Abstract
Pathogen challenge can trigger an integrated set of signal transduction pathways, which ultimately leads to a state of “high alert,” otherwise known as systemic or induced resistance in tissue remote to the initial infection. Although large-scale gene expression during systemic acquired resistance, which is induced by salicylic acid or necrotizing pathogens has been previously reported using a bacterial pathogen, the nature of systemic defense responses triggered by an incompatible necrotrophic fungal pathogen is not known. We examined transcriptional changes that occur during systemic defense responses in Arabidopsis plants inoculated with the incompatible fungal pathogen Alternaria brassicicola. Substantial changes (2.00-fold and statistically significant) were demonstrated in distal tissue of inoculated plants for 35 genes (25 up-regulated and 10 down-regulated), and expression of a selected subset of systemically expressed genes was confirmed using real-time quantitative polymerase chain reaction. Genes with altered expression in distal tissue included those with putative functions in cellular housekeeping, indicating that plants modify these vital processes to facilitate a coordinated response to pathogen attack. Transcriptional up-regulation of genes encoding enzymes functioning in the β-oxidation pathway of fatty acids was particularly interesting. Transcriptional up-regulation was also observed for genes involved in cell wall synthesis and modification and genes putatively involved in signal transduction. The results of this study, therefore, confirm the notion that distal tissue of a pathogen-challenged plant has a heightened preparedness for subsequent pathogen attacks.
Plants have evolved a number of defense strategies to protect themselves from pathogen invasion. The hypersensitive response at the site of attempted infection is one of the most common features of disease resistance response in incompatible plant-microbe interactions. In many instances, the onset of this initial response also activates a signaling process that makes the plant not only locally, but also systemically, more refractory to subsequent infections by a broad spectrum of pathogens. This response is known as systemic acquired resistance (SAR) and is accompanied with activation of many plant genes (Ryals et al., 1996). The defense signal, salicylic acid (SA) is one key modulator of SAR. Although the specific role of SA as a systemic signal is still not clear, its accumulation in local and distal (uninoculated) tissues is accompanied with coordinated expression of a specific subset of defense genes (Malamy et al., 1990; Métraux et al., 1990; Maleck et al., 2000). Pathogen challenge also leads to the synthesis of other signaling molecules such as jasmonates and ethylene, which along with SA regulate distinct but overlapping patterns of defense gene expression (Reymond and Farmer, 1998; Feys and Parker, 2000; Schenk et al., 2000; Devadas et al., 2002). Recent evidence also suggests that there is an extensive overlap between the gene expression triggered by pathogen inoculation, defense-signaling compounds, and other physiological processes such as senescence (Quirino et al., 1999; Morris et al., 2000; He et al., 2002).
It has been hypothesized that systemic resistance may result from induced tissues being “primed” to provide a rapid and strong defense response following subsequent pathogen challenge (Conrath et al., 2002). A part of this process appears to be the development of a “primed state” (Conrath et al., 2002) or “maintenance period” at later stages of induction of SAR (e.g. at 48 h after infection with Pseudomonas syringae; Maleck et al., 2000). Previously, it has been shown that inoculation of Arabidopsis with incompatible strains of the fungus Alternaria brassicicola results in a strong systemic response leading to the sustained induction of genes such as PR1 and PDF1.2 (Penninckx et al., 1996), which are respective markers for the salicylate and jasmonate defense-signaling pathways (Ryals et al., 1996; Penninckx et al., 1996; Manners et al., 1998). Interestingly, although inoculation of Arabidopsis with A. brassicicola induces SA marker gene PR1 (Penninckx et al., 1996), a growing body of evidence indicates that basal resistance to this pathogen requires a functional jasmonate/ethylene pathway. For example, coi1 and ein2 mutants defective in the jasmonate/ethylene signaling exhibit higher susceptibility to A. brassicicola infection (Thomma et al., 1998, 1999), whereas the wild-type resistance against this fungus was restored by methyl jasmonate (MJ) treatment in the pad3 mutant defective for camalexin biosynthesis (Thomma et al., 1998). In addition, compatible isolates of A. brassicicola are not sensitive to SAR induced by isonicotinic acid, a functional SA-analog (Ton et al., 2002). All of these make this particular Arabidopsis-pathogen interaction very interesting to study in regard to systemic events resulting either independently or in conjunction with the better-studied SAR response. However, so far, very little is known about genes that may be induced in distal noninoculated leaves of Arabidopsis after inoculation with A. brassicicola.
We previously reported gene induction in Arabidopsis leaves locally challenged by an incompatible isolate of A. brassicicola and after treatment with defense regulators (Schenk et al., 2000). In this paper, using microarray analysis, we specifically examined the transcriptional changes that occur in uninoculated tissue after local inoculation of Arabidopsis leaves with A. brassicicola. In addition to known defense genes, we identified a number of genes and presumed biochemical functions that have not been previously associated with systemic or local defense responses in plants. We expect that further characterization of these genes and functions will extend our understanding on how plants defend themselves from pathogen attack and help to design improved crop protection strategies.
RESULTS AND DISCUSSION
Time Course of PDF1.2 Expression in Distal and Local Tissue
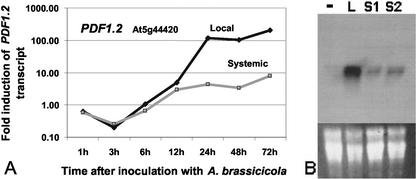
The aim of this study was to gain insights into transcriptional changes that occur in distal tissue during defense responses triggered by an incompatible fungal pathogen. Because this type of systemic response can potentially last several days, we first set out to identify a representative time point where full spectrum of systemic gene expression could be studied (“systemic maintenance period”). We hypothesized that the expression from known defense genes should be relatively high during this period to provide the plant a long-lasting and a broad spectrum of resistance. A well-established marker gene for the defense response in Arabidopsis is the PDF1.2 gene that encodes an antimicrobial peptide. A time-course study after inoculation with A. brassicicola was performed for this gene using real-time quantitative reverse transcriptase-PCR (RT-qPCR) and showed that induction occurred as early as 6 to 12 h after inoculation (Fig. 1A). Interestingly, no difference in the timing of the first induction of PDF1.2 was observed between local and distal tissue, suggesting that the signal for systemic activation of this gene travels quite rapidly. However, accumulation of the PDF1.2 transcript in time points after 12 h was much higher in local tissue (118-fold induction at 24 h) than that in distal tissue (4.3-fold induction at 24 h). The highest induction ratios of PDF1.2 compared with uninoculated control plants were found at 72 h for both local (202-fold induction) and distal (7.8-fold induction) tissue. Northern-blot analysis of the PDF1.2 gene confirmed significant expression at 72 h in locally inoculated and in distal leaf tissue of two independent inoculation experiments (Fig. 1B). Thus, a time point corresponding to 72 h after fungal inoculation was chosen to study the systemic responses during the maintenance period. It is important to note that genes that show specific induction during the early stages of systemic defense responses in this interaction may not be represented in our analysis.
Figure 1.
Expression profile of the Arabidopsis PDF1.2 gene after inoculation with Alternaria brassicicola in locally inoculated (Loc) or remaining distal (Sys) leaf tissue. A, The time-course study using RT-qPCR shows induction ratios at various time points after inoculation. A northern-blot analysis was carried out to confirm RT-qPCR results at 72 h after inoculation (lower part shows applied RNA amounts) from local (L) or distal tissue (two experimental replicates, S1 and S2, are shown), or from leaves from uninoculated control plants (–).
cDNA Microarray Analysis of Systemic Gene Expression after Inoculation by A. brassicicola
Three independent inoculation experiments were performed, and total RNA was isolated from locally inoculated and uninoculated (distal) leaves as well as from uninoculated control plants, converted to cDNA, labeled with separate fluorescence dyes, and hybridized onto cDNA microarrays in binary comparisons of samples to equivalent controls in each replicate experiment. The cDNA microarray we used was previously described and contains 2,375 Arabidopsis expressed sequence tags (ESTs) with a bias toward putative defense-related and regulatory genes (Schenk et al., 2000).
A total of 100 genes that showed differential expression with a mean change in expression greater than 2-fold in distal tissue as compared with equivalent tissue from uninoculated control plants were identified after analysis of microarray data, and also passed all other selection criteria for spot quantifications (see “Materials and Methods”). These included 83 induced and 17 repressed genes (Table I; supplementary table, which can be viewed at www.plantphysiol.org). Statistical analyses of microarray data across the three independent experiments showed that 35 of these genes had t test probability values higher than 95% (P < 0.05), suggesting that the expression values observed for these particular genes in different biological replicates were the most robust. Of these genes, 25 were up-regulated and 10 were down-regulated in the distal leaves (Table I). The PR1 and the PDF1.2 marker genes were among these 35 significantly altered genes and showed 2.55- and 2.86-fold increases in expression, respectively, with high significance values (P < 0.01). The induction ratios and other data for the remaining identified genes that showed probability values below 95% (P > 0.05) but otherwise met all other microarray data analysis criteria are shown in the supplementary table.
