Abstract
Ethanolic fermentation is classically associated with flooding tolerance when plant cells switch from respiration to anaerobic fermentation. However, recent studies have suggested that fermentation also has important functions in the presence of oxygen, mainly in germinating pollen and during abiotic stress. Pyruvate decarboxylase (PDC), which catalyzes the first step in this pathway, is thought to be the main regulatory enzyme. Here, we characterize the PDC gene family in Arabidopsis. PDC is encoded by four closely related genes. By using real-time quantitative polymerase chain reaction, we determined the expression levels of each individual gene in different tissues, under normal growth conditions, and when the plants were subjected to anoxia or other environmental stress conditions. We show that PDC1 is the only gene induced under oxygen limitation among the PDC1 gene family and that a pdc1 null mutant is comprised in anoxia tolerance but not other environmental stresses. We also characterize the expression of the aldehyde dehydrogenase (ALDH) gene family. None of the three genes is induced by anoxia but ALDH2B7 reacts strongly to ABA application and dehydration, suggesting that ALDH may play a role in aerobic detoxification of acetaldehyde. We discuss the possible role of ethanolic fermentation as a robust back-up energy production pathway under adverse conditions when mitochondrial function is disturbed.
The ethanolic fermentation pathway branches off the main glycolytic pathway at pyruvate. In the first step, pyruvate is the substrate of pyruvate decarboxylase (PDC), yielding CO2 and acetaldehyde. Subsequently, acetaldehyde is reduced to ethanol with the concomitant oxidation of NADH to NAD+ by alcohol dehydrogenase (ADH). In the present day aerobic atmosphere, ethanolic fermentation is used only by specialized organisms or under particular conditions. In plants, it has been studied because of its relevance in flooding tolerance where plant cells switch from aerobic respiration to anaerobic fermentation (for review, see Drew, 1997).
ADH has been the subject of numerous genetic studies, and adh mutants have been reported for a number of species, including maize (Zea mays; for review, see Freeling and Bennett, 1985), tobacco (Nicotiana tabacum; Rousselin et al., 1990), and Arabidopsis (Jacobs et al., 1988). Maize adh null mutants are sensitive to strict anoxia but no obvious phenotype is apparent in acclimated plants (Johnson et al., 1994). Experiments with isogenic maize lines differing in ADH activity over a approximately 200-fold range indicated that ADH activity does not limit the capacity for energy production by ethanolic fermentation unless there is a reduction in activity to less than 1% of wild-type levels (Roberts et al., 1989). These results suggest that ADH is present in large excess and are inconsistent with the idea of ADH as the regulatory enzyme of this pathway.
Overexpression of a bacterial PDC in transgenic tobacco resulted in constitutive high and active protein levels under both normoxic and anoxic conditions (Bucher et al., 1994). The acetaldehyde produced by the bacterial and the endogenous PDC, was converted efficiently to ethanol by the endogenous ADH. This suggests a key regulatory role for the first enzyme of ethanolic fermentation, PDC. Overexpression of Arabidopsis genes for the enzymes ADH and PDC improved the tolerance of Arabidopsis roots to low oxygen conditions (Dennis et al., 2000; Shiao et al., 2002). Loss-of-function mutations in PDC would provide a valuable tool to understand the importance of ethanolic fermentation in flooding tolerance, but such mutants have not yet been reported in plants.
A new and exciting aspect of ethanolic fermentation is the suggested involvement in stress signaling and response to environmental stresses other than low oxygen (Tadege et al., 1999). The micro-array expression profiles from stressed plants often reveal increases in ADH and/or PDC expression (Desikan et al., 2001; Seki et al., 2002). Furthermore, specific analysis of the ADH gene from rice (Oryza sativa), maize, and Arabidopsis showed ADH to be induced by cold (Christie et al., 1991), wounding (Kato-Noguchi, 2001), dehydration (Dolferus et al., 1994), and the phytohormone abscisic acid (ABA; Hwang and VanToai, 1991; de Bruxelles et al., 1996), in line with the observation from the micro-array experiments. Preliminary data on expression of the PDC genes from Arabidopsis during abiotic stresses have been reported (Dolferus et al., 1997). Thus it is conceivable that ethanolic fermentation is part of a general response to environmental stress.
Additional evidence indicating a role in biotic stress is provided by experiments with transgenic potato plants overexpressing bacterial PDC. These plants accumulated acetaldehyde and showed a lesion-mimic phenotype (Tadege et al., 1998). Furthermore, several markers normally associated with plant defense were expressed, and lesion formation was accompanied by a significant resistance to a fungal pathogen. The cell death was developmentally and environmentally regulated and the drastic effect on sugar export suggested a link between carbohydrate metabolism and disease susceptibility.
Although PDC and ADH gene induction has been demonstrated, ethanol and acetaldehyde production as a result of stress treatment has only been reported for red pine (Pinus resinosa) and birch (Betula spp.) seedlings exposed to sulfur dioxide, water deficiency, freezing, and ozone (Kimmerer and Kozlowski, 1982). A comprehensive investigation of PDC and ADH gene induction and the determination of acetaldehyde and ethanol production during stress treatments would provide valuable information on how ethanolic fermentation is involved in the response to abiotic or biotic stress.
One problem during aerobic stress is toxicity of reactive acetaldehyde. One of the pathways for the detoxification of aldehydes to less reactive forms is the oxidation to carboxylic acids by aldehyde dehydrogenase (ALDH). ALDH converts acetaldehyde to acetate concomitantly reducing NAD+ to NADH. In plants, ALDH is strongly expressed in germinating pollen of tobacco (op den Camp and Kuhlemeier, 1997), and it is essential for the restoration of fertility in male sterile maize plants (Cui et al., 1996; Liu et al., 2001). During anoxia, however, results from tobacco leaves indicated that ALDH is not induced (op den Camp and Kuhlemeier, 1997). In contrast to these results, ALDH2B5 is anaerobically induced in rice seedlings (Nakazono et al., 2000). Enhanced activity of ALDH would deplete the NAD+ pool in the cytosol and consequently block glycolysis. Therefore, speculations about the possible function of ALDH during anoxia need to take into account the requirement for NAD+ regeneration during glycolysis. We therefore investigated whether Arabidopsis ALDH is induced during anoxia as compared with other environmental stresses.
In this paper, we present results demonstrating the requirement for ethanolic fermentation during anoxia and its involvement during adaptation to other environmental stresses in Arabidopsis. Expression levels of the relevant mRNA from PDC, ADH, and ALDH were determined quantitatively. The identification of a pdc1 mutant by T-DNA insertional mutagenesis allowed us to study the function for PDC1 in Arabidopsis.
RESULTS
Characterization of the PDC Gene Family in Arabidopsis
Screening several gene and protein databases for Arabidopsis Columbia (Col-0) sequences with the protein sequence of tobacco PDC2 (Bucher et al., 1995) revealed four open reading frames with high homology to tobacco PDC2. Two open reading frames had very high homology with the GenBank entries PDC1 (U71121) and PDC2 (U71122) cloned from the accessions C24 and Landsberg erecta (Ler), respectively. The corresponding open reading frame in the accession Col-0 were annotated in the Arabidopsis Genome sequence as PDC1, PDC2, PDC3, and PDC4 (Table I). Arabidopsis PDC1 is located on chromosome 4, PDC2 is located on chromosome 5, and PDC3 and PDC4 genes are positioned at the end of chromosome 5 and are separated by only 1,600 bp. An alignment of the four Arabidopsis PDC and the tobacco PDC2 protein sequences revealed a high overall consensus in all the functional motifs including the catalytic site (Fig. 1). Two anaerobic response elements can be found in the promoter of PDC1 (Hoeren et al., 1998). No G-Box (ABA-responsive element) could be found in the promoter of PDC1 in contrast to the Arabidopsis ADH gene (Dolferus et al., 1994). No known stress-related elements were identified in the promoter regions of PDC2, PDC3 and PDC4.
