both NRT1.1 and NIA1 transcription. Another hypothesis might be that the two NIA isoforms display subtle differences in their functional properties in vivo, which are not revealed by our in vitro measurements. Finally, because of the inverse correlation between shoot NRA and expression of NRT1.1 and NIA1 in roots, a negative control exerted by shoots, mediated by a long-distance signal, has also to be considered as an alternative hypothesis.
The Cause of NRT1.1 and NIA1 Overexpression in NR-Deficient Plants: Decreased NO3– Reduction or Increased NO3– Accumulation?
Up-regulation of gene expression in NR-deficient plants has been proposed to result from “overinduction” by NO3– which accumulates at very high levels in the absence of active NR (Scheible et al., 1997; Forde, 2000). This may hold true for NRT1.1 and NIA1, known to be inducible by NO3–. However, several lines of evidence do not support this hypothesis. First, variation of the accumulation of NO3– in NR-deficient or -overexpressing plants mostly occurs in the shoot, not in the roots (Quilleré et al., 1994; Gojon et al., 1998; Lejay et al., 1999). Second, increasing the level of NO3– supply does not stimulate expression of NRT1.1 and NIA1 either in WT or in the G′4-3 double mutant. The increase in external NO3– concentration results in an amplification of the overexpression of NRT1.1 and NIA1 in the G′4-3 NR-deficient mutant compared with WT (Fig. 4). However, this amplification is not due to the increased accumulation of NRT1.1 and NIA1 transcripts in the roots of G′4-3 plants but to a decrease in the level of these transcripts in the WT. These results are fully consistent with the hypothesis of a feedback repression exerted by NO3– reduction in the WT, because in this genotype, NO3– reduction rate is expected to go up with increasing NO3– supply.
Feedback Repression by NO2–
Because expression of NRT1.1 is not downregulated by NH4+, the effect of two other direct products of NO3– reduction, namely NO2– and OH–, has been considered. Stimulation of NRT1.1 expression by the acidification of the external medium has been described previously (Tsay et al., 1993). Our results do not support the hypothesis that a lowered OH– production in NR-deficient plants may be the cause for overexpression of NRT1.1 and NIA1 in NR-deficient plants. First, even when WT and G′4-3 plants are maintained at the same external pH, NRT1.1 and NIA1 transcripts still accumulate at higher levels in the roots of the mutant. Second, acidification of the external medium from pH 6 to 5 triggers opposite responses of NRT1.1 and NIA1, indicating that the common up-regulation of both genes in NR mutants is unlikely to be explained by a pH effect. However, the effect of external pH on NRT1.1 expression indicates that pH may interact with specific NR-dependent factors to repress the gene in wild type. On the other hand, these data do not rule out a possible role of changes of cytoplasmic pH. Such investigations to test this hypothesis will require direct measurements in root cells expressing NRT1.1 and NIA1 with H+-specific microelectrodes (Walker et al., 1996) or 31P-NMR techniques (Bligny and Douce, 2001).
The strong reduction of both NIA1 and NRT1.1 root transcript levels in response to the addition of NO2– in the nutrient solution in absence of N limitation supports the hypothesis of NO2– acting as a repressor of the expression of both genes. NO2– has been shown to inhibit NO3– uptake in barley (Hordeum vulgare; King et al., 1993), but the explanation usually provided to account for this effect is the competition between the two anions for the activity of the uptake systems. In our experiments, the decrease of NRT1.1 gene expression is correlated with a reduction of NO3– influx in both the high- and low-affinity ranges. This is consistent with the fact that NRT1.1 plays a major role in both high- and low-affinity NO3– uptake under conditions similar to those used in our study, in particular in presence of NH4NO3 as a N source (Touraine and Glass, 1997; Wang et al., 1998; Liu et al., 1999). In addition to the absence of symptoms on treated plants, the possibility that this down-regulation of both NRT1.1 and NIA1 expression results from a toxic effect of NO2– is contradicted by at least six arguments: (a) The repression is rapid (less than 3 h); (b) the effect is fully reversible; (c) the treatments promote relatively modest increases of root NO2– content (2 orders of magnitude less than NO3–), most probably because of the high affinity of the NiR enzyme for its substrate (Beevers and Hageman, 1980); (d) no changes are observed in shoots of plants exposed to NO2–; (e) other genes such as EF1α, NiR, NRT2.1, and AMT1 are not affected; and (f) unlike NO3– uptake, NH4+ uptake is not affected. The hypothesis of NO2– being a repressor of NIA1 gene is somehow in contradiction with the fact that leaves of NiR antisense tobacco transformants cultivated on NO3– as the sole nitrogen source display an increase of NO2– accumulation as well as NR transcript level (Vaucheret et al., 1992). Because these tobacco plants were expected to be seriously N limited, the feedback repression exerted by down-stream N metabolites was most probably released, and therefore it cannot be excluded that a repression by NO2– might have been overcome by the stimulation of gene expression triggered by the N limitation. Also, whether shoots and roots display similar or different responses upon an increasing NO2– accumulation remains to be investigated.