Table I.
Expression profiles of Arabidopsis genes significantly induced or repressed in distal leaf tissue at 72 h (systemic maintenance period) after inoculation with A. brassicicola
| Putative Function
|
Accession cDNA Microarray Systemic Response
|
Quantitative RT-PCR
|
Additional Independent Replicates
|
Previously Reported Responses
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ch1
|
SD1
|
Ch2
|
SD2
|
NNDiff
|
Ratio
|
P
|
Local
|
Systemic
|
Local
|
Systemic
|
||||||||||||||
| Ratio | se | Ratio | se | 48 h | se | 72 h | se | 48 h | se | 72 h | se | |||||||||||||
| Antimicrobial | ||||||||||||||||||||||||
| PDF1.2 (At5g44420; see Fig. 1) | T04323 | 0.265 | 0.04 | 0.757 | 0.02 | -0.96 | 2.86 | <0.01 | 109.19 | 45.20 | 8.09 | 1.79 | 143.1 | 39.16 | 128.8 | 73.47 | 3.46 | 0.07 | 5.88 | 1.90 | Loc | SA | MJ | Eth |
| PR-1 (At2g14610) | M90508 | 0.063 | 0.00 | 0.161 | 0.01 | -0.87 | 2.55 | <0.01 | Loc | SA | ||||||||||||||
| Putative endochitinase (At2g43610) | R89996 | 4.977 | 0.69 | 1.890 | 0.91 | 0.90 | -2.63 | 0.03 | SA | MJ | ||||||||||||||
| Disease resistance | ||||||||||||||||||||||||
| Leu-rich repeat protein (At5g21090) | Z34187 | 0.966 | 0.10 | 2.309 | 0.05 | -0.82 | 2.39 | <0.01 | 1.62 | 0.24 | 1.37 | 0.42 | Loc | |||||||||||
| Similar to downy mildew resistance protein (At1g72930) | N65692 | 0.688 | 0.19 | 2.005 | 0.15 | -0.98 | 2.91 | 0.01 | 1.44 | 0.66 | 2.93 | 1.01 | -1.47 | 0.42 | -1.02 | 0.20 | -1.36 | 0.06 | 1.74 | 0.58 | ||||
| Putative disease resistance protein (At1g33590) see Fig. 2 | N65549 | 0.054 | 0.04 | 0.216 | 0.07 | -1.20 | 4.00 | 0.01 | 1.71 | 0.21 | 1.93 | 0.37 | 1.95 | 0.40 | 2.13 | 0.66 | 1.56 | 0.18 | 1.16 | 0.15 | ||||
| Putative disease resistance protein (At1g33600) | 1.23 | 0.22 | 4.35 | 2.44 | ||||||||||||||||||||
| Disease resistance protein (At4g33300) | T45626 | 0.059 | 0.05 | 0.196 | 0.10 | -1.08 | 3.34 | 0.05 | MJ | |||||||||||||||
| Regulatory | ||||||||||||||||||||||||
| ARR1-like putative protein (At3g46640) | Z34539 | 0.094 | 0.02 | 0.256 | 0.02 | -0.93 | 2.72 | 0.01 | 1.48 | 0.38 | 3.30 | 1.26 | 1.50 | 0.29 | 1.62 | 0.22 | 1.21 | 0.05 | 2.14 | 0.31 | SA | MJ | ||
| Basic helix/loop/helix 6 protein (At1g32640) | N96133 | 0.176 | 0.12 | 1.678 | 1.20 | -1.62 | 9.52 | 0.05 | 1.50 | 0.88 | 7.46 | 3.06 | 4.22 | 0.44 | 1.77 | 0.18 | 4.44 | 0.38 | 1.17 | 0.03 | Loc | MJ | ||
| MAP kinase 3* (ATMPK3) (At3g45640; see Fig. 2) | H36168 | 0.723 | 0.27 | 2.653 | 1.67 | -1.14 | 3.67 | 0.06 | 1.21 | 0.23 | 3.91 | 1.06 | 2.50 | 0.34 | 1.32 | 0.41 | 1.84 | 0.10 | -1.12 | 0.18 | Loc | SA | MJ | |
| Heat shock transcription factor (At5g45710) | H37587 | 4.707 | 0.84 | 1.534 | 0.46 | 1.02 | -3.07 | 0.02 | Loc | SA | MJ | |||||||||||||
| Receptor-like protein kinase (At5g49660) | T44735 | 8.998 | 1.09 | 3.351 | 1.45 | 0.91 | -2.69 | 0.02 | Loc | SA | MJ | |||||||||||||
| Receptor kinase (At5g58300) | T44469 | 3.752 | 0.78 | 1.460 | 0.21 | 0.88 | -2.57 | 0.03 | SA | MJ | ||||||||||||||
| Zinc finger protein (At1g66140) | T22622 | 3.666 | 1.00 | 1.269 | 0.13 | 1.00 | -3.00 | 0.03 | -1.23 | 0.05 | 1.07 | 0.42 | -2.76 | 0.30 | -1.70 | 0.36 | -2.51 | 0.26 | -2.19 | 0.69 | SA | MJ | ||
| Cell wall modifications | ||||||||||||||||||||||||
| Polygalacturonase inhibitor AtPGIP2 (At5g06870) | Z33873 | 0.178 | 0.02 | 0.620 | 0.10 | -1.11 | 3.49 | 0.01 | 2.07 | 0.43 | 1.91 | 0.28 | Loc | MJ | ||||||||||
| Polygalacturonase inhibitor AtPGIP1 (At5g06860) | 8.31 | 2.10 | 1.54 | 0.21 | ||||||||||||||||||||
| Putative pectin methylesterase (At1g11580) | T88068 | 0.062 | 0.03 | 0.172 | 0.05 | -0.94 | 2.78 | 0.02 | Loc | SA | MJ | Eth | ||||||||||||
| Putative cellulose synthase AtCSLB3 (At2g32530) | T26B15_9 | 0.079 | 0.02 | 0.216 | 0.09 | -0.94 | 2.76 | 0.03 | MJ | |||||||||||||||
| Putative pectate lyase* (At1g04680; see Fig. 2) | AA006024 | 0.216 | 0.12 | 0.557 | 0.41 | -0.88 | 2.58 | 0.12 | 1.03 | 0.15 | -1.28 | 0.09 | 1.87 | 0.70 | 2.34 | 0.16 | 2.29 | 0.23 | 2.27 | 0.27 | Loc | SA | MJ | |
| Arabinogalactan-like protein (At2g20520) | T88396 | 0.869 | 0.12 | 0.315 | 0.14 | 0.94 | -2.76 | 0.03 | SA | MJ | ||||||||||||||
| Fatty acid metabolism (β-oxidation; see Fig. 3) | ||||||||||||||||||||||||
| Acyl-CoA oxidase-like protein (At4g16760) | N38332 | 0.377 | 0.13 | 1.015 | 0.34 | -0.92 | 2.69 | 0.02 | 1.74 | 0.40 | 1.24 | 0.03 | 3.04 | 1.05 | 1.46 | 0.21 | 1.87 | 0.43 | -1.14 | 0.03 | Loc | |||
| Putative 3-ketoacyl-CoA thiolase protein (At2g33150) | T12940 | 0.035 | 0.01 | 0.101 | 0.02 | -0.98 | 2.92 | 0.03 | 1.71 | 0.31 | 1.31 | 0.25 | 2.16 | 0.44 | 2.29 | 0.68 | 1.05 | 0.19 | 1.44 | 0.52 | SA | |||
| Catalase 3 (At1g20620) | H76812 | 0.746 | 0.44 | 1.500 | 0.30 | -0.67 | 2.01 | 0.04 | 2.24 | 0.17 | 1.66 | 0.28 | 2.72 | 0.42 | 2.47 | 0.14 | 1.80 | 0.07 | 1.25 | 0.14 | Loc | MJ | ||
| Enoyl-CoA hydratase* (Multifunctional protein; At5g43280) | T43247 | 0.13 | 0.02 | 0.222 | 0.03 | -0.38 | 1.69 | 0.01 | 1.17 | 0.12 | 1.31 | 0.14 | 1.68 | 0.54 | 1.03 | 0.03 | 1.55 | 0.13 | 1.04 | 0.03 | SA | Eth | ||
| Acyl-CoA synthetase* (At4g23850) | N38362 | 0.286 | 0.07 | 0.875 | 0.55 | -1.02 | 3.06 | 0.07 | 1.31 | 0.05 | 1.54 | 0.11 | 1.74 | 0.05 | 1.32 | 0.31 | 1.38 | 0.05 | 1.22 | 0.36 | Loc | SA | MJ | |
| Secondary metabolism | ||||||||||||||||||||||||
| Cytochrome P450 monooxygenase CYP83B1 (At4g31500) | T75944 | 0.317 | 0.02 | 1.186 | 0.07 | -1.16 | 3.75 | <0.01 | 2.45 | 0.01 | 2.25 | 0.32 | 2.56 | 1.07 | 2.21 | 0.53 | 1.44 | 0.35 | 1.19 | 0.12 | Loc | MJ | ||
| Peroxidase 6 (At1g71695) | T04797 | 0.243 | 0.03 | 0.552 | 0.05 | -0.78 | 2.28 | <0.01 | Loc | MJ | ||||||||||||||
| Putative cytochrome P450 CYP71B7 (At1g13110) | T04814 | 0.434 | 0.07 | 0.924 | 0.20 | -0.72 | 2.13 | 0.01 | Loc | MJ | ||||||||||||||
| Desacetoxyvindoline 4-hydroxylase (At1g06620) | Z34689 | 0.087 | 0.04 | 0.241 | 0.10 | -0.94 | 2.77 | 0.03 | 1.22 | 0.08 | -1.16 | 0.06 | 2.71 | 0.68 | 2.09 | 0.34 | 2.88 | 0.35 | 1.36 | 0.43 | MJ | |||