Table I.
Comparison of amino acid identity for PDC proteins from tobacco and Arabidopsis
Nos. indicate the percentage of amino acid identity done by a global alignment with a Blossum 35 matrix. Gene accession nos.: NtPDC2 (X81855), AtPDC1 (AT4G33070), AtPDC2 (AT5G54960), AtPDC3 (AT5G01330), and AtPDC4 (AT5G01320).
| NtPDC2 | AtPDC1 | AtPDC2 | AtPDC3 | |
|---|---|---|---|---|
| NtPDC2 | - | |||
| AtPDC1 | 80.4 | - | ||
| AtPDC2 | 78.9 | 81.5 | - | |
| AtPDC3 | 80.6 | 85.2 | 77.8 | - |
| AtPDC4 | 80.6 | 88.6 | 80.6 | 89.8 |
Figure 1.
Partial comparison of the amino acid sequences of PDC proteins from Arabidopsis and orthologs in other species. Partial amino acid sequence alignments of PDC proteins from Zymomonas mobilis (ZymPDC; GenBank no. M15368), tobacco (NtPDC2; GenBank no. X81855), and Arabidopsis (AtPDC1, AT4G33070; AtPDC2, AT5G54960; AtPDC3, AT5G01330; and AtPDC4, AT5G01320). Active site is highlighted with asterisk. Alignments were generated using the Multalin software v5.4.1 with the Blossum 62-12-2 matrix (Corpet, 1988). Black boxes indicate high consensus levels (100%); gray boxes indicate low consensus levels (> 50%).
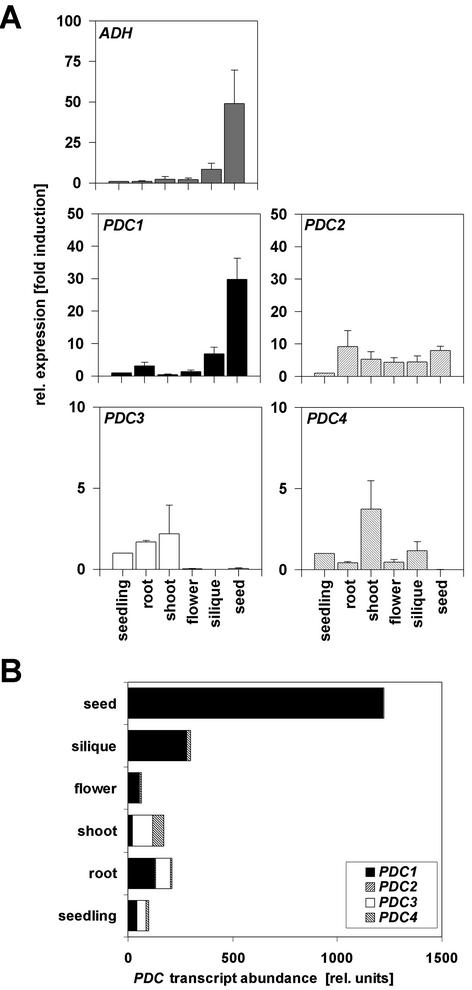
Expression of PDC in Various Organs Determined by Real-Time Quantitative Reverse Transcriptase (RT)-PCR
Preliminary experiments indicated that under standard growth conditions, each of the four PDC genes was expressed at low levels, at or below the limit of detection by RNA gel-blot analysis (data not shown). To analyze the mRNA accumulation of the four different genes coding for PDC, we used real-time quantitative RT-PCR technology (see “Materials and Methods” and supplementary material, which can be viewed at http://www.plantphysiol.org). RNA was isolated from various organs, and the abundance of the four PDC mRNA and the single-ADH mRNA in these tissues was quantified based on separate RNA isolations derived from three independent experiments (Fig. 2). ADH was expressed in all organs. The highest expression was observed in imbibed seeds (50-fold higher compared with 14-d-old seedlings). The expression profile of PDC1 closely matched ADH expression. Expression of PDC2, PDC3, and PDC4 could also be seen in most organs but, in contrast to ADH and PDC1, expression was not particularly high in imbibed seeds (Fig. 2A). The real-time RT-PCR method can also be used to compare absolute transcript abundances of genes in mRNA preparations (Fig. 2B). PDC1 transcripts were the dominant PDC mRNAs in roots, flowers, siliques, and seeds. In siliques and seeds, more than 96% of all PDC transcripts belonged to the PDC1 class. PDC3 was abundant in whole seedlings, roots, and shoots, whereas PDC2 and PDC4 transcripts contributed overall only minor proportions to the total PDC mRNA population.
Figure 2.
Relative expression level and transcript abundance of fermentation genes in aerobic conditions during development. A, Quantification of mRNA levels was achieved by using a quantitative real-time PCR system (see “Materials and Methods”). ADH and PDC expression levels were normalized with respect to the internal control ACT2 and are plotted relative to the expression from whole seedlings. Data bars represent the mean ± se level of transcripts from three experiments with independent RNA extractions. Note different scales as y axis. B, PDC transcript abundance in different organs. Total PDC mRNA in seedlings = 100. Contribution from individual genes according to A is represented.
PDC1 Is Strongly Induced during Anoxia
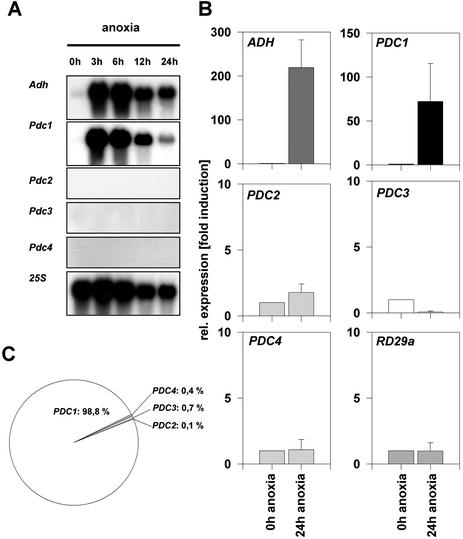
In maize and Arabidopsis, strong induction of fermentation genes takes place in anaerobic conditions (Dolferus et al., 1985; Bailey-Serres et al., 1988; Conley et al., 1999). RNA gel blotting was performed with specific probes from the 3′-untranslated regions of the four PDC genes of Arabidopsis. A strong hybridization signal from ADH and PDC1 appeared after 3 h of anoxia and declined slowly during prolonged anoxia (Fig. 3A). Expression of PDC2, PDC3, and PDC4 was not detected, consistent with the weak expression previously observed for these genes (Fig. 2). These results suggest that PDC1 is the only gene induced in anaerobiosis among the PDC gene family in Arabidopsis.
Figure 3.