To our knowledge, this work is the first report pointing out the ability of NO2– to repress genes involved in N acquisition in higher plants. Such a role is unexpected for NO2–, which is believed to be toxic and present at very low levels within the cell, because the activity of the NiR enzyme measured in vitro is in large excess. However, recent results obtained on transgenic Arabidopsis plants overexpressing a spinach (Spinacia oleracea) NiR cDNA suggest that reduction of NO2– may be a rate-limiting step (Takahashi et al., 2001). This is compatible with the hypothesis of NO2– acting as a regulatory signal. Such hypothesis has already been proposed in the unicellular algae Chlamydomonas reinhardtii. In this organism, NO2– represses genes encoding NR and NO3– transporters (Loppes et al., 1999). The mode of action of NO2– remains unknown. It may act directly, or it may act indirectly through the involvement of a related metabolite such as NO. NO is now considered to be a regulatory signal involved various in plant responses to environmental stimuli (Beligni and Lamattina, 2001; Wendehenne et al., 2001; Desikan et al., 2002; Murgia et al., 2002). NO might be produced chemically by decomposition of HNO2 or enzymatically by NR from NO2– as a substrate when it accumulates in tissues (Dean and Harper, 1988; Yamasaki and Sakihama, 2000; Rockel et al., 2002). Interestingly, NO has been proposed to inhibit root NRA in lettuce (Lactuca sativa) plants (Hufton et al., 1996).
All together, our data support a model postulating that the NO2– (or NO) produced by NR represses NRT1.1 and NIA1 expression in the roots. This regulation, which appears to be independent of the nitrogen status of the plant, corresponds to a mechanism for coordinating NO3– uptake and assimilation. It has the particularity to be specific for NO3– nutrition, as opposed to feedback repression by reduced N metabolites (NH4+ and/or amino acids), that targets NO3– as well as NH4+ acquisition. This regulation is not common to all genes involved in NO3– assimilation. Although the NiR gene is up-regulated in NR-deficient mutants, it is not repressed upon NO2– addition, indicating that NiR is probably not under the same control as NRT1.1 and NIA1.
Further studies are required to understand the physiological significance of this regulation. One hypothesis may be related to the adaptation to root anoxia, from which plants suffer during flooding periods. The ability of roots to accumulate and to excrete NO2– under hypoxia has been extensively used to assay NRA in vivo (Radin, 1974). Under anoxic conditions, NO2– can be produced in the roots following uptake and reduction of NO3–, whereas NiR activity is strongly lowered by the shortage of reduced ferredoxin availability. It can then be speculated that repression of both NO3– uptake and assimilatory systems by NO2– might provide a mechanism to prevent toxic accumulation of NO2– by limiting its production in hypoxic cells.
MATERIALS AND METHODS
Plant Material and Culture Conditions
Five genotypes of Arabidopsis ecotype Col were used: G′4-3 and G5 (Wilkinson and Crawford, 1991, 1993), chl2, chl4, and chl6 (Braaksma and Feenstra, 1982; LaBrie et al., 1992). The nia1::Ds mutant (Parinov et al., 1999) is of the ecotype Ler. All mutants were provided by Arabidopsis Biological Resource Center (Ohio State University, Columbus) and Nottingham Arabidopsis Stock Center (Nottingham, UK). Plants at the vegetative stage were grown hydroponically under standard or sterile conditions. Basal nutrient solution without nitrogen contained 1 mm KH2PO4, 1 mm MgSO4, 250 μm CaCl2, 0.1 mm Na-Fe-EDTA, 50 μm KCl, 50 μm H3BO3, 5 μm MnSO4, 1 μm ZnSO4, 1 μm CuSO4, and 0.1 μm (NH4)6Mo7O24, pH adjusted to 6 with KOH. Under non-sterile conditions, plants were cultivated in a 10-L tank as previously described (Lejay et al., 1999) with the following environmental parameters: light/dark cycle, 8 h/16 h; light intensity, 300 μmol s-1 m-2 photosynthetically active radiation; temperature, 22°C/20°C; and 70% hygrometry. The nutrient solution was renewed twice a week during the first part of the culture and daily the last week before the experiment. Under sterile conditions, plants were grown in the same basal medium in presence of 10 g L-1 Suc and 2 mm MES-Tris, pH 6, according to Touraine and Glass (1997). Plants were held by a nylon grid (480-μm gauge) on the top of a membrane raft (three to four plants per box) floating on 60 mL of nutrient solution and were renewed twice during the 1st week of culture and daily thereafter. For all experiments, plants of the same age cultivated in various conditions were harvested at the same time in the middle of the light period. When necessary, treatments were initiated for various times before the harvest.