| Cytochrome C1 precursor MHK7 (At5g40810) | Z25972 | 0.065 | 0.03 | 0.188 | 0.03 | -0.96 | 2.92 | 0.03 | Loc | SA | ||||||||||||||
| Cytochrome P450 monooxygenase CYP71B2 (At1g13080) | T43466 | 0.828 | 0.13 | 0.330 | 0.08 | 0.86 | -2.51 | 0.02 | MJ | |||||||||||||||
| Cell maintenance/development/other | ||||||||||||||||||||||||
| Triose phosphate translocator (At5g46110) | N96984 | 0.082 | 0.00 | 0.227 | 0.00 | -0.94 | 2.78 | <0.01 | ||||||||||||||||
| Malic enzyme-like protein (At5g11670) | H76074 | 0.643 | 0.07 | 1.807 | 0.28 | -0.95 | 2.81 | 0.01 | MJ | |||||||||||||||
| Putative OsNAC6 protein (At1g01720; see Fig. 2) | R84102 | 0.146 | 0.02 | 0.403 | 0.15 | -0.93 | 2.75 | 0.02 | 1.45 | 0.25 | 3.72 | 1.19 | 7.62 | 1.38 | 3.78 | 0.60 | 5.14 | 1.36 | 1.74 | 0.33 | ||||
| Putative cold acclimation protein (dehydrin) (At1g20450) | H37695 | 0.500 | 0.19 | 2.454 | 0.59 | -1.32 | 4.91 | 0.02 | 3.19 | 0.01 | 5.16 | 1.98 | 1.68 | 0.29 | 1.61 | 0.32 | 1.63 | 0.12 | 1.39 | 0.09 | ||||
| Similar to cold-regulated protein cor47 (At1g20440) | 5.48 | 1.58 | 6.85 | 1.89 | 1.34 | 0.57 | 1.62 | 0.23 | -1.04 | 0.23 | 2.12 | 0.58 | ||||||||||||
| Unknown protein (At4g38820) | T44509 | 0.075 | 0.02 | 0.152 | 0.02 | -0.68 | 2.04 | 0.03 | SA | |||||||||||||||
| Unknown protein (At5g19250) | N37872 | 0.763 | 0.27 | 2.055 | 0.97 | -0.92 | 2.69 | 0.05 | 1.29 | 0.38 | 1.86 | 0.52 | ||||||||||||
| Ser carboxypeptidase precursor (At3g10410) | H37490 | 3.972 | 0.13 | 1.791 | 0.16 | 0.76 | -2.22 | <0.01 | Loc | SA | ||||||||||||||
| GAST1-like protein (At1g74670) | H36867 | 2.739 | 0.50 | 1.259 | 0.31 | 0.74 | -2.17 | 0.01 | Loc | SA | ||||||||||||||
| Pyruvate dehydrogenase E1 α subunit (At1g24180) | R65310 | 6.641 | 0.33 | 2.467 | 0.64 | 0.92 | -2.69 | 0.01 | SA | MJ | ||||||||||||||
Shown for each gene are the normalized fluorescent cDNA microarray mean intensities above local background of the control (Ch1) and the treated (Ch2) sample including SD (SD1 and SD2), as well as the corresponding normalized difference (NNDiff), ratio, and probability values (t test). Signals that were higher than twice the average background plus 2 × SD of the other channel in each independent experiment are considered significant. Only genes with probability values (P < 0.05) and three additional genes of interest (*) are shown. Genes were assigned to putative functional groups and sorted by probability values. In addition, results from quantitative RT-PCR analysis are shown as mean values and se for locally-inoculated (local) and distal tissue. RNA for RT-qPCR was either obtained from the same three replicates used in microarray experiments (72 h after inoculation with A. brassicicola) or from two additional independent replicate experiments (48 and 72 h after inoculation). Previously reported responses by Schenk et al. (2000; for induced or repressed ratios > 2.00) to A. brassicicola in local tissue (Loc) and responses to plant defense signaling compounds SA, MJ, and ethylene (Eth) are also shown. A supplementary table including genes differentially expressed in distal tissue with lower probability values but that passed all other selection criteria is available at http://www.plantphysiol.org
Twenty-seven of the genes with significantly altered systemic expression have been previously shown to be affected by SA and/or jasmonate treatment, and 15 of the significantly altered systemic genes were also altered in expression in the locally inoculated tissues at 72 h after inoculation (Schenk et al., 2000; see also Table I). This further suggested that the genes identified in this analysis were relevant to plant defense responses. Interestingly, there was a strong relationship between the direction of up- or down-regulation by these signals and the direction of change in the distal tissue. This is consistent with the results of Kazan et al. (2001) and Chapman et al. (2002), who have used cluster and principal component analysis, respectively, to examine the extent of overlap in pathogen- and chemically (SA, MJ, and ethylene) induced gene expression. These analyses have shown that there was a close relationship between jasmonate and ethylene responses. In addition, the systemic response showed a strong correlation with both SA- and MJ-induced responses (Kazan et al., 2001; Chapman et al., 2002), suggesting that both pathways play significant roles during the systemic response triggered by A. brassicicola inoculation.
We also compared the genes induced in Arabidopsis-A. brassicicola interaction with those reported by Maleck et al. (2000), who have studied gene expression patterns in Arabidopsis during SAR triggered by bacterial infection. Of the 35 genes that we showed to be significantly altered in expression in distal tissue after A. brassicicola challenge, only three genes (encoding PR1, catalase 3, and a putative cytochrome P450 monooxygenase (At4g31500; CYP83B) were observed to be similarly systemically altered by Maleck et al. (2000) at 48 h after inoculation with the bacterial pathogen P. syringae. The low level of overlap in gene expression observed between these two studies that have used different EST collections is not surprising, considering that resistance against this bacterial pathogen and against biotrophic pathogens such as Peronospora parasitica is thought to occur mainly through SA-mediated defense responses. In contrast, resistance against necrotrophic pathogens (e.g. Botrytis cinerea and the A. brassicicola isolate used in this study) appears to depend on jasmonate/ethylene-mediated defense responses (Thomma et al., 2001). Further supporting this conclusion, approximately 20% of the genes identified in this study were also altered after chitin (elicitor) treatment in Arabidopsis using the same microarray as used in this study (Ramonell et al., 2002). A. brassicicola is known to contain chitin in its cell wall, and this may explain the extent of overlap observed in gene expression between these two studies.