Relative expression level and transcript abundance of fermentation genes during anoxia. A, RNA gel-blot analysis from seedlings treated with different periods of anoxia. Hybridization probes specific to each of the PDC genes were used. 25S hybridization was used as control. B, Quantification of ADH and PDC transcripts from seedlings submitted to 24-h anoxia using the a quantitative real-time PCR system. Expression of RD29a was used as a control. Expression levels were normalized with respect to the internal control ACT2 and are plotted relative to the expression at 0-h anoxia. Data bars represent the mean ± se level of transcripts from three experiments with independent RNA extractions. Note different scales as y axis. C, PDC transcript abundance in anoxia-treated seedlings. Contribution from individual genes according to A is represented.
To settle the question of whether PDC2, PDC3, and PDC4 are induced during anoxia to levels too low to be detected by classical RNA gel-blot analysis, quantitative RT-PCR technology was used. As a positive internal control we amplified the ADH RNA, which is known to be strongly induced by anoxia (Dolferus et al., 1985). As expected, ADH showed a approximately 200-fold induction of transcript amounts after a 24-h anoxia treatment of seedlings compared with the nontreated control (Fig. 3B). Expression analysis of the well-characterized stress-gene RD29a (Gilmour and Thomashow, 1991; Nordin et al., 1991; Yamaguchi-Shinozaki and Shinozaki, 1994) under an anoxic atmosphere showed this gene not to be induced. As already seen in the RNA gel-blot analysis, PDC1 was strongly (approximately 70-fold) induced by anoxia. PDC2, PDC3, and PDC4 were not induced by anoxia, but low levels of transcripts were detected. The relative transcript abundance is very low for PDC2 (approximately 1,600 times less), PDC3 (approximately 130 times less), and PDC4 (approximately 250 times less) compared with the transcript abundance of PDC1 during anoxia. The proportion of PDC1 transcripts in anoxia-treated cells is about 99%. The mRNA abundance of PDC2, PDC3, and PDC4 represent under anoxia only about 1% of all PDC transcripts (Fig. 3C).
Identification and Molecular Analysis of the pdc1 Mutant
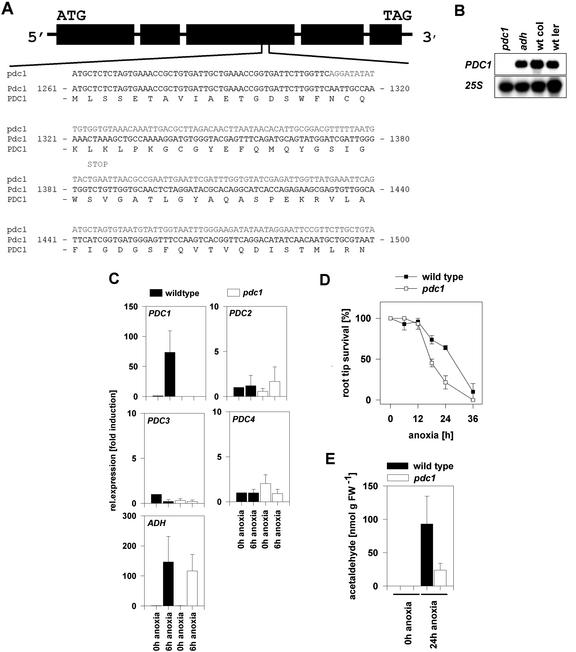
The expression analysis revealed PDC1 to be the only strongly expressed gene of the PDC gene family during anoxia. PDC2, PDC3, and PDC4 are dramatically less abundant than PDC1 transcripts and we therefore focused further on the PDC1 gene. To study the role of PDC1 in Arabidopsis, the T-DNA insertion collection of the Torrey Mesa Research Institute was screened. A multiple sequence alignment with the PDC1 sequence against the database of the T-DNA-derived flanking sequences was performed, which resulted in an alignment representing a high probability of identity with a particular line in the collection. Using a primer from the T-DNA left border plus a gene-specific primer, a single PCR product was identified in this line, and subsequent sequencing of the fragment revealed an insertion in the third exon of PDC1, at position 1,312 bp after the ATG in the coding sequence (Fig. 4A). This mutant line was designated pdc1.
Figure 4.
Interference of a T-DNA insertion in PDC1 with gene expression and survival under anoxia. A, Structure of the Arabidopsis PDC1 gene. DNA sequences of the wild type and the mutant line showing the T-DNA-derived sequences in bold and the wild-type amino acid sequence. A potential stop codon in the reading frame of the T-DNA insertion in PDC1 is indicated. B, Expression level in pdc1 and adh seedlings treated for 6 h under anoxia. Total RNA (10 μg) from wild-type seedlings of the accessions Col-0 and Ler and from pdc1 (Col-0) and adh (Ler) were subjected to RNA gel-blot analysis with a specific probe from the 3′-untranslated region of PDC1. 25S labeling indicated equal RNA loading. C, Quantification of mRNA levels of PDC and ADH genes in wt and pdc1 mutant seedlings under normoxia (0-h anoxia) or after 6-h anoxia. Expression levels were normalized with respect to the internal control ACT2 and are plotted relative to the expression at 0-h anoxia. Data bars represent the mean ± se level of transcripts from three experiments with independent RNA extractions. D, Survival of pdc1 and wt seedlings under anoxia. The survival of the existing root tip was scored from seedlings subjected to different periods of anoxia and a 3-d-long recovering time. Black squares represent wild-type seedlings; white squares represent mutant seedlings. Data represent the average of three survival experiments with independent plant material ± se. E, Production of acetaldehyde in roots submitted to 24-h anoxia measured by gas chromatography. Detached roots were infiltrated with 50 mm Glc and 0.1 mm CaSO4 and were incubated for 24 h in an anoxic bench or in normal atmosphere. Values represent the average of between three and five measurements of different populations of seedlings ± se.
At position 1,385 bp in the inserted T-DNA sequence of pdc1, a potential stop codon was identified. Furthermore, the thiamine pyrophosphate-binding site (an essential cofactor of the enzyme) is located 85 bp after the insertion site in the wild-type coding sequence. This analysis indicated a complete disruption of the PDC1 gene. To confirm this, RNA was isolated from Col-0 and Ler wild type, and pdc1 (Col-0) and the adh mutant (Ler) were subjected to a 6-h anoxic period (Fig. 4B) and analyzed by RNA gel-blot analysis. PDC1 gene expression was observed in the two different wild-type accessions and in the adh mutant, but pdc1 mutant plants showed no transcripts at all after 6 h of anaerobic conditions.
To investigate whether other members of the PDC gene family might compensate for PDC1, real-time RT-PCR analysis was performed. PDC1 transcripts in pdc1 mutants were not detectable either in normoxic or in anoxic conditions, confirming the data from the RNA gel-blot analysis (Fig. 4C). None of the other three genes showed increased transcript levels in pdc1 mutant seedlings treated by anoxia compared with the wild type. Thus, the three PDC genes do not compensate for a complete loss of PDC1 transcripts in pdc1 mutant plants by induced expression. ADH expression is not influenced in the pdc1 mutant. Conversely, PDC1 expression is not influenced in the adh mutant (Fig. 4B).