15N Uptake and Assimilation Studies
Root influxes of NO3– and NH4+ were assayed according to Delhon et al. (1995) and Gazzarrini et al. (1999), respectively. Plants were sequentially transferred to 0.1 mm CaSO4 for 1 min and to the complete nutrient solution containing either NO3– or NH4+ (99% atom excess 15N) for 5 min. At the end of the labeling, roots were washed for 1 min in 0.1 mm CaSO4 and were separated from shoots. The organs were dried at 70°C for 48 h, weighed, and analyzed for total 15N content using a continuous-flow isotope ratio mass spectrometer coupled with a carbon nitrogen elemental analyzer (ANCAMS, PDZ Europa, Crewe, UK) as described by Clarkson et al. (1996). Preliminary influx studies were performed using a labeling solution containing 1 mm 15NO3– and various mixtures of 1 mm NO2– or 1 mm NH4+. No significant differences were observed between the results measured with these various solutions, indicating that in our conditions, the presence of NH4+ and NO2– in the labeling solution has a negligible effect on NO3– influx (data not shown). NO2– was extracted by boiling fresh tissue for 15 min in water. Total NRA and NO2– were assayed according to Robin (1979).
Northern Blot
Total RNAs were isolated by phenol-guanidine extraction followed by lithium chloride precipitation according to Lobreaux et al. (1992). RNAs (20 μg per lane) were resolved by electrophoresis on MOPS-formaldehyde agarose gels, blotted on to Biotrans (+) nylon membranes, and covalently linked to the filter by UV cross-linking (Ausubel et al., 1988). Hybridization to a randomly primed radiolabeled probe was done at 42°C in 50% (v/v) formamide, 1% (w/v) sarkosyl, 5× SSC (0.75 m NaCl, and 0.075 m Na3 citrate, pH 7), and 100 μg mL-1 salmon sperm DNA. Membranes were washed twice at 42°C in 0.1% (w/v) SDS and 2× SSC for 15 min and then twice in 0.1% (w/v) SDS and 0.1× SSC for 15 min. Quantification of radioactive signals were achieved using a PhosphoImager (Storm, Molecular Dynamics, Sunnyvale, CA). Blots were stripped in 0.01× SSC for 5 min at 100°C. Gene-specific probes used in this work corresponded to the full-length cDNA of NRT1.1 (Tsay et al., 1993), the 1.6-kb EcoRI-HindIII fragment of the pAtc46 plasmid carrying a partial sequence of the NIA2 cDNA (Crawford et al., 1988), the expressed sequence tag (EST) 134 J7/T7 carrying a partial sequence of the NIA1 cDNA, and the EST 177N14 carrying a partial sequence of the NiR cDNA (ESTs were provided by the Arabidopsis Biological Resource Center stock center). Two probes were systematically used as controls: the full-length cDNA of the elongation factor EF1αA3 as a gene nondirectly related to N metabolism (Axelos et al., 1989), and a 25S rDNA fragment (Choumane and Heizman, 1988) to monitor the equal loading of blots.
Acknowledgments
We thank Françoise Cellier, Siobhan Staunton, Tim Tranbarger, and Sabine Zimmermann for critical reading of the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.018523.