Confirmation of Microarray Results by RT-qPCR
To test whether induction of gene expression in distal tissue could be confirmed by another method, we performed RT-qPCR assays on a subset of 22 induced and 1 repressed genes selected from each putative functional group given in Table I. Cross-hybridization from closely related members of the gene families may be problematic in microarray experiments, whereas because of the use of specific primers, expression detected from qPCR experiments should be a true reflection of gene expression measured. These RT-qPCR experiments used RNA from three biological replicates that were previously used for microarray hybridizations and additional independent biological experiments (Table I). To allow a direct comparison of the response in inoculated and distal leaf tissue, RNA obtained from inoculated tissue was used in addition to the RNA from distal tissue of the same plants and the equivalent uninoculated control plants grown in parallel. Results from these RT-qPCR experiments generally confirmed the induction patterns of genes in distal tissue, which was usually accompanied by a similar response in locally inoculated tissue (Table I). Altered expression of three genes (At1g66140, At1g04680, and At1g06620) was not confirmed at 72 h after inoculation using RNA previously used for microarray analysis. However, further analysis of independent replicates by RT-qPCR confirmed altered expression of these genes in distal tissue. In addition, differences in the induction values obtained by these two methods of gene expression analysis were observed. In general, -fold induction values measured by RT-qPCR were lower than those measured by cDNA microarray hybridization. However, five genes, including three highly expressed genes (encoding PDF1.2, ATMPK3, and a putative cold acclimation protein), showed lower induction ratios in microarray experiments, possibly due to saturation of the hybridization signals measured. To obtain further expression data for individual members of gene families, three additional genes were selected that had close DNA sequence homology to the ESTs used in microarray hybridization experiments. These included three pairs of tandemly linked genes encoding putative disease resistance proteins (At1g33590 and At1g33600), polygalacturonase inhibitors (At5g06860 and At5g06870), and cold stress-associated proteins (At1g20440 and At1g20450). These experiments showed induction of all three gene pairs in both local and distal tissues (Table I). However, there were substantial differences in the levels of induction between different family members. For example, At1g33600 showed significantly higher induction (4.35-fold) in distal tissue than the gene with closest DNA sequence homology (84%) to the EST used in microarray hybridizations (At1g33590; 1.93-fold). It appears likely, therefore, that transcripts of At1g33600 cross-hybridized to the cDNA of this EST on the microarrays, contributing to the overall induction ratio of 4.00 (Table I).
Time-Course Analysis of Expression Patterns of Selected Genes
Although it was not possible to test the expression patterns of all genes identified by RT-qPCR due to time and cost involved, some genes from each functional group were chosen for further expression profiling. For this purpose, two additional inoculation experiments were carried out (independent from previous microarray experiments). In these experiments, samples of locally inoculated and distal leaf tissue together with tissue from control plants were collected at 48 and 72 h after inoculation with A. brassicicola. Additional samples were collected for one replicate at 1, 3, 6, 12, and 24 h to allow for further characterization of selected genes which also included three genes (encoding putative pectate lyase [At1g04680], acyl-CoA synthetase [At4g23850], and a multifunctional protein [At5g06860]) that were of particular interest but that did not pass the initial microarray data analysis criteria. The results obtained by RT-qPCR from these independent replicates confirmed the induction patterns in systemic tissue of the genes tested at either 48 or 72 h after inoculation (Table I). However, expression for some genes was higher at 48 h than at 72 h (e.g. genes encoding a basic helix-loop-helix protein or desacetoxyvindoline 4-hydroxylase), suggesting that some variability of gene expression occurred for these independent experiments despite careful control of the experimental procedures, including the plant growth and incubation environment. Variability observed in gene induction values between the replicates may have also contributed to the finding that only 35 of 100 genes that showed a mean of 2-fold alteration in gene expression were significant at 95% probability in statistical analysis. This indicates the importance of substantial experimental replication to obtain reliable results, an issue that has been overlooked in some studies.
Functional Identities of Genes Expressed in Distal Tissue
Putative Disease Resistance and Regulatory Genes
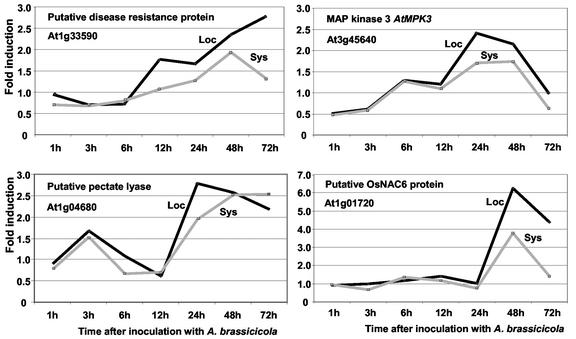
Five homologs of disease resistance genes were induced in distal tissue after inoculation with A. brassicicola (Table I). Similarly, the transcript abundance of seven genes with potential roles in signal transduction and transcriptional regulation was also altered in distal tissue (three induced and four repressed). The observation that the expression of disease resistance gene homologs and signaling proteins is enhanced is consistent with a plant condition that can more readily perceive and respond to pathogen challenge. Expression of one of the putative disease resistance genes identified in this group, At1g33590, was studied further by RT-qPCR in a time-course study using RNA from another independent replicate after inoculation with A. brassicicola. An induction of this gene between 6 and 12 h in the locally inoculated leaves was accompanied by a gradual but weaker increase in the distal tissue where maximum induction (1.93-fold) was reached at 48 h (Fig. 2). Interestingly, another gene (At1g33600) encoding a putative disease resistance protein located directly adjacent to At1g33590 showed a much stronger induction in distal tissue (4.35-fold; Table I), suggesting that different members of this family may be regulated differently.
Figure 2.
Expression profiles of selected Arabidopsis genes during a time course in local (Loc) and distal (Sys) leaf tissue after inoculation with A. brassicicola. Shown are induction ratios (y axes) obtained by RT-qPCR experiments over time after inoculation (x axes). Note that the y axis scale is different for the gene encoding a putative OsNAC6 protein.
Strong induction in distal tissue was also observed for several putative regulatory genes, e.g. encoding a putative Arabidopsis response regulator (ARR1-like) and a basic helix-loop-helix protein. Recently, we have undertaken functional studies on the latter gene showing that this gene is likely to play a role in the regulation of the jasmonate-dependent defense responses (J.P. Anderson, P.M. Schenk, K. Kazan, and J.M. Manners, unpublished data). We also identified two genes encoding putative mitogen-activated protein (MAP) kinases (AtMPK3 and AtMEKK1) induced in distal tissue but at relatively low significance (P = 0.06 and P = 0.12, respectively; Table I; supplementary table). MAP kinases are known to be involved in transducing extracellular stimuli into intercellular responses and confer resistance to both bacterial and fungal pathogens via a signaling cascade that has recently been found to be remarkably conserved among plants, insects, and mammals (Kovtun et al., 2000; Zhang and Klessig, 2001; Asai et al., 2002). AtMEKK1 has recently been reported to be regulated by pathogen infection and flagellin 22 protein and possibly acts upstream from AtMPK3 (Asai et al., 2002). It is also hypothesized that AtMEKK1 may be involved in the activation of PDF1.2 and THI2.1 genes through AtMEK1 and AtMPK4 proteins. AtMPK4 has also been shown to down-regulate SA-mediated responses, possibly by blocking SA synthesis (Zhang and Klessig, 2001). To test the kinetics of AtMPK3 induction, we further investigated expression of this gene in a time-course study after A. brassicicola inoculation. Maximum induction values were observed at 24 and 48 h in local and distal tissue, respectively, before an apparently strong down-regulation at 72 h (Fig. 2). This transient expression of AtMPK3 may coincide with the signaling cascade required to activate the innate immune response where AtMAPK3 protein plays a key role (Asai et al., 2002).
Cell Wall Modifications
Genes associated with plant cell wall synthesis (e.g. a gene encoding catalytic subunits of cellulose synthase), degradation (e.g. putative genes encoding pectin methyl esterase and pectate lyase) and secondary modification (peroxidase) also showed differential expression (Table I). The induction of peroxidase activity in distal plant tissues is well known and peroxidase has often been used as an enzymatic marker in early SAR studies (Kogel et al., 1994; Young et al., 1995; Rasmussen et al., 1995). Enhanced peroxidase activity has been shown to enhance disease resistance in transgenic plants (Kazan et al., 1998; Way et al., 2000). Two other ESTs for cellulose synthase (accession nos. T45414 and N96707) were among the 83 genes that were also induced 2-fold but at lower P values (0.06 and 0.08, respectively; supplementary table). The expression profile of a gene (At1g04680) encoding a putative pectate lyase has been studied in a time course where a similar pattern was monitored in both local and distal tissue (Fig. 2). After an initial transient induction of 1.67- and 1.53-fold at 3 h after inoculation in local and distal tissue, respectively, a maximum level of transcriptional induction in distal tissue was reached between 24 and 72 h (1.95- and 2.53-fold induction, respectively). Pectate lyase is involved in degradation of pectin in the plant cell wall. Interestingly, transcriptional changes detected in genes for cell wall modification were higher represented at the time points investigated in the systemic defense response (13% of all genes induced 2-fold) than that in tissue locally inoculated with A. brassicicola or treated with signaling compounds SA, MJ, or ethylene (each approximately 6%; Schenk et al., 2000). This further suggests a key role for cell wall metabolism in the systemic defense responses. Involvement of pectate lyase and cellulose synthase in plant defense has recently been shown (Ellis et al., 2002; Vogel et al., 2002).
