Abstract
The PDF1.2 gene of Arabidopsis encoding a plant defensin is commonly used as a marker for characterization of the jasmonate-dependent defense responses. Here, using PDF1.2 promoter-deletion lines linked to the β-glucoronidase-reporter gene, we examined putative promoter elements associated with jasmonate-responsive expression of this gene. Using stably transformed plants, we first characterized the extended promoter region that positively regulates basal expression from the PDF1.2 promoter. Second, using promoter deletion constructs including one from which the GCC-box region was deleted, we observed a substantially lower response to jasmonate than lines carrying this motif. In addition, point mutations introduced into the core GCC-box sequence substantially reduced jasmonate responsiveness, whereas addition of a 20-nucleotide-long promoter element carrying the core GCC-box and flanking nucleotides provided jasmonate responsiveness to a 35S minimal promoter. Taken together, these results indicated that the GCC-box plays a key role in conferring jasmonate responsiveness to the PDF1.2 promoter. However, deletion or specific mutations introduced into the core GCC-box did not completely abolish the jasmonate responsiveness of the promoter, suggesting that the other promoter elements lying downstream from the GCC-box region may also contribute to jasmonate responsiveness. In other experiments, we identified a jasmonate- and pathogen-responsive ethylene response factor transcription factor, AtERF2, which when overexpressed in transgenic Arabidopsis plants activated transcription from the PDF1.2, Thi2.1, and PR4 (basic chitinase) genes, all of which contain a GCC-box sequence in their promoters. Our results suggest that in addition to their roles in regulating ethylene-mediated gene expression, ethylene response factors also appear to play important roles in regulating jasmonate-responsive gene expression, possibly via interaction with the GCC-box.
Diseases caused by fungal, bacterial, and viral pathogens result in enormous reductions in yield and quality from agricultural crops around the world. However, although plants are continuously challenged by pathogenic microorganisms, the development of disease is a relatively rare event. It is clear, therefore, that plants possess efficient defense systems. Effective induction of the defense response requires pathogen recognition followed by a network of signal transduction processes, resulting in the rapid activation of defense gene expression. A number of secondary signal molecules such as salicylic acid (SA), jasmonic acid (JA) and its methyl ester, methyl jasmonate (MeJA), and ethylene act to amplify and regulate defense responses after initial activation (Dong, 1998; Reymond and Farmer, 1998; Schenk et al., 2000). Interestingly, the SA- and jasmonate/ethylene-dependent signaling pathways appear to modulate plant defense responses against different classes of pathogens (for review, see Thomma et al., 2001). Treatment with MeJA provides Arabidopsis plants with protection against subsequent inoculation with a range of necrotrophic fungi, but not against the biotrophic pathogen Peronospora parasitica (Thomma et al., 1998). Similarly, several studies of Arabidopsis mutants including ein2, coi1, jar1, and fad3/7/8, in which the jasmonate/ethylene-response is disrupted, have indicated that resistance to a variety of necrotrophic pathogens requires a functional jasmonate/ethylene-signaling pathway (Staswick et al., 1998; Vijayan et al., 1998; Thomma et al., 1999). In contrast, resistance to the biotroph P. parasitica but not to the necrotroph Alternaria brassicicola is enhanced by pretreatment with isonicotinic acid, a functional analog of SA (Thomma et al., 1998).
A number of classes of regulatory proteins and transcription factors are known to play important roles in relaying the pathogen-initiated signals to downstream components for the activation of plant defense responses. Among these, ethylene response factor domain-containing transcription factors (ERFs) are implicated as key regulators of plant defense responses. For example, the tomato (Lycopersicon esculentum) gene Pti4 encodes an ERF protein that is induced by defense-signaling molecules and by the bacterial pathogen Pseudomonas syringae pv. tomato (Thara et al., 1999; Gu et al., 2000). Pti4 and other tomato ERFs Pti5 and Pti6 are phosphorylated by Pto kinase, and this provides a direct link between R-gene mediated pathogen recognition and subsequent defense gene activation (Zhou et al., 1997; Gu et al., 2000). Transgenic expression of ERFs such as ERF1 of Arabidopsis, Pti4 of tomato, and Tsi1 of tobacco (Nicotiana tabacum) activates defense gene expression and provides enhanced resistance against fungal and bacterial pathogens (Park et al., 2001; Berrocal-Lobo et al., 2002; Gu et al., 2002). Expression of ERFs may also provide resistance to abiotic stresses (e.g. osmotic stress), indicating that multiple signaling pathways may converge on ERFs to regulate gene expression in response to a number of biotic and abiotic stresses (Fujimoto et al., 2000; Park et al., 2001; Memelink et al., 2001).