The pdc1 Mutant Is More Susceptible to Anoxia
Development of pdc1 mutant plants and survival during anoxia were investigated. Under standard growing conditions, no morphological phenotype was apparent (data not shown). To reveal the possible consequences of a null mutation in PDC1, attention was focused on the survival of plants under anoxia. Studies on the differential adaptation of shoots and roots of Arabidopsis to low oxygen indicated that ethanol fermentation is essential in roots but not in shoots (Ellis et al., 1999). We therefore analyzed the ability of the root tip of pdc1 mutants to survive prolonged anoxia (Fig. 4D). After 12 h of anoxia, viability declined in wild-type and pdc1 root tips. This susceptibility to anoxia was more pronounced in the mutant line, which revealed a significant decrease (P < 0.025) of the survival rate of pdc1 root tips after 18 h relative to wild-type plants. After 24 h, the survival ability of the wild type was more obvious; 64% of wild-type plants still had healthy root tips, whereas pdc1 root tips survived significantly less well (P < 0.05). After 36 h of complete anoxia, pdc1 roots were all dead, whereas some wild-type roots still survived. To analyze the consequences of the lack of PDC1, we determined the product of the PDC enzyme, acetaldehyde, in mutant roots during anoxia. Mutant pdc1 roots produced considerably less acetaldehyde than the wild type (26%; Fig. 4E), but the decrease was less than the reduction in total PDC transcript levels (1% of wild-type mRNA level). No production of acetaldehyde was observed in aerobic conditions. Taken together, the lack of PDC1 results in a 99% reduction in PDC mRNA, in a 74% reduction in acetaldehyde concentration, and in a pronounced susceptibility to anoxia in root tips.
Induction of Ethanolic Fermentation by Environmental Stresses Other Than Anoxia
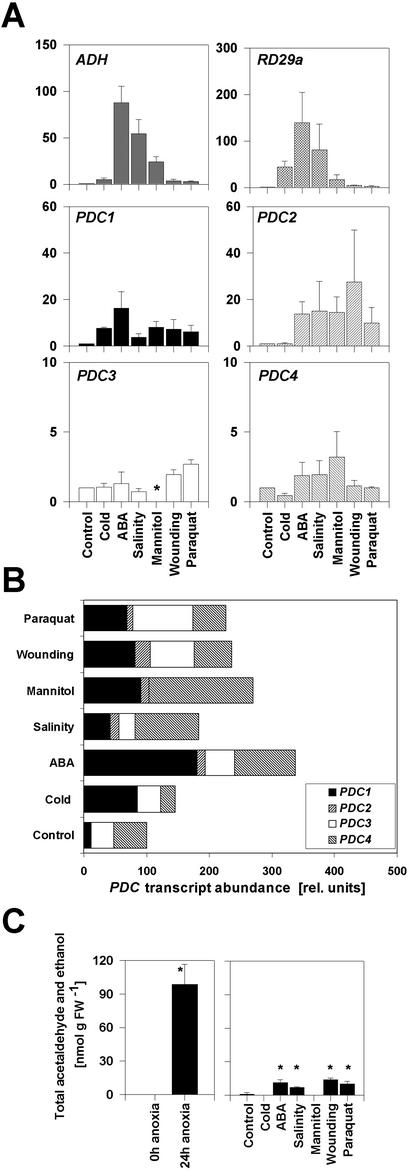
The induction of expression of the ADH gene in several species by environmental conditions such as cold, osmotic stress, or wounding suggested a role for PDC in these conditions. Therefore, we analyzed the behavior of PDC transcripts of Arabidopsis in seedlings subjected to a range of abiotic stresses. Seedlings were treated with cold, mannitol, ABA, salinity, wounding, and paraquat (a herbicide that exacerbates O2- radical production and is used to study oxidative stress; Penninckx et al., 1998), and gene expression was quantified. The Arabidopsis ADH gene is known to be induced by cold, mannitol, and ABA treatment (Dolferus et al., 1994; de Bruxelles et al., 1996). Our analysis confirmed these earlier results and demonstrated ADH induction in response to salt stress, wounding, and paraquat treatments (Fig. 5A). The transcript pattern obtained for the known stress-inducible gene RD29a was in agreement with the literature (Gilmour and Thomashow, 1991; Nordin et al., 1991; Yamaguchi-Shinozaki and Shinozaki, 1994). RD29a was strongly induced by ABA, salinity, cold, and dehydration, whereas wounding and paraquat treatments induced the RD29a gene only weakly.
Figure 5.
Relative expression levels of fermentation genes under different abiotic stresses and total acetaldehyde and ethanol production by leaves submitted to anoxia or other stress treatments. A, Quantification of mRNA of ADH, PDC, and RD29a transcripts with the a quantitative real-time PCR system from seedlings treated with different abiotic stresses. Expression levels were normalized with respect to the internal control ACT2 and are plotted relative to the control treatment. Data bars represent the mean ± se level of transcripts from three experiments with independent RNA extractions. Asterisk indicates no detection of amplification. Note different scales as y axis. B, PDC transcript abundance in different treatments. Total PDC mRNA in control-treated seedlings = 100. Contribution from individual genes according to A is represented. C, Leaves were subjected to the indicated treatments. Measurements were made by gas chromatography. Values represent the mean ±se of between three and five measurements of different leaves. Asterisks indicate significant differences relative to the control treatment, P < 0.005).
Next, we analyzed the stress-induced expression levels for all four PDC genes of Arabidopsis. PDC1 was induced by all treatments; the strongest induction was observed by ABA application (23-fold compared with the control treatment). Cold, salinity, mannitol, wounding, and paraquat induced PDC1 to comparable levels (8- to 10-fold). PDC2 was induced by several stresses compared with the control treatment (Fig. 5A). PDC3 and PDC4 mRNA levels were not markedly affected by any treatment. However, the absolute transcript abundance of PDC2 in the different stress-treated plants was always lower compared with PDC1 (Fig. 5B). PDC3 and PDC4 transcripts are in several treatments more abundant than PDC1, although these genes are not markedly induced by these conditions compared with the control treatment (Fig. 5, A and B).
Because both PDC and ADH genes are inducible under stress conditions, we decided to measure the products of the pathway, acetaldehyde and ethanol, by gas chromatography. We found acetaldehyde production to be stimulated by ABA and paraquat treatment and during salt and wounding stress, but not during cold and mannitol treatments (Fig. 5C). In all cases, the amount of acetaldehyde and ethanol produced was much less than under anoxia.
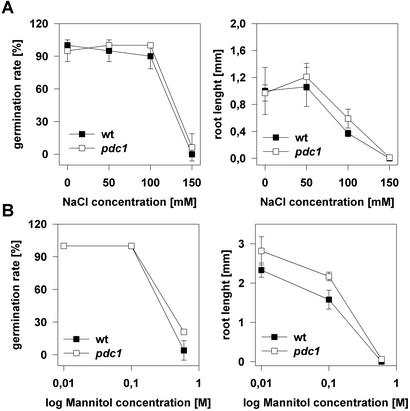
The expression data show that PDC1 is strongly inducible under abiotic stress conditions, that PDC2 contributed only marginally to total PDC levels, and that PDC3 and PDC4 show no induction. Detection of acetaldehyde and ethanol during stress treatments demonstrated that ethanolic fermentation was occurring during these conditions. To see whether the loss of PDC1 results in an enhanced susceptibility of the pdc1 mutant to environmental stress conditions, we determined the germination rate and the root elongation on agar plates containing different concentrations of sodium chloride and mannitol, which represent a salt stress and a dehydration stress, respectively. In both assays, no considerable differences were observed between wild-type and mutant plants in the germinating rate, and only slight effects were observed on root elongation (Fig. 6), indicating that germination under osmotic stress conditions is presumably independent of PDC1.