Our microarray analysis also identified a gene (AtPGIP2, At5g06870; Table I) encoding a polygalacturonase inhibitor, an enzyme specifically inhibiting fungal endogalactorunases and located within the Arabidopsis cell wall (Ferrari et al., 2003). We subsequently confirmed the response of this and another polygalactorunase inhibitor gene (AtPGIP1, At5g06860) to A. brassicicola inoculation by RT-qPCR (Table I). These two genes (AtPGIP1 and AtPGIP2) have very close sequence similarity and are tandemly linked/duplicated on chromosome 5 (see also Ferrari et al., 2003). Induction of both of these genes by fungal inoculation suggested that their protein products would have inhibitory effects on polygalactorunases potentially secreted by fungal pathogens. Ferrari et al. (2003) have recently reported that the enzymes encoded by these two genes are the inhibitors of polygalactorunases from B. cinerea, another necrotrophic fungal pathogen. Moreover, overexpression of these genes in Arabidopsis reduced the disease symptoms caused by B. cinerea (Ferrari et al., 2003), providing direct evidence for the roles of these two genes in plant-necrotrophic pathogen interactions.
Fatty Acid Metabolism
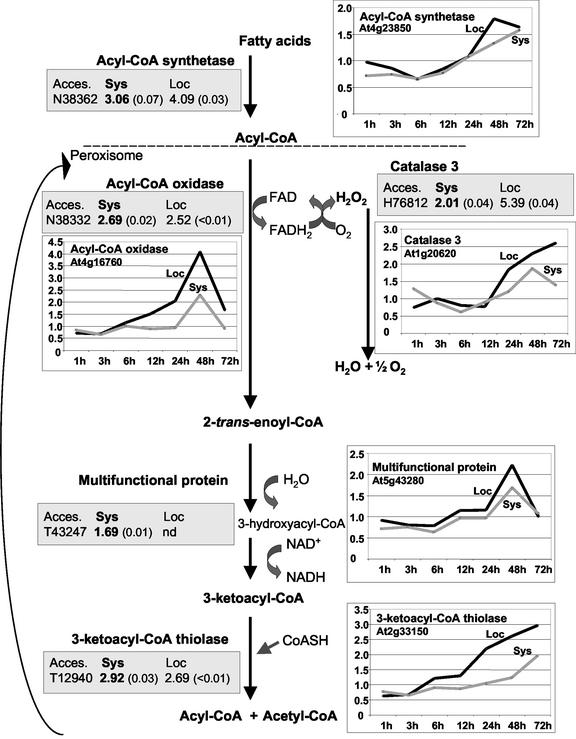
Several housekeeping genes were altered in expression in distal leaves. Most noticeably, transcripts encoding enzymes for all steps of the β-oxidation of fatty acids were coordinately up-regulated in distal tissue (Table I; Fig. 3). These were acyl-CoA synthetase, acyl-CoA oxidase, catalase, multifunctional protein (2-transenoyl-CoA hydratase and l-3-hydroxyacyl-CoA dehydrogenase), and 3-ketoacyl-CoA thiolase. Among these genes, only a homolog of the Arabidopsis 3-ketoacyl-CoA thiolase in tomato (Lycopersicon esculentum) has recently been shown to be activated during Pto-mediated disease resistance (Mysore et al., 2002). Amino acid sequence analysis of the predicted proteins by pSORT (http://psort.nibb.ac.jp/) showed that all enzymes, including the catalase 3, were targeted to peroxisomes (except acyl-CoA synthetase, which is located in the cytoplasm). The EST (T43247) listed for the multifunctional protein did not meet the mean induction ratio cutoff of at least 2.00. However, its high probability value (P = 0.01) indicates robustness of the microarray data obtained for this gene. RT-qPCR using two additional biological replicates confirmed the microarray data for these genes (Table I), even though, as observed for other genes, induction ratios were relatively lower when measured by RT-qPCR than by microarray hybridizations. Again, this could be due to cross-hybridization from the other up-regulated members of the gene family with close homology to the EST used in microarray analysis.
Figure 3.
Overview of the β-oxidation of saturated fatty acids in plants (modified from Graham and Eastmond, 2002) and Arabidopsis genes functioning in this pathway, which were shown to be induced in distal tissue after inoculation with A. brassicicola. Shown for each enzyme are the transcriptional changes as induction ratios (with probability value) of the corresponding ESTs with GenBank Accession numbers (Acces.) for microarray analysis. Ratios obtained for distal tissue (Sys) were compared with responses (previously reported by Schenk et al. [2000]) for local tissue (Loc). RT-qPCR was used to reveal expression profiles (shown as induction values) at different times after inoculation. y axes scales differ for each gene.
We further examined the activation of the genes encoding enzymes involved in the β-oxidation of fatty acids by RT-qPCR analysis in both local and distal tissue using time-course studies (Fig. 3). Increased expression was measured for all genes involved in this pathway with similar induction patterns at both local and distal tissue. The earliest induction was observed at 12 h after inoculation (2.57-fold in local tissue and 1.41-fold in distal tissue) for the gene encoding acyl-CoA-oxidase and peaked at 48 h after inoculation (Fig. 3). The time-course expression study of the gene encoding catalase-3 (CAT3) revealed up-regulation at 24 to 72 h after inoculation with A. brassicicola, in both local and distal tissue (Fig. 3).
Heightened fatty acid metabolism may be significant in priming the synthesis of plant defense-signaling molecules such as jasmonic acid (JA) and other oxylipins (Graham and Eastmond, 2002; Howe and Schilmiller, 2002) and merits further study (see below). A. brassicicola-inoculated and distal leaves have been shown to have elevated levels of JA (Penninckx et al., 1996). The induction of the genes involved in β-oxidation of fatty acids in local tissue (Table I; Fig. 3) suggests that this response was not specific to systemic defense responses. Interestingly, activation of this pathway was not specific to pathogen inoculation, because the induction of these genes after treatment with defense-signaling molecules had also been observed (Schenk et al., 2000). This was confirmed by RT-qPCR analysis where, most notably, SA treatment specifically induced the induction of acyl-CoA-oxidase (4.17-fold) and 3-ketoacyl-CoA thiolase (4.2-fold) at 12 h after treatment (P.M. Schenk, K. Kazan, and J.M. Manners, unpublished data). Furthermore, it has previously been shown that genes involved in the β-oxidation pathway are also activated during senescence and upon exposure to certain other stresses. For instance, He et al. (2002) recently reported the up-regulation of transcript levels of 3-keto-acyl-thiolase by senescence. Similarly, Seki et al. (2002) found elevated transcript levels of acyl-CoA oxidase and 3-ketoacyl-CoA thiolase in Arabidopsis plants exposed to drought, cold, or high-salinity stress. It is also becoming clear that SA, JA, ethylene treatments, and pathogen challenge can all promote responses associated with stress and/or senescence (John et al., 1995; Butt et al., 1998; Morris et al., 2000; He et al., 2002). Therefore, it is possible that the increased transcription from the genes involved in β-oxidation may be due to the senescence/stress-promoting effects of the plant-signaling compounds or pathogen.
To further explore the relationship between β-oxidation, senescence, and the plant defense responses, we examined the transcript levels of these genes in the Arabidopsis mutant constitutive expressor of pathogenesis-related genes/hypersenescence1 (cpr5/hys1; Bowling et al., 1997; Yoshida et al., 2002). This mutant shows an enhanced accumulation of reactive oxygen species, elevated levels of gene expression associated with both SA- and JA-dependent defense responses, and a hyper-senescence phenotype in the absence of any treatment/inoculation (Bosch et al., 1998; Clarke et al., 2000; Yoshida et al., 2002). Our RT-qPCR analysis data showed higher levels of transcripts of the genes encoding acyl-CoA oxidase (1.52-fold), 3-ketoacyl-CoA thiolase (1.75-fold), and catalase (1.47-fold) in untreated cpr5 plants compared with wild-type plants grown in parallel. This further suggests that an overlap exists for the β-oxidation pathway between plant defense and stress responses, including senescence. Up-regulation of the components of the β-oxidation pathway by multiple signals could provide additional insights for the overlap observed between plant defense and other stress responses (Hanfrey et al., 1996; Quirino et al., 1999; Chen et al., 2002; Swidzinski et al., 2002).