References
- Ausubel FM, Brent R, Kinston RE, Moore DD, Seidmann IG, Smith JA, Struhl K (1988) Current Protocols in Molecular Biology. Greene/Wiley Publishers, New York
- Axelos M, Bardet C, Liboz T, Le Van Thai A, Curie C, Lescure B (1989) The gene family encoding the Arabidopsis thaliana translation elongation factor EF-1 alpha: molecular cloning, characterization and expression. Mol Gen Genet 219: 106-112 [DOI] [PubMed] [Google Scholar]
- Beevers L, Hageman RH (1980) Nitrate and nitrite reduction. In BJ Mifin, ed, The Biochemistry of Plants. Academic Press, New York, pp 115-168
- Beligni MV, Lamattina L (2001) Nitric oxide in plants: the history is just beginning. Plant Cell Environ 24: 267-278 [Google Scholar]
- Bligny R, Douce R (2001) NMR and plant metabolism. Curr Opin Plant Biol 4: 191-196 [DOI] [PubMed] [Google Scholar]
- Braaksma FJ, Feenstra WJ (1982) Isolation and characterization of nitrate reductase-deficient mutants of Arabidopsis thaliana. Theor Appl Genet 64: 83-90 [DOI] [PubMed] [Google Scholar]
- Cerezo M, Tillard P, Filleur S, Munos S, Daniel-Vedele F, Gojon A (2001) Major alterations of the regulation of root NO3– uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol 127: 262-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CL, Acedo GN, Cristinsin M, Conkling MA (1992) Sucrose mimics the light induction of Arabidopsis nitrate reductase gene transcription. Proc Natl Acad Sci USA 89: 1861-1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CL, Acedo GN, Dewdney J, Goodman HM, Conkling MA (1991) Differential expression of the two Arabidopsis nitrate reductase genes. Plant Physiol 96: 275-279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choumane W, Heizman P (1988) Structure and variability of nuclear ribosomal genes in the genus Helianthus. Theor Appl Genet 76: 481-489 [DOI] [PubMed] [Google Scholar]
- Clarkson DT, Gojon A, Saker LR, Wiersema PK, Purves JV, Tillard P, Arnold GM, Paams AJM, Waalburg W, Stulen I (1996) Nitrate and ammonium influxes in soybean (Glycine max) roots: direct comparison of 13N and 15N tracing. Plant Cell Environ 19: 859-868 [Google Scholar]
- Coruzzi G, Bush DR (2001) Nitrogen and carbon nutrient and metabolite signaling in plants. Plant Physiol 125: 61-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM, Glass ADM (1998) Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci 3: 389-395 [Google Scholar]
- Crawford NM, Smith M, Bellissimo D, Davis RW (1988) Sequence and nitrate regulation of the Arabidopsis thaliana mRNA encoding nitrate reductase, a metalloflavoprotein with three functional domains. Proc Natl Acad Sci USA 85: 5006-5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean JV, Harper JE (1988) The conversion of nitrite to nitrogen oxide(s) by the constitutive NAD(P) H-nitrate reductase enzyme from soybean. Plant Physiol 88: 389-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhon P, Gojon A, Tillard P, Passama L (1995) Diurnal regulation of NO3– uptake in soybean plants: I. Changes in NO3– influx, efflux, and N utilization in the plant during the day/night cycle. J Exp Bot 46: 1585-1594 [Google Scholar]
- Deng M, Moureaux T, Caboche M (1989) Tungstate, a molybdate analog inactivating nitrate reductase, deregulates the expression of the nitrate structural gene. Plant Physiol 91: 304-309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Griffiths R, Hancock J, Neill S (2002) A new role for an old enzyme: nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 16314-16318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filleur S, Daniel-Vedele F (1999) Expression analysis of a high-affinity nitrate transporter isolated from Arabidopsis thaliana by differential display. Planta 207: 461-469 [DOI] [PubMed] [Google Scholar]
- Filleur S, Dorbe MF, Cerezo M, Orsel M, Granier F, Gojon A, Daniel- Vedele F (2001) An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett 489: 220-224 [DOI] [PubMed] [Google Scholar]
- Forde BG (2000) Nitrate transporters in plants: structure, function and regulation. Biochim Biophys Acta 1465: 219-235 [DOI] [PubMed] [Google Scholar]
- Forde BG (2002) Local and long-range signaling pathways regulating plant responses to nitrate. Annu Rev Plant Physiol Plant Mol Biol 53: 203-224 [DOI] [PubMed] [Google Scholar]
- Frommer WB, Hummel S, Rentsch D (1994) Cloning of an Arabidopsis histidine transporting protein related to nitrate and peptide transporters. FEBS Lett 347: 185-189 [DOI] [PubMed] [Google Scholar]