Several members of the ERF family bind specifically to the sequence AGCCGCC through the conserved ERF domain (Hao et al., 1998). This cis-acting sequence, known as the GCC-box, is found in the promoters of many pathogen-responsive genes such as PDF1.2, Thi2.1, and PR4 (basic chitinase; Zhou et al., 1997; Manners et al., 1998). The GCC-box has been shown to function as an ethylene-responsive element that is necessary, and in some cases sufficient, for the regulation of transcription by ethylene (Ohme-Takagi and Shinshi, 1995; Shinshi et al., 1995). In addition, the GCC-box is implicated in ozone-responsive expression of tobacco basic-type pathogenesis-related PR-1 protein gene via ethylene-dependent signaling (Grimmig et al., 2003).
More recently, an ERF subfamily of transcription factors called octadecanoid-responsive Catharanthus AP2s (ORCAs) has been implicated in jasmonate-regulated expression of secondary metabolic pathways in Madagascar periwinkle (Catharanthus roseus; Menke et al., 1999a; van der Fits and Memelink, 2000). MeJA treatment and fungal elicitors rapidly induce transcript levels of Orca2 and Orca3. The ORCA2 and ORCA3 proteins regulate overlapping but distinct sets of genes associated with secondary metabolism via specific binding to a promoter element called the jasmonate- and elicitor-responsive element, which contains a core GCC-box (Menke et al., 1999a; van der Fits and Memelink, 2000).
The PDF1.2 gene of Arabidopsis encodes a plant defensin, and its expression is induced by pathogen challenge both locally at the site of inoculation by incompatible fungal pathogen and systemically in remote noninoculated regions of the plant (Penninckx et al., 1996). This activation occurs via a jasmonate/ethylene-mediated signaling pathway, rather than via the SA-dependent pathway of defense gene activation (Penninckx et al., 1996, 1998). Although the promoter of the PDF1.2 gene contains several putative cis-acting sequence elements, such as a GCC-box (Manners et al., 1998), the roles of these elements and their respective trans-acting factors in regulation of the PDF1.2 gene by jasmonates have not been investigated directly. We previously reported on the jasmonate and pathogen-mediated activation of the PDF1.2 promoter as promoter-reporter transgene that mimicked the expression patterns of the native PDF1.2 gene (Manners et al., 1998; Mitter et al., 1998). Examination of the 1,183-bp region upstream of the transcription start site of the PDF1.2 gene revealed several putative motifs with homology to known cis-elements involved in the transcriptional regulation of other genes, including those with particular relevance for jasmonate-mediated expression. Two of these motifs, located between -277 and -495 of the PDF1.2 promoter, included the jasmonate-responsive element, TGACG (-392 to -396) present in the Lox1 gene of barley (Hordeum vulgare; Rouster et al., 1997), and a 9-bp sequence, AAATGTTGT (-410 to -419), similar to a sequence within the jasmonate-responsive region of the promoter of the soybean (Glycine max) gene VspB (Mason et al., 1993).
In the present study, we set out to delineate the regions of the PDF1.2 promoter that control jasmonate responsiveness. We report here on the identification of regions of the PDF1.2 promoter including a GCC-box that actively modulate the expression of this gene in response to jasmonates. In addition, we identified a jasmonate-, ethylene-, and pathogen-responsive Arabidopsis ERF transcription factor whose transgenic overexpression in Arabidopsis resulted in transcriptional activation of the PDF1.2 gene.