Genes Involved in Secondary Metabolism, Cell Maintenance, and Development
We observed significant induction of two genes encoding cytochrome P450 enzymes with yet unknown functions. The gene CYP83B1 (At4g31500; see Table I) encodes a cytochrome P450 enzyme involved in glucosinolate synthesis (Bak and Feyereisen, 2001; Bak et al., 2001). Recent evidence indicates that glucosinolates have roles in resistance of Arabidopsis to various necrotrophic pathogens (Tierens et al., 2001). More interestingly, Hemm et al. (2003) have also showed that the CYP83A1 gene encoding another cytochrome P450 that is very closely related to our CYP83B1 gene functions in the cross road of pathways involved in the biosynthesis of glucosinolates and phenylpropanoids in Arabidopsis, further suggesting defensive roles for the genes identified in our analysis.
Interestingly, a gene putatively involved in indole metabolism (desacetoxyvindoline 4-hydroxylase) was induced significantly. Additionally, at a lower significance (P = 0.07), Trp decarboxylase was also induced. Although involvement of Trp decarboxylase in the synthesis of the Arabidopsis phytoalexin (camalexin) is not yet known, this enzyme is involved in the biosynthesis of pharmaceutically important monoterpenoid indole alkaloids in opium poppy (Papaver somniferum) and Madagascar periwinkle (Catharanthus roseus; Verpoorte and Memelink, 2002), its expression is induced by fungal elicitors in Madagascar periwinkle (Ouwerkerk and Memelink, 2001), and activation of this gene by AP2/ERF-domain transcription factor ORCA3 occurs via a jasmonate-responsive element (van der Fits and Memelink, 1999).
A gene encoding a malic enzyme isoform was among the housekeeping genes that were induced after inoculation with A. brassicicola. NADP-malic enzymes from maize (Zea mays) and cucumber (Cucumis sativus) have recently been found to accompany other plant defense responses (Havelda and Maule, 2000; Maurino et al., 2001). Interestingly, induced expression of NADP(+) malic enzyme was found in uninfected cucumber cells of the plants locally infected with cucumber mosaic virus. This, together with the induction of genes for triose phosphate translocation from the chloroplast and a cytochrome C subunit, suggests that metabolic activities leading to a higher energy state may be a part of the systemic gene programming. Involvement of a high-energy state with plant defense responses have also recently been implicated by Scheideler et al. (2002).
A gene showing homology the rice (Oryza sativa) OsNAC6 gene, which is thought to play a role in plant development (Kikuchi et al., 2002), was strongly up-regulated in both local and distal tissue after fungal inoculation (Table I). A time-course study using RT-qPCR showed that induction of this gene in distal tissue was restricted to 48 to 72 h (Fig. 2). Currently, the exact role of this gene in plant defense is not clear. However, identification of a OsNAC6 homolog in tomato, which is coordinately regulated during a Pto-mediated disease resistance, suggests a defensive role for this gene (Mysore et al., 2002).
In conclusion, the results of the microarray experiments described here supports the notion that plant defense is a complex physiological event with potential involvement of many genes. This complexity is further enhanced by the fact that resistance against most necrotrophic fungal pathogens does not follow a gene for gene-type interaction but rather is influenced by many genes with relatively smaller effects. In this context, some of the genes reported here might have more direct or specific effects on the final outcome of the interaction between Arabidopsis and A. brassicicola, whereas induction of other genes may be as a result of an overall stress response triggered by the pathogen inoculation. Our results suggest that alterations that occur in the transcription of genes involved in pathogen perception, signal transduction, cell wall modification, fatty acid metabolism, and secondary metabolism may be key processes associated with the systemically activated primed state.
MATERIALS AND METHODS
Inoculation Experiments
Arabidopsis plants were grown to eight- to 12-leaf stage in controlled environment rooms (24°C–20°C day and night temperature) and a photo-period of 8 h light (170 μE m-2 s-1) under transparent covers (Yates, Brisbane, Australia) and were treated with either defense-inducing chemical signal compounds or fungal pathogens. All treatments were carried out at 1 h after the start of the illumination period. To identify genes whose expression changes during plant defense responses in uninoculated (distal) tissue, we inoculated three to four leaves on one side of the rosette of 6-week-old Arabidopsis cv Columbia plants (200 plants replicate-1) with 5-μL drops of a freshly prepared spore suspension of (105 spores mL-1) Alternaria brassicicola (isolate UQ4273; freshly grown on agar plates containing clarified V8 vegetable juice (Campbell Soup Company, Camden, NJ). The remaining leaves of the rosette were left uninoculated, and plants were incubated under a transparent dome to provide high humidity. Uninoculated distal leaves of the same age as the locally inoculated leaves were collected opposite of the inoculation side. This inoculation system ensured that no accidental cross-contamination of distal uninoculated leaves with A. brassicicola spores occurred. Uninoculated control plants were grown in parallel under the same conditions, and leaf tissue was collected at the same time as for the inoculated plants. A total of five completely independent experiments were performed on separate occasions to provide replicate RNA samples for either statistical analysis of microarray data or RT-qPCR assays.
Microarray Hybridization and Analysis of Expression Data
The methods of total RNA isolation, northern-blot analysis, preparation of probes, hybridizations, and scanning of slides have been described previously (Schenk et al., 2000). Stringent control measures were applied for all steps of data analysis so that all results on gene induction and repression presented were reproduced and had signals that were within the window of resolution of the microarray hybridization method. Gene expression data was normalized using a set of custom Perl scripts (Schenk et al., 2000; also available at http://www.tpp.uq.edu.au/microarray/pdmd.htm). Overall background for each experiment was calculated using a set of 118 control spots on each slide. Data points where expression was not greater than two sds above the overall background for at least one channel were discarded. Because weak hybridization signals are less reproducibly quantified than stronger signals, an additional background criterion was applied. Therefore, at least one signal had to be two times higher than critical background (average background plus 2× sds) of that channel and also at least 2-fold higher than the critical background of the other channel to be considered for further data analysis. As shown in Table I and the supplementary table, statistical analyses of quantified and normalized data from both fluorescent signals were carried out for each gene by calculating the sds for each signal, and statistical significances were determined using Student's t test.
To monitor and assess differential gene expression, normalized signal intensities were used to calculate induction or repression ratios as well as normalized differences using Perl scripts. Genes that showed induction or repression ratios of at least 2.00, normalized differences of at least 0.500, and t test probabilities of at least 80% are reported (Table I; supplementary data). The genes that did not meet these criteria have been removed from the data set (except for a gene encoding a multifunctional protein [T43247] whose induction ratio [1.69] was below 2.00 but was highly significant [P < 0.01]). It is, therefore, probable that due to the stringent criteria used in data analysis, our results may underestimate the extent of altered gene expression in distal tissue.
RT-qPCR Analysis of Transcripts
Results from microarray experiments were also validated for a subset of selected genes by RT-qPCR experiments using either the same RNA samples used for microarray experiments or RNA isolated from two additional biological replicates (e.g. plants grown and inoculated separately from microarray experiments). In these experiments, we collected Arabidopsis leaf tissue samples for RNA isolations at 48 and 72 h after inoculation. Additional samples were taken for one biological replicate at 1, 3, 6, 12, and 24 h after inoculation. Leaf tissue from 50 plants each of Arabidopsis cpr5 mutant and wild-type cv Columbia (grown in parallel) plants was collected at the eight- to 12-leaf stage for RNA preparation.