Figure 6.
Survival rate of germinating pdc1 and wild-type seeds on plates containing different concentrations of sodium chloride or mannitol. Surface-sterilized pdc1 and wild-type seeds were placed on 0.85% (w/v) agar plates containing the indicated concentrations of either NaCl or mannitol. After an incubation at 4°C for 3 d to break seed dormancy, plates were transferred to the growth room under long-day conditions (23°C ± 2°C). Percentage of germination was scored after 1 week, and root length of germinated seeds was measured. Data represent the mean ± se of four replicate experiments. A, Germination on NaCl-containing plates. B, Germination on mannitol-containing plates.
ALDH Transcript Levels Do Not Increase during Anoxia But during Environmental Stress
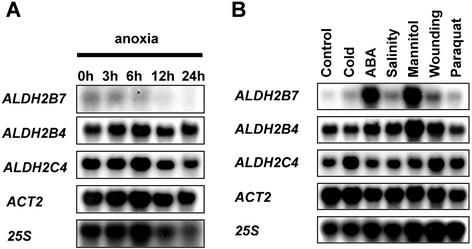
Acetaldehyde is a potentially toxic molecule that can be detoxified through the action of ALDH. We therefore determined the transcript profiles of each of the three Arabidopsis ALDH genes (Skibbe et al., 2002). RNA gel-blot analysis of anoxia-treated seedlings was carried out for each of the three genes ALDH2B4, ALDH2B7, and ALDH2C4 using gene-specific probes. The results showed no measurable increases of Arabidopsis ALDH transcript levels during anoxia (Fig. 7A).
Figure 7.
Relative expression level of ALDH genes during anoxia and during other stresses. A, RNA gel-blot analysis from seedlings subjected to different periods of anoxia. Hybridization probes specific to the ALDH genes were used. ACT2 and 25S hybridization were used as controls. Accession numbers: ALDH2B7 (AT1G23800), ALDH2B4 (AT3G48000), and ALDH2C4 (GenBank no. AF349448). B, RNA gel-blot analysis of ALDH transcripts from seedlings treated with different abiotic stresses. ACT2 and 25S hybridization were used as controls.
To reveal whether ALDH genes are induced by environmental stresses other than anoxia, we performed RNA gel-blot analysis with RNAs derived from seedlings treated with different stresses. ABA and mannitol treatments strongly induced the transcription of the putative mitochondrial ALDH2B7 gene (Fig. 7B). Wounding and salinity induced the gene to a lesser extent, whereas cold and paraquat had no effect on the expression of ALDH2B7. These results indicate that there may be a function for ALDH2B7 during an abiotic stress response. The expression pattern of the putative cytosolic gene ALDH2C4 and the putative mitochondrial gene ALDH2B4 was only marginally influenced by stress treatments. The housekeeping gene ACTIN2 is barely regulated by any of the stresses, validating its use as the reference gene for the quantitative real-time PCR experiments.
DISCUSSION
Dominance of PDC1 Transcript Abundance during Anoxia
The four PDC genes annotated in the Arabidopsis Genome Sequence Bank are highly homologous to each other and to other plant PDC genes (Fig. 1; Table I). All functional motifs are conserved, and thus they are likely to encode proteins with bona fide PDC enzymatic activity. This is supported by the reduced accumulation of the product of the enzyme, acetaldehyde, in the pdc1 null mutant (Fig. 4E). No conserved stress-related cis-elements were present in any of the other PDC genes.
The transcript abundance of all PDC genes under normal conditions was at the detection limit of classical RNA gel-blot analysis preventing accurate quantitation. Quantitative real-time RT-PCR enabled us to determine transcript levels of each of the PDC genes independently. The careful analysis of the mRNA populations with three independent experiments and RNA extraction for the real-time RT-PCR analysis made it possible to obtain reliable expression profiles. We could demonstrate that among the four PDC genes only PDC1 showed appreciable variation in expression between different tissues and high levels of induction by anoxia. Previous sequence inspection reported two anaerobic response elements in the 5′ upstream region of PDC1 from Arabidopsis (Hoeren et al., 1998). In siliques and seeds, PDC1 transcripts alone make up more than 96% of the total PDC mRNA. Imbibed seeds and developing or ripening fruits are compact tissues, and it is conceivable that they experience hypoxia. Thus, the high PDC1 expression in anoxic seedlings, seeds, and fruits may reflect activation of a common signaling pathway. However, the regulation of the PDC genes during development might depend on a separate signaling pathway than the one involved in response to anoxia, as demonstrated for the regulation of ADH expression by ABA and during anoxia (Dolferus et al., 1994; de Bruxelles et al., 1996).
Under normal conditions PDC1 transcripts make up about 10% to 40% of the total PDC mRNA in seedlings (Figs. 2B and 5B). Under oxygen limitation, PDC1 transcripts are strongly induced during anoxia and represent in these cells about 99% of the total PDC mRNA (Fig. 3C). This dominant status of one PDC gene in Arabidopsis is different from tobacco. In tobacco, at least two copies exist for PDC (Bucher et al., 1995), one of which is induced anaerobically in vegetative tissues, whereas the other is constitutively expressed in reproductive organs. This is an additional difference with Arabidopsis, where PDC1, PDC2, and PDC4 are expressed in flowers (Fig. 2A).
Mutation of PDC1 Results in Susceptibility to Anoxia
Root tips of pdc1 mutant seedlings are less tolerant to oxygen deprivation than the wild type (Fig. 4D), indicating that the reduced PDC levels compromise survival. It should be noted that for up to 12 h, there is no significant difference, but prolonged anoxia results in the accelerated death of the root tip in mutant plants. The real-time PCR analysis showed that under anoxia, the total PDC mRNA concentration in pdc1 mutants was reduced to approximately 1% of wild-type levels. No compensation of the loss of pdc1 expression through induction of the other genes was observed (Fig. 4C). The absence of the PDC1 transcript in the pdc1 mutant had no effect on the level of expression of ADH, and respectively, the absence of ADH transcripts in the adh null-mutant did not influence the expression of PDC1 (Fig. 4, B and C). This indicates, that the regulation of the expression of these two genes is independent of the presence of the other. The reduction of PDC mRNA levels is accompanied by a reduced production of acetaldehyde and ethanol. However, there is a quantitative difference between the reduction in mRNA and enzyme product levels, with acetaldehyde accumulating to 26% of wild-type levels (Fig. 4E). We envisage three possibilities for the comparatively high acetaldehyde production in pdc1 knock-out roots. First, acetaldehyde production may be the result of the activity from the enzymes encoded by PDC2, PDC3, and PDC4. The latter mRNAs may be more efficiently translated, or the proteins be more stable or have a higher specific activity. Second, in the anoxically induced wild type, PDC enzymes may be in large excess, and the flux through the pathway may be limited by other factors than enzyme abundance. Third, acetaldehyde in pdc1 roots may be the product of a yet unknown PDC-independent pathway. In any case, knock-out of the major gene, PDC1, compromises survival under oxygen deprivation.