- Gansel X, Munos S, Tillard P, Gojon A (2001) Differential regulation of the NO3– and NH4+ transporter genes AtNrt2.1 and AtAmt1.1 in Arabidopsis: relation with long-distance and local controls by N status of the plant. Plant J 26: 143-155 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Lejay L, Gojon A, Ninnemann O, Frommer WB, von Wirén N (1999) Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11: 937-947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass AD, Britto DT, Kaiser BN, Kinghorn JR, Kronzucker HJ, Kumar A, Okamoto M, Rawat S, Siddiqi MY, Unkles SE et al. (2002) The regulation of nitrate and ammonium transport systems in plants. J Exp Bot 53: 855-864 [DOI] [PubMed] [Google Scholar]
- Gojon A, Dapoigny L, Lejay L, Tillard P, Rufty TW (1998) Effects of genetic modification of nitrate reductase expression of 15NO3– uptake and reduction in Nicotiana plants. Plant Cell Environ 21: 43-53 [Google Scholar]
- Guo FQ, Wang R, Chen M, Crawford NM (2001) The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is activated and functions in nascent organ development during vegetative and reproductive growth. Plant Cell 13: 1761-1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FQ, Wang R, Crawford NM (2002) The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is regulated by auxin in both shoots and roots. J Exp Bot 53: 835-844 [DOI] [PubMed] [Google Scholar]
- Huang NC, Chiang CS, Crawford NM, Tsay YF (1996) CHL1 encodes a component of the low-affinity nitrate uptake system in Arabidopsis and shows cell type-specific expression in roots. Plant Cell 8: 2183-2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang NC, Liu KH, Lo HJ, Tsay YF (1999) Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11: 1381-1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufton CA, Besford RT, Wellburn AR (1996) Effect of NO pollution on growth, nitrate reductase activities and associated protein contents in glasshouse lettuce grown hydroponically in winter with CO2 enrichment. New Phytol 133: 495-501 [Google Scholar]
- King BJ, Siddiqi MY, Ruth TJ, Warner RL, Glass ADM (1993) Feedback regulation of nitrate influx in barley roots by nitrate, nitrite, and ammonium. Plant Physiol 102: 1279-1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Fraisier V, Scheible WR, Quesada A, Gojon A, Stitt M, Caboche M, Daniel-Vedele F (1998) Expression studies of Nrt2;1Np, a putative high-affinity nitrate transporter: evidence for its role in nitrate uptake. Plant J 14: 723-731 [Google Scholar]
- LaBrie ST, Wilkinson JQ, Tsay YF, Feldmann KA, Crawford NM (1992) Identification of two tungstate-sensitive molybdenum cofactor mutants, chl2 and chl7, of Arabidopsis thaliana. Mol Gen Genet 233: 169-176 [DOI] [PubMed] [Google Scholar]
- Lejay L, Tillard P, Lepetit M, Olive F, Filleur S, Daniel-Vedele F, Gojon A (1999) Molecular and functional regulation of two NO3– uptake systems by N- and C-status of Arabidopsis plants. Plant J 18: 509-519 [DOI] [PubMed] [Google Scholar]
- Lin Y, Cheng CL (1997) A chlorate-resistant mutant defective in the regulation of nitrate reductase gene expression in Arabidopsis defines a new HY locus. Plant Cell 9: 21-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Huang CY, Tsay YF (1999) CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11: 865-874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobreaux S, Massenet O, Briat JF (1992) Iron induces ferritin synthesis in maize plantlets. Plant Mol Biol 19: 563-575 [DOI] [PubMed] [Google Scholar]
- Loppes R, Radoux M, Ohresser MC, Matagne RF (1999) Transcriptional regulation of the NIA1 gene encoding nitrate reductase in Chlamydomonas reinhardtii: effects of various environmental factors on the expression of a reporter gene under the control of the Nia1 promoter. Plant Mol Biol 41: 701-711 [DOI] [PubMed] [Google Scholar]
- Murgia I, Delledonne M, Soave C (2002) Nitric oxide mediates ironinduced ferritin accumulation in Arabidopsis. Plant J 30: 521-528 [DOI] [PubMed] [Google Scholar]
- Ninnemann O, Jauniaux JC, Frommer WB (1994) Identification of a high affinity NH4+ transporter from plants. EMBO J 13: 3464-3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M, Krapp A, Daniel-Vedele F (2002) Analysis of the NRT2 nitrate transporter family in Arabidopsis: structure and gene expression. Plant Physiol 129: 886-896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, De Y, Yang WC, Kumaran M, Sundaresan V (1999) Analysis of flanking sequences from dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11: 2263-2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouteau S, Cherel I, Vaucheret H, Caboche M (1989) Nitrate reductase mRNA regulation in Nicotiana plumbaginifolia nitrate reductase-deficient mutants. Plant Cell 1: 1111-1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilleré I, Dufossé C, Roux Y, Foyer CH, Caboche M, Morot-Gaudry JF (1994) The effect of deregulation of NR gene expression on growth and nitrogen metabolism of Nicotiana plumbaginifolia plants. J Exp Bot 45: 1205-1211 [Google Scholar]