For RT-qPCR experiments, 5 μg of total RNA was denatured at 70°C for 5 min followed by quick chill on ice in a 13-μL reaction containing 10 ng of anchored oligo(dT) 23-mers, 4.5 ng of random hexamer primers (Invitrogen, Carlsbad, CA), and 0.5 μL of 20 mm dNTPs. After the addition of 4 μL of 5× reaction buffer (Invitrogen) and 2 μL of 0.1 m dithiothreitol, the reaction was preheated to 42°C for 2 min before adding 1 μL (200 units) of Superscript II reverse transcriptase (Invitrogen) followed by incubation at 42°C for another 50 min. After terminating the reaction at 70°C for 15 min, the resulting cDNA was subsequently taken up in a volume of 500 μL, and SYBR green-labeled PCR fragments were amplified by using gene-specific primers designed from the coding sequence and over an RNA splice junction (if available) of each gene using the Primer Express 1.5 software (Applied Biosystems, Foster City, CA). RT-qPCR using the ABI PRISM 7700 sequence detector and SYBR Green Master mix (Applied Biosystems) was carried out using primers (listed in Table II) at a final concentration of 0.28 μm each and 1 μL (the equivalent of 10 ng total RNA) of cDNA as template. PCR-cycling conditions comprised an initial polymerase activation step at 95°C for 10 min, followed by 45 cycles at 95°C for 15 s and 59°C for 1 min. Real-time DNA amplification was monitored and analyzed using the Sequence Detector 1.7 program (Applied Biosystems). Differences in cycle numbers during the linear amplification phase between samples containing cDNA from treated and untreated plants were used to determine differential gene expression, each cycle representing a 2-fold change in template abundance. ses of the means were calculated using the JMP In statistics software (SAS Institute Inc.). Expression detected from three β-actin genes of Arabidopsis, β-actin-2 (At3g18780), β-actin-7 (At5g09810), and β-actin-8 (At1g49240) with universal actin forward primer 5′-AGTGGTCGTACAACCGGTATTGT-3′ and specific reverse primers 5′-GATGGCATGGAGGAAGAGAGAAAC-3′, 5′-GAGGAAGAGCATTCCCCTCGTA-3′, and 5′-GAGGATAGCATGTGGAACTGAGAA-3′, respectively, were used as combined internal standards to normalize small differences in template amounts.
Table II.
Forward and reverse primers used for RT-qPCR
| Encoded Protein | Primers Used for Quantitative PCR (5′-3′) |
|---|---|
| PDF1.2 (At5g44420) | TTGCTGCTTTCGACGCA |
| TGTCCCACTTGGCTTCTCG | |
| Leu-rich repeat protein (At5g21090) | TGCTCACATTCCTTTACAGAACTTTG |
| TGCAGTTAGTGTCGTAGCTTGCA | |
| Similar to downy mildew resistance protein (At1g72930) | AAAGTTCTTAAATGGAGGCAAGCA |
| AGCTTCGAGTCATCATCACCTGA | |
| Putative disease resistance protein (At1g33590) | CTGCTCTTCTCTTAAATTGTTAGTTTGTCTC |
| GCTTATTAGCCTCATGCTTTAAAATCTTGA | |
| Putative disease resistance protein (At1g33600) | CGGAGAGCCGCAGAGACAG |
| AACTTGAGTCATAACCATCTTTGTGG | |
| ARR1-like putative protein (At3g46640) | GGCGGAAGAAGGAGATTCAGG |
| TGAGCTACAACGTCAACGAATCTCT | |
| Basic helix-loop-helix 6 protein (At1g32640) | AGGAGGCTGACGGTTGTCG |
| CCACCAAAACCTCACTGAGGAA | |
| MAP kinase 3 (ATMPK3) (At3g45640) | GACAGAGTTGCTTGGCACACC |
| GCTAAGGGCTGACGTGGGA | |
| Zinc finger protein (At1g66140) | GGTCTCTTCATCATCCTAACTATATAAGAAAAAG |
| TTTCGAGGTCTAATATTGGTCTCATG | |
| Putative pectate lyase (At1g04680) | CATCCCACCAACCAAGTTAACG |
| TTCCCCGGCTCTGTTGC | |
| Acyl-CoA oxidase-like protein (At4g16760) | CGTTCCTTATACACTCAGGTCCG |
| TCGTAACGGCCAAGAACCG | |
| Putative 3-ketoacyl-CoA thiolase protein (At2g33150) | TGGGCGCTACAGGAGCG |
| TCCCCGTCCCAATGCA | |
| Catalase 3 (At1g20620) | ACACCAGAGAGGGAAACTTTGATCT |
| TCCCATCACGGATGAAGAACA | |
| Enoyl-CoA hydratase (Multifunctional protein; At5g43280) | GGAAGTGAAGCAAAGGATCCTTGG |
| AGACTTGCGTCCTATTCCTTC | |
| Acyl-CoA synthetase (At4g23850) | CCAGTGCCATTTGACATGGA |
| CATTTCGTCGATCACACTTGGTAG | |
| Cytochrome P450 monooxygenase CYP83B1 (At4g31500) | TGATGCAGATCTACAAAGACCAACC |
| CGTGTCAGTTCCCGGCAC | |
| Polygalacturonase inhibitor AtPGIP2 (At5g06870) | GTAGGAACAAGCTTACAGGTCCGA |
| CAGAGAGCTGGTTGTGTGATAGGA | |
| Polygalacturonase inhibitor AtPGIP1 (At5g06860) | AACAAACTTACAGGTTCCATAACCAGA |
| TGATAGGCGAAGGTCAGGGAC | |
| Desacetoxyvindoline 4-hydroxylase (At1g06620) | TTCCGGAGATTTGTAGGGATATTATG |
| CCCTAGAGCTTCTGATAAAAGCTCG | |
| Putative OsNAC6 protein (At1g01720) | CATGGGAGCTTCCTGGTTTAGC |
| GGACCGGTTAGGACGCGA | |
| Putative cold acclimation protein (dehydrin) (At1g20450) | AGCTCTTCTTCCTCTTCGAGTGATG |
| CCACTGTTTTCACATGATCTCCTTC | |
| Similar to cold-regulated protein cor47 (At1g20440) | CTTCTTCCTCTTCGAGCGATGA |
| CCACTAGTCCTTTCTTATCTTCCTCTCC | |
| Unknown protein (At5g19250) | GCTTATGGAGCCGAAGGTCAC |
| CTTTCGCATCGGTGGTAGTGG |
The sensitivity and accuracy of transcript abundance detection was examined by using a dilution series of 1-, 1.5-, 2-, and 3-fold template, set up as triplicates to detect transcript levels of At2g28040 encoding a receptor protein kinase (primers 5′-CTGCTCTTCTCTTAAATTGTTAGTTTGTCTC-3′ and 5′-GCTTATTAGCCTCATGCTTTAAAATCTTGA-3′) Template changes of 1.33-, 1.50-, 2.00-, and 3.00-fold were detectable as 1.36 ± 0.13, 1.53 ± 0.08, 2.07 ± 0.09, and 3.34 ± 0.09, respectively (shown as average ± sds, assuming that each amplification cycle difference corresponded to a 2-fold change).
Supplementary Material
Acknowledgments
We thank Dr. Chris Somerville for kind provision of the Arabidopsis EST collection, Dr. Todd Richmond for use of the Perl scripts, and the Arabidopsis Biological Resource Center for supplying seeds of cpr5 mutant. We are grateful to Drs. Paul Ebert and Luis Oñate-Sanchez for critical reading of the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.021683.
This research was partly supported by the Grains Research and Development Corporation of Australia (to J.M.M.) and by the U.S. Department of Energy and the Carnegie Institution of Washington (to S.C.S.).
The online version of this article contains Web-only data. The supplemental material is available at http://www.plantphysiol.org.