PDC1 Is Induced by Stress
Increase of PDC expression has been reported by cold and mannitol treatments in Arabidopsis (Dolferus et al., 1997; Conley et al., 1999) and by cold treatments in rice and maize (Christie et al., 1991). Our results also indicate a strong induction of the PDC1 gene and an increased production of fermentation metabolites by a broad range of stresses other than anoxia (Fig. 5C). The increase of expression, however, was lower than during anoxia. In addition, we were able to detect the production of fermentation metabolites in stress-treated seedlings, although to a lesser extend than during anoxia. The measured production of fermentation metabolites might be an underestimation, because acetaldehyde and ethanol might have been remetabolized.
Taken together, these results suggest the involvement of ethanolic fermentation in response to abiotic stress, as indicated both at the transcriptional level and by the accumulation of ethanolic fermentation products. Abiotic stress can disrupt respiratory activities and it has been proposed that compensation for impaired mitochondrial function will occur by shifting the metabolism form aerobic to anaerobic fermentation (Levitt, 1980).
However, germination assays with the pdc1 knock-out plants on osmotic pressure producing medium, indicated, that PDC1 seems not to be essential for survival in these conditions (Fig. 6). Thus, ethanolic fermentation might be a part of abiotic stress adaptation but how much it contributes to stress-tolerance has to be investigated.
Arabidopsis ALDHs: A Function in Detoxification of Products Derived from Reactive Oxygen Species?
None of the three ALDH genes from Arabidopsis were induced during anoxia (Fig. 7A). This is in agreement with the situation in tobacco leaves (op den Camp and Kuhlemeier, 1997) and in contrast to the results of Nakazono et al. (2000), who showed increased expression of ALDH2B5 in submerged rice seedlings, although there was incongruence between the pattern of ALDH2B5 protein and ALDH2B5 mRNA data. Activity of ALDH during anoxia would deplete the essential NAD+ pool in any compartment and thus adversely affect anoxia tolerance. Any model postulating the operation of ALDH during anoxia would have to explain how NAD+ is regenerated under these conditions.
ALDH2B7 expression is strongly induced by cold and mannitol treatments (Fig. 7B). This increase in transcripts suggests a specific function or regulation in stress response. Op den Camp and Kuhlemeier (1997) in tobacco and Skibbe et al. (2002) in Arabidopsis showed that ALDHs convert acetaldehyde to acetate in vitro. In addition, Liu and Schnable (2002) demonstrated through an in-depth analysis a functional specialization of the two maize mitochondrial ALDHs. Similarly, the putative mitochondrial located ALDH2B7 of Arabidopsis might be involved in an ABA-mediated response to dehydration. Removing toxic aldehydes may prevent membrane damage in the mitochondria. This pathway might operate in addition to the cytosolic activity of ADH in detoxification of acetaldehyde. A similar detoxification pathway was also put forward by Møller (2001) to explain how ALDH could prevent pollen abortion in T-cytoplasm maize plants. Interestingly, another Arabidopsis ALDH gene, which clusters to the class 3 ALDHs (oxidizing aromatic aldehydes and fatty aldehydes) is also induced in response to dehydration and ABA treatment (Kirch et al., 2001). ALDH activity during stress might occur independent of ethanol fermentation. We envisage therefore a possible role of ethanolic fermentation as a robust energy production pathway in the cytosol in conditions where the mitochondrial ATP machinery is damaged.
MATERIALS AND METHODS
Plant Growth Conditions
Seeds of Arabidopsis, accessions Col-0 or Ler were surface sterilized and plated on 0.5× Murashige and Skoog medium containing 10% (w/v) Suc, vitamins (1 × 10-4% myo-inositol, 1 × 10-6% nicotinic acid, 1 × 10-6% pyrodoxin-HCl, 1 × 10-5% thiamine-HCl, and 2 × 10-6 % Gly) and 0.85% (w/v) agar. Plates were incubated at 4°C for 3 d to break seed dormancy and then transferred to the growth room (23°C ±2°C, 16-h-light/8-h-dark cycles) in vertical position for 2 to 3 weeks as described by Chung and Ferl (1999).
Identification of pdc1
The pdc1 mutant was identified in the T-DNA population (accession Col-0) originating at the Torrey Mesa Research Institute (San Diego) by a random sequencing based screen of the T-DNA left border (line no. 688.D02). Genetic analysis by PCR using primers from the left border (5′-TAG CAT CTG AAT TTC ATA ACC AAT CTC GAT ACA C-3′) and a PDC1-specific primer (5′-ACA TTC AGA AGA TGC TCT CTA GTG AAA C-3′) identified homozygous mutant lines and enabled the isolation of a fragment covering the insertion site for sequencing. Homozygous pdc1 mutant plants were further examined. The adh mutant in the background Ler was obtained directly from the Nottingham Arabidopsis Stock Center online catalog (35 N8095; http://nasc.nott.ac.uk/home.html).
Stress Treatments
For anoxic treatments, 2- to 3-week-old plantlets on plates were placed in an anoxia work bench (Forma Scientific, Mariatta, OH) in the dark. Cold and mannitol treatments were performed as previously described by Dolferus et al. (1994) with the following modifications: Before the beginning of the treatments, seedlings were transferred to petri dishes containing 12 mL of liquid 0.5× Murashige and Skoog solution for 24 h (16-h-light/8-h-dark cycle) on a shaker at 70 rpm. For the cold treatment, the plates were then transferred to 11°C in the dark for 24 h on a shaker. For the salt stress, the seedlings were shaken in liquid 0.5× Murashige and Skoog solution containing 300 mm NaCl for 4 h in the light. For the wounding treatments, leaves were wounded three times each with forceps, kept in light and harvested after 4 h. For paraquat (methyl viologen, Sigma-Aldrich, St. Louis) treatment, a 25 μm solution was sprayed and the plant kept in light, and seedlings were harvested after 4 h. For ABA treatment ([+]-cis,trans-ABA, Sigma-Aldrich), the plants were incubated with 10 μm ABA as described by de Bruxelles et al. (1996) in the light.
Acetaldehyde Measurements in the Gas Phase
Roots of hydroponically grown plants or leaves from plants grown on soil were prepared as follows. Material was washed three times with distilled water. Roots or leaves were then infiltrated with a 50 mm Glc and 0.1 mm CaSO4 solution three times for 2.5 min under vacuum. Infiltrated samples were briefly blotted on tissue paper, placed in 10-mL gas-tight glass-bottles, and sealed after transfer either to the anaerobic working bench and kept in the dark according to Bucher and Kuhlemeier (1993) or to aerobic atmosphere, also kept in the dark. After 24 h, 2.5-mL samples of head spaces were taken with a gas-tight syringe (Hamilton, Bonaduz, Switzerland) and immediately injected and analyzed by gas chromatography as described by Bucher et al. (1994) with the following modifications: column temperature, 180°C; injector temperature, 190°C; and detector temperature, 220°C.
Survival Assay for Anoxia, Dehydration, and Salt Stress
Survival assay for anoxia was done according to Ellis et al. (1999) with the following modifications: Before anoxic conditions, seedlings were transferred for 24 h to plates containing a liquid Murashige and Skoog solution. The medium was removed and substituted with fresh medium, which was gassed for 1 h with N2. After the incubation in the anoxic bench, seedlings were placed on recovery plates, and the positions of the root tip were scored on the back of the plates. The recovery phase lasted for 3 d where the plates were incubated vertically under diffuse light conditions. Root tip survival was scored as the ability of the main root tip to extend beyond the mark scored after the treatment.