- Radin JW (1974) Distribution and development of nitrate reductase activity in germinating cotton seedlings. Plant Physiol 53: 458-463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin P (1979) Etude de quelques conditions d'extraction de la nitrate réductase des racines et des feuilles de plantules de maïs. Physiol Vég 17: 45-54 [Google Scholar]
- Rockel P, Strube F, Rockel A, Wildt J, Kaiser WM (2002) Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J Exp Bot 53: 103-110 [PubMed] [Google Scholar]
- Scheible WR, Gonzales-Fontes A, Lauerer M, Mm̈ller-Röber B, Caboche M, Stitt M (1997) Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9: 783-798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner HY, Song W, Zhang L, Naider F, Becker JM, Stacey G (1994) An Arabidopsis peptide transporter is a member of a new class of membrane transport proteins. Plant Cell 6: 1289-1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M (1999) Nitrate regulation of metabolism and growth. Curr Opin Plant Biol 2: 178-186 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Sasaki Y, Ida S, Morikawa H (2001) Nitrite reductase gene enrichment improves assimilation of NO2– in Arabidopsis. Plant Physiol 126: 731-741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Ida S, Irifune K, Oeda K, Morikawa H (1994) Nucleotide sequence of a gene for nitrite reductase from Arabidopsis thaliana. DNA Seq 5: 57-61 [DOI] [PubMed] [Google Scholar]
- Touraine B, Glass AD (1997) NO3– and ClO3– fluxes in the chl1-5 mutant of Arabidopsis thaliana: Does the CHL1-5 gene encode a low-affinity NO3– transporter? Plant Physiol 114: 137-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM (1993) The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705-713 [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Chabaud M, Kronenberger J, Caboche M (1990) Functional complementation of tobacco and Nicotiana plumbaginifolia nitrate reductase deficient mutants by transformation with the WT alleles of the tobacco structural genes. Mol Gen Genet 220: 468-474 [Google Scholar]
- Vaucheret H, Kronenberger J, Lepingle A, Vilaine F, Boutin JP, Caboche M (1992) Inhibition of tobacco nitrite reductase activity by expression of antisense RNA. Plant J 2: 559-569 [PubMed] [Google Scholar]
- Walker DJ, Leigh RA, Miller AJ (1996) Potassium homeostasis in vacuolate plant cells. Proc Natl Acad Sci USA 93: 10510-10514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Liu D, Crawford N (1998) The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc Natl Acad Sci USA 95: 15134-15139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendehenne D, Pugin A, Klessig DF, Durner J (2001) Nitric oxide: comparative synthesis and signaling in animal and plant cells. Trends Plant Sci 6: 177-183 [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Crawford NM (1991) Identification of the Arabidopsis CHL3 gene as the nitrate reductase structural gene NIA2. Plant Cell 3: 461-471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JQ, Crawford NM (1993) Identification and characterization of a chlorate-resistant mutant of Arabidopsis thaliana with mutations in both nitrate reductase structural genes NIA1 and NIA2. Mol Gen Genet 239: 289-297 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Sakihama Y (2000) Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: in vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett 468: 89-92 [DOI] [PubMed] [Google Scholar]
- Yu X, Sukumaran S, Marton L (1998) Differential expression of the Arabidopsis NIA1 and NIA2 genes: Cytokinin-induced nitrate reductase activity is correlated with increased NIA1 transcription and mRNA levels. Plant Physiol 116: 1091-1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Jennings A, Barlow PW, Forde BG (1999) Dual pathways for regulation of root branching by nitrate. Proc Natl Acad Sci USA 96: 6529-6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo D, Okamoto M, Vidmar JJ, Glass ADM (1999) Regulation of a putative high-affinity nitrate transporter (Nrt2;1At) in roots of Arabidopsis thaliana. Plant J 17: 563-568 [DOI] [PubMed] [Google Scholar]