References
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu W-L, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signaling cascade in Arabidopsis innate immunity. Nature 415: 977-983 [DOI] [PubMed] [Google Scholar]
- Bak S, Feyereisen R (2001) The involvement of two P450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol 127: 108-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen R (2001) CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 101-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch J, Verbsky ML, Robertson TL, Larkin JC, Kunkel BN (1998) Analysis of resistance gene-mediated defense responses in Arabidopsis thaliana plants carrying a mutation in cpr5. Mol Plant-Microbe Interact 11: 1196-1206 [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent NPR1-independent resistance. Plant Cell 9: 1573-1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt A, Mousley C, Morris K, Beynon J, Can C, Holub E, Greenberg JT, Buchanan-Wollaston V (1998) Differential expression of a senescence-enhanced metallothioenin gene in Arabidopsis in response to isolates of Peronospora parasitica and Pseudomonas syringae. Plant J 16: 209-221 [DOI] [PubMed] [Google Scholar]
- Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X (2000) Roles of salicylic acid jasmonic acid and ethylene in cpr induced resistance in Arabidopsis. Plant Cell 12: 2175-2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman S, Schenk PM, Kazan K, Manners JM (2002) Using biplots to interpret gene expression patterns in plants. Bioinformatics 18: 202-204 [DOI] [PubMed] [Google Scholar]
- Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA et al. (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14: 559-574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U, Pieterse CM, Mauch-Mani B (2002) Priming in plant-pathogen interactions. Trends Plant Sci 7: 210-216 [DOI] [PubMed] [Google Scholar]
- Devadas SK, Enyedi A, Raina R (2002) The Arabidopsis hrl1 mutation reveals novel overlapping roles for salicylic acid jasmonic acid and ethylene signaling in cell death and defence against pathogens. Plant J 30: 467-480 [DOI] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner J (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557-1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Vairo D, Ausubel FM, Cervone F, De Lorenzo G (2003) Tandemly duplicated Arabidopsis genes that encode polygalacturonase-inhibiting proteins are regulated coordinately by different signal transduction pathways in response to fungal infection. Plant Cell 15: 93-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Parker JE (2000) Interplay of signaling pathways in plant disease resistance. Trends Genet 16: 449-455 [DOI] [PubMed] [Google Scholar]
- Graham IA, Eastmond PJ (2002) Pathways of straight and branched fatty acid catabolism in higher plants. Prog Lipid Res 41: 156-181 [DOI] [PubMed] [Google Scholar]
- Hanfrey C, Fife M, Buchanan-Wollaston V (1996) Leaf senescence in Brassica napus: expression of genes encoding pathogenesis-related proteins. Plant Mol Biol 30: 597-609 [DOI] [PubMed] [Google Scholar]
- Havelda Z, Maule AJ (2000) Complex spatial responses to cucumber mosaic virus infection in susceptible Cucurbita pepo cotyledons. Plant Cell 12: 1975-1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Fukushige H, Hildebrand DF, Gan S (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128: 876-884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemm MR, Ruegger MO, Chapple C (2003) The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 15: 179-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Schilmiller AL (2002) Oxylipin metabolism in response to stress. Curr Opin Plant Biol 5: 230-236 [DOI] [PubMed] [Google Scholar]
- John I, Drake R, Farrell A, Cooper W, Lee P, Horton P, Grierson D (1995) Delayed leaf senescence in ethylene deficient ACC-oxidase antisense-tomato plants: molecular and physiological analysis. Plant J 7: 483-489 [Google Scholar]
- Kazan K, Goulter KC, Way HM, Manners JM (1998) Expression of a pathogenesis-related peroxidase of Stylosanthes humilis in transgenic tobacco and canola and its effect on disease development. Plant Sci 136: 207-217 [Google Scholar]
- Kazan K, Schenk PM, Wilson I, Manners JM (2001) DNA microarrays: new tools in the analysis of plant defence responses. Mol Plant Pathol 2: 177-185 [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Ueguchi-Tanaka M, Yoshida KT, Nagato Y, Matsusoka M, Hirano HY (2002) Molecular analysis of the NAC gene family in rice. Mol Gen Genet 262: 1047-1051 [DOI] [PubMed] [Google Scholar]
- Kogel K-H, Beckhove U, Dreschers J, Munch S, Romme Y (1994) Acquired resistance in barley. Plant Physiol 106: 1269-1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J (2000) Functional analysis of oxidative stress-associated mitogen-activated protein kinase cascades in plants. Proc Natl Acad Sci USA 97: 373-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J, Carr JP, Klessig DF, Raskin I (1990) Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250: 1002-1004 [DOI] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton K, Dangl JL, Dietrich RA (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet 26: 403-409 [DOI] [PubMed] [Google Scholar]
- Manners JM, Penninckx IAMA, Vermaere K, Kazan K, Brown R, Morgan A, Maclean DJ, Curtis MD, Cammue BPA, Broekaert WF (1998) The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol Biol 38: 1071-1080 [DOI] [PubMed] [Google Scholar]
- Maurino VG, Saigo M, Andreo CS, Drincovich MF (2001) Non photosynthetic “malic enzyme” from maize: a constitutively expressed enzyme that responds to plant defence inducers. Plant Mol Biol 45: 409-420 [DOI] [PubMed] [Google Scholar]
- Métraux JP, Signer H, Ryals J, Ward E, Wyss-Benz M, Gaudin J, Raschdorf K, Schmid E, Blum W, Inverardi B (1990) Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250: 1004-1006 [DOI] [PubMed] [Google Scholar]
- Morris KA-H, Mackerness S, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V (2000) Salicylic acid has a role in regulating gene expression during senescence. Plant J 23: 677-685 [DOI] [PubMed] [Google Scholar]
- Mysore KS, Crasta OR, Tuori RP, Folkerts O, Swirsky PB, Martin GB (2002) Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J 32: 299-315 [DOI] [PubMed] [Google Scholar]
- Ouwerkerk PB, Memelink J (2001) Elicitor-responsive promoter regions in the tryptophan decarboxylase gene from Catharanthus roseus. Plant J 25: 43-53 [DOI] [PubMed] [Google Scholar]
- Penninckx IAMA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Buchala A, Metraux JP, Manners JM, Broekaert WF (1996) Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8: 2309-2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino BF, Normanly J, Amasino RM (1999) Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defence-related genes. Plant Mol Biol 40: 267-278 [DOI] [PubMed] [Google Scholar]
- Ramonell KM, Zhang B, Ewing RB, Chen Y, Dong X, Stacey G, Somerville S (2002) Microarray analysis of chitin elicitation in Arabidopsis thaliana. Mol Plant Pathol 3: 301-311 [DOI] [PubMed] [Google Scholar]
- Rasmussen JB, Smith JA, Williams S, Burkhart W, Ward E, Somerville SC, Ryals J, Hammerschmidt R (1995) cDNA cloning and systemic expression of acidic peroxidases associated with systemic acquired-resistance to disease in cucumber. Physiol Mol Plant Pathol 46: 389-400 [Google Scholar]
- Reymond P, Farmer EE (1998) Jasmonate and salicylate as global signals for defence gene expression. Curr Opin Plant Biol 1: 404-411 [DOI] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner H-Y, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8: 1808-1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheideler M, Schlaich NL, Fellenberg K, Beissbarth T, Hauser NC, Vingron M, Slusarenko AJ, Hoheisel JD (2002) Monitoring the switch from housekeeping to pathogen defense metabolism in Arabidopsis thaliana using cDNA arrays. J Biol Chem 277: 10555-10561 [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by cDNA microarray analysis. Proc Natl Acad Sci USA 97: 11655-11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T et al. (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279-292 [DOI] [PubMed] [Google Scholar]
- Swidzinski JA, Sweetlove LJ, Leaver CJ (2002) A custom microarray analysis of gene expression during programmed cell death in Arabidopsis. Plant J 30: 431-446 [DOI] [PubMed] [Google Scholar]
- Thomma BP, Penninckx IA, Broekaert WF, Cammue BP (2001) The complexity of disease signaling in Arabidopsis. Curr Opin Immunol 13: 63-68 [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF (1998) Separate jasmonate and salicylate-dependent defense response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95: 15107-15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Tierens KF, Broekaert WF (1999) Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol 121: 1093-2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierens KF, Thomma BP, Brouwer M, Schmidt J, Kistner K, Porzel A, Mauch-Mani B, Cammue BP, Broekaert WF (2001) Study of the role of antimicrobial glucosinolate-derived isothiocyanates in resistance of Arabidopsis to microbial pathogens. Plant Physiol 125: 1688-1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, Van Pelt JA, Van Loon LC, Pieterse CM (2002) Differential effectiveness of salicylic acid-dependent, and jasmonate-ethylene dependent induced resistance in Arabidopsis. Mol Plant-Microbe Interact 15: 27-34 [DOI] [PubMed] [Google Scholar]
- van der Fits L, Memelink J (1999) The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Novartis Found Symp 223: 150-157 [DOI] [PubMed] [Google Scholar]
- Verpoorte R, Memelink J (2002) Engineering secondary metabolite production in plants. Curr Opin Biotechnol 13: 181-187 [DOI] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Schiff C, Somerville SC (2002) PMR6 a pectate lyase gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14: 2095-2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way H, Kazan K, Goulter KG, Birch R, Manners JM (2000) Expression of Shpx2 gene from Stylosanthes confers resistance to Phytophthora parasitica and Cercospora nicotiana in transgenic tobacco. Mol Plant Pathol 1: 223-232 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Ito M, Nishida I, Watanabe A (2002) Identification of a novel gene HYS1/CPR5 that has a repressive role in the induction of leaf senescence and pathogen-defence responses in Arabidopsis thaliana. Plant J 29: 427-437 [DOI] [PubMed] [Google Scholar]
- Young SA, Guo A, Guikema JA, White F, Leach J (1995) Rice cationic peroxidase accumulates in xylem vessels during incompatible interactions with Xanthomonas oryzae pv oryzae. Plant Physiol 107: 1333-1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF (2001) MAPK cascades in plant defense signaling. Trends Plant Sci 6: 520-527 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.