Survival assay for dehydration and salt stress was performed on Murashige and Skoog plates containing the indicated concentrations of either mannitol or NaCl. Seeds of wild type and pdc1 were surface sterilized and placed on the indicated plates. These were then incubated at 4°C for 2 d and then transferred to the growth room (23°C ±2°C, 16-h-light/8-h-dark cycles) in vertical position for 1 week. Germination rate and root length were scored after this period.
RNA Extraction and Northern-Blot Analysis
Total RNA was isolated from seedlings of control or stress treatments or mutants and transferred to nylon membranes as described by Caderas et al. (2000). Hybridizations were carried out under standard conditions with randomly labeled probes (Sambrook et al., 1989). ALDH probes were generated from a cDNA by PCR from using the following primers: ALDH2C4, 5′-GTT ACC GGA GAT CAA ATT CAC CAA G and 5′-CAG TCA AGA TCA AAC CCG AAG TAG C; ALDH2B4, 5′-TCA GCT TCC TCT CCC TTA CTG TTT CG and 5′-ACT TCG TCT CGT TCG CCC TCT TTA TC; and ALDH2B7, 5′-AAG ATA CAG TAA CCT CGC TGC TGC TG and 5′-CCA ATG CCA CTC ATC TTA ACC CTC C. No cross-hybridization could be detected among the probes (data not shown). To determine expression of the housekeeping gene ACTIN2 (EMBL no. ATU41998), blots were hybridized with an ACT2 probe generated by RT-PCR (forward primer, 5′-ATT CAG ATG CCC AGA AGT CTT GTT-3′; reverse primer, 5′-GAA ACA TTT TCT GTG AAC GAT TCC T-3′
Reverse Transcription and PCR Optimization
Total RNA (2 μg per reaction) was DNase I treated. First-strand synthesis of cDNA was performed by using oligo(dT) primer and avian myeloblastosis virus RT. The following primers were used for RT-PCR experiments: ADH forward primer, 5′-AGT TGT GGT TTG TCT ACT GGG TTA G-3′, and reverse primer, 5′-AGA GTC CTC TCA TTC AAG AAA TTC A-3′; PDC1 forward primer, 5′-CTC GTT GAC GCC ATT CAT AAC-3′, and reverse primer, 5′-CCA TGA TAA AGC GTA CAT GGA A-3′; PDC2 forward primer, 5′-TTT GGT AGT GTC TTC ACC GTT C-3′, and reverse primer, 5′-TTC TTG GGA TGG GAT CTC AAC-3′; PDC3 and PDC4 forward primer, 5′-CTG GTC TTG TCG ATG CTA TTC A-3′, and reverse primer, 5′-AAA CTT TGT CAA CAA GGG GTT C-3′; PDC4 reverse primer, 5′-CAC CAT CAA TGG TAA TGG TAC A-3′; RD29a forward primer, 5′-GTG GAG AAG ATC TCT ACC GAG AAG G-3′, and reverse primer, 5′-CAT CAA AGA CGT CAA ACA AAA CAC A-3′; and ACT2 forward primer, 5′-ATT CAG ATG CCC AGA AGT CTT GTT-3′, and reverse primer, 5′-GAA ACA TTT TCT GTG AAC GAT TCC T-3′. Primers were optimized for amplification in a gradient cycler with various annealing temperature from 47.5°C to 63.1°C.
Quantitative Real-Time PCR
For quantitative real-time PCR experiments, the LightCycler system (Roche Diagnostics, Mannheim, Germany) was used. For PCR-reactions, a mastermix of the following reaction components was prepared (the end-concentration is indicated in parentheses): 12 μL of water, 2 μL of MgCl2 (2.5 mm), 1 μL of forward primer (0.5 μm), 1 μL of reverse primer (0.5 μm), and 2 μL of LightCycler (Fast Start DNA Master SYBR Green I, Roche Diagnostics). LightCycler mastermix was filled in the LightCycler glass capillaries, and 2 μL of cDNA was added as PCR template. Capillaries were closed, centrifuged, and placed into the LightCycler rotor. The following LightCycler experimental run protocol was used: denaturation program (95°C for 10 min), amplification and quantification program repeated 45 to 55 times (95°C for 15 s, annealing temperature: 63°C for 15 s for all primer combinations except for PDC3 amplification:58°C, 72°C for 20 s with a single fluorescence measurement), melting curve program (65°C–95°C with heating rate of 0.1°C s-1 and a continuous fluorescence measurement), and finally a cooling step at 40°C.
For relative quantification, PCR efficiencies for each gene were determined as follows: Standard curves for each gene was performed using the cDNA with the highest abundance of the gene to cover the range of all template concentrations. For PDC2, an external standard curve was performed on a purified PDC2-PCR fragment. Real-time PCR efficiencies (E) were calculated from the given slopes in the LightCycler software of the standard curves according to the equation: E = 10 [-1/slope]. PCR efficiencies for the used target and reference genes: ADH, 1.95; ACT2, 1.99; RD29a, 1.90; PDC1, 1.88; PDC2, 1.70; PDC3, 1.57; and PDC4, 1.92 (see also supplementary materials; they can be viewed at www.plantphysiol.org). Crossing points, defined as the point at which the fluorescence rises above the background fluorescence was determined using the “Fit Point Method” in the LightCycler software 3.5.3 (Roche Diagnostics). cDNA abundance of the PDC genes was calculated by crossing point differences between amplification of the different genes after a baseline adjustment. Gene-specific PCR efficiency was used to calculate the induction level. A mathematical model, which was shown recently to hold in experimental environments, was used to quantify the expression of target genes relative to the expression of a reference gene (Pfaffl, 2001). The relative expression ratio of the target genes were calculated based on their efficiencies (E) and the crossing points (CP) deviation of an unknown sample versus a control and expressed in comparison to the reference gene ACT2 as shown in equation 1.
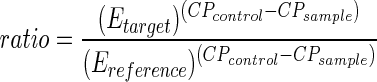 |
1 |
where relative expression [fold induction] means x-fold induction of the expression of gene A relative to the standard gene B compared by a control experiment.
Supplementary Material
Acknowledgments
We thank A. Sessions and P. Ho (Torrey Mesa Research Institute, San Diego) for providing the seeds of the pdc1 mutant. We also thank the Nottingham Arabidopsis Stock Center (Nottingham, UK) for providing the seeds of the adh mutant. We furthermore thank R. Brändle and S. Zeeman for critical reading of the manuscript and our colleagues in the laboratory for stimulating discussions and general support.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.016907.
The online version of this article contains Web-only data. The supplemental material is available at http://www.plantphysiol.org.
References
- Bailey-Serres J, Kloeckener-Gruissem B, Freeling M (1988) Genetic and molecular approaches to the study of the anaerobic response and tissue specific gene expression in maize. Plant Cell Environ 11: 351-357 [Google Scholar]
- Bucher M, Kuhlemeier C (1993) Long-term anoxia tolerance. Plant Physiol 103: 441-448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher M, Brändle R, Kuhlemeier C (1994) Ethanolic fermentation in transgenic tobacco expressing Zymomonas mobilis pyruvate decarboxylase. EMBO J 13: 2755-2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher M, Brander KA, Sbicego S, Mandel T, Kuhlemeier C (1995) Aerobic fermentation in tobacco pollen. Plant Mol Biol 28: 739-750 [DOI] [PubMed] [Google Scholar]
- Caderas D, Muster M, Vogler H, Mandel T, Rose JKC, McQueen-Mason S, Kuhlemeier C (2000) Limited correlation between expansin gene expression and elongation growth rate. Plant Physiol 123: 1399-1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Hahn M, Walbot V (1991) Low-temperature of alcohol dehydrogenase-1 mRNA and protein activity in maize and rice seedlings. Plant Physiol 95: 699-706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HJ, Ferl RJ (1999) Arabidopsis alcohol dehydrogenase expression in both shoots and roots is conditioned by root growth environment. Plant Physiol 121: 429-436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley TR, Peng HP, Shih MC (1999) Mutations affecting induction of glycolytic and fermentative genes during germination and environmental stresses in Arabidopsis. Plant Physiol 119: 599-607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 22: 10881-10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Wise RP, Schnable PS (1996) The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science 272: 1334-1336 [DOI] [PubMed] [Google Scholar]
- de Bruxelles GL, Peacock WJ, Dennis ES, Dolferus R (1996) Abscisic acid induces the alcohol dehydrogenase gene in Arabidopsis. Plant Physiol 111: 381-391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis ES, Dolferus R, Ellis M, Rahman M, Wu Y, Hoeren FU, Grover A, Ismond KP, Good AG, Peacock WJ (2000) Molecular strategies for improving waterlogging tolerance in plants. J Exp Bot 51: 89-97 [PubMed] [Google Scholar]
- Desikan R, Machkerness SAH, Hancock JT, Neill SJ (2001) Regulation of the Arabidopsis transcriptsome by oxidative stress. Plant Physiol 127: 159-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolferus R, Ellis M, de Bruxelles G, Trevaskis B, Hoeren F, Dennis ES, Peacock WJ (1997) Strategies of gene action in Arabidopsis during anoxia. Ann Bot 79: 21-31 [Google Scholar]
- Dolferus R, Jacobs M, Peacock WJ, Dennis ES (1994) Differential interactions of promoter elements in stress responses of the Arabidopsis Adh Gene. Plant Physiol 105: 1075-1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolferus R, van den Bossche D, Jacobs M (1985) Alcohol dehydrogenase in Arabidopsis: analysis of the induction phenomenon in plants and tissue cultures. Mol Gen Genet 199: 256-264 [Google Scholar]
- Drew MC (1997) Oxygen deficiency and root metabolism: injury and acclimation under hypoxia and anoxia. Annu Rev Plant Physiol Plant Mol Biol 48: 223-250 [DOI] [PubMed] [Google Scholar]
- Ellis MH, Dennis ES, Peacock WJ (1999) Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiol 119: 57-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Bennett DC (1985) Maize Adh1. Annu Rev Genet 19: 297-323 [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Thomashow MF (1991) Cold acclimation and cold-regulated gene expression in ABA mutants of Arabidopsis thaliana. Plant Mol Biol 45: 113-141 [DOI] [PubMed] [Google Scholar]
- Hoeren FU, Dolferus R, Wu Y, Peacock WJ, Dennis ES (1998) Evidence for a role of AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics 149: 479-490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SY, VanToai TT (1991) Abscisic acid induces anaerobiosis tolerance in corn. Plant Physiol 97: 593-597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M, Dolferus R, Van den Bosche D (1988) Isolation and biochemical analysis of ethyl methanesulfonate-induced alcohol dehydrogenase null mutants of Arabidopsis thaliana (L.) Heynh. Biochem Genet 26: 105-122 [DOI] [PubMed] [Google Scholar]
- Johnson JR, Cobb JB, Drew MC (1994) Hypoxic induction of anoxia tolerance in roots of Adh1 null Zea mays L. Plant Physiol 105: 61-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato-Noguchi H (2001) Wounding stress induces alcohol dehydrogenase in maize and lettuce seedlings. Plant Growth Regul 35: 285-288 [Google Scholar]
- Kimmerer TW, Kozlowski TT (1982) Ethylene, ethane, acetaldehyde and ethanol production under stress. Plant Physiol 69: 840-847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch HH, Nair A, Bartels D (2001) Novel ABA- and dehydration–inducible aldehyde dehydrogenase genes isolated from the resurrection plant Craterostigma plantgineum and Arabidopsis thaliana. Plant J 28: 555-567 [DOI] [PubMed] [Google Scholar]
- Levitt J (1980) Responses of plants to environmental stresses: chilling, freezing, and high temperature stresses. In TT Kozlowski, eds, Physiological Ecology: A Series of Monographs, Texts and Treaties, Ed 2, Vol 1. Academic Press, New York, pp 23-64 [Google Scholar]
- Liu F, Cui X, Horner HT, Weiner H, Schnable PS (2001) Mitochondrial aldheyde dehydrogenase activity is required for male fertility in maize. Plant Cell 13: 1063-1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Schnable PS (2002) Functional specialization of maize mitochondrial aldehyde dehydrogenases. Plant Physiol 130: 1657-1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller IM (2001) A more general mechanism of cytoplasmic male fertility? Trends Plant Sci 6: 560. [DOI] [PubMed] [Google Scholar]
- Nakazono M, Tsuji H, Li Y, Saisho D, Arimura S, Tsutsumi N, Hirai A (2000) Expression of a gene encoding mitochondrial aldheyde dehydrogenase in rice increases under submerged conditions. Plant Physiol 124: 587-598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordin K, Heino P, Palva ET (1991) Separate signal pathways regulate the expression of a low-temperature-induced gene in Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 115: 875-879 [DOI] [PubMed] [Google Scholar]
- op den Camp R, Kuhlemeier C (1997) Aldehyde dehydrogenase in tobacco pollen. Plant Mol Biol 35: 355-365 [DOI] [PubMed] [Google Scholar]
- Penninckx IA, Thomma BP, Buchala A, Metraux JP, Broekaert WF (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10: 2103-2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: 2003-2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JKM, Chang K, Webster C, Callis J, Walbot V (1989) Dependence of ethanolic fermentation, cytoplasmic pH regulation, and viability on the activity of alcohol dehydrogemase in hypoxic maize root tips. Plant Physiol 89: 1275-1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousselin P, Lepingle A, Faure JD, Bitoun R, Caboche M (1990) Ethanol-resistant mutants of Nicotiana plumbaginifolia are deficient in the expression of pollen and seed alcohol dehydrogenase activity. Mol Gen Genet 222: 409-415 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T et al. (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279-292 [DOI] [PubMed] [Google Scholar]
- Shiao TL, Ellis MH, Dolferus R, Dennis ES, Doran PM (2002) Overexpression of alcohol dehydrogenase or pyruvate decarboxylase improves growth of hairy roots at reduced oxygen concentrations. Biotechnol Bioeng 77: 455-461 [DOI] [PubMed] [Google Scholar]
- Skibbe DS, Liu F, Wen TJ, Yandeau MD, Cui X, Cao J, Simmons CR, Schnable PS (2002) Characterisation of the aldehyde dehydrogenase gene family of Zea mays and Arabidopsis. Plant Mol Biol 48: 751-764 [DOI] [PubMed] [Google Scholar]
- Tadege M, Bucher M, Stähli W, Suter M, Dupuis I, Kuhlemeier C (1998) Activation of plant defense responses and sugar efflux by expression of pyruvate decarboxylase in potato leaves. Plant J 16: 661-671 [Google Scholar]
- Tadege M, Dupuis I, Kuhlemeier C (1999) Ethanolic fermentation: new functions for an old pathway. Trends Plant Sci 4: 320-325 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.