RESULTS
Production of Transgenic Lines Carrying PDF1.2 Promoter Deletion Constructs and Identification of a Positive Regulatory Element in the PDF1.2 Promoter
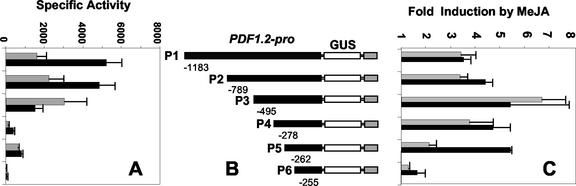
We generated a series of 5′ deletions of the PDF1.2 promoter fused in frame with the β-glucoronidase (GUS, UidA) reporter gene in a binary vector for use in plant transformation (Fig. 1C). The locations of the putative cis sequences that may be associated with jasmonate responses were taken into account when designing the 5′-deletion series. The deletion series included, in addition to the complete P1 construct (-1,183), three constructs P2 (-789), P3 (-495), and P4 (-278), in which relatively large deletions of 394, 688, and 905 nucleotides from the P1 construct were made. The constructs P5 (-262) and P6 (-255) removed only 16 and 23 nucleotides from the previous smallest construct P4 and were designed to examine the role of the GCC-box (GCCGCC between -255 to -261). We generated between seven and 12 independent transgenic lines stably transformed for each of the PDF1.2-promoter deletion constructs. We randomly selected five independently transformed lines for each PDF1.2 promoter deletion construct and analyzed GUS activity in homogenates from whole sterile-grown seedlings as described in “Materials and Methods.” Average GUS expression data from five transgenic lines carrying the same construct revealed that basal expression (i.e. absence of pathogen challenge and MeJA treatment) of the reporter genes in plants carrying the longer P1, P2, and P3 PDF1.2 promoter deletions was significantly higher (P < 0.01) than that observed for plants carrying the shorter P4, P5, and P6 constructs (Fig. 1A, shaded bars). Comparison of average GUS activity in untreated plants carrying the P3 and P4 deletion constructs indicated that deletion of the -495 to -279 region of the PDF1.2 promoter resulted in a 97% reduction in expression. This suggested that a putative positive regulatory element is apparently located within the -495 to -278 region of the PDF1.2 promoter.
Figure 1.
Basal and MeJA-responsive expression of GUS activity from the PDF1.2 promoter deletion lines. A, Average basal GUS activity (fluorescence units per milligram of protein) in each of the PDF1.2 promoter deletion constructs. Each bar represent data from five independent transgenic lines derived from six replicates in two independent experiments. Shaded bars, Plants grown in sterile conditions; black bars, plants grown in soil. B, The PDF1.2 promoter reporter constructs that were placed in the binary vector pBI101.3 used to generate transgenic plant lines. C, Average induction values from each promoter deletion construct in MeJA-treated plants. Induction data were generated from five independent transgenic lines for sterile-grown seedlings, whereas two representative lines were used for soil-grown plants. GUS activity assays were performed on 20 T2 plants for each transgenic line. Bars indicate se.
We also examined the basal expression of the reporter gene in rosette leaves of soil-grown 6-week-old plants (two lines for each PDF1.2 promoter deletion construct; Fig. 1A, black bars). Despite some differences in the basal expression levels driven by the PDF1.2 promoter deletions P1, P2, and P3 between the two growth conditions, the overall effect of the putative enhancer element revealed in the seedling studies was still readily detectable in the soil-grown plants
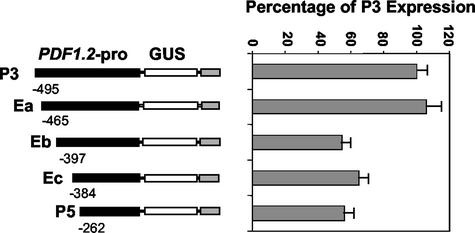
To further delineate the presence of a putative positive regulatory element within the -495 to -278 region of the PDF1.2 promoter, we performed transient expression assays. In these assays, we cobombarded the leaves of wild-type Arabidopsis plants with three constructs, which in addition to each promoter deletion construct included the 35S-green fluorescent protein (GFP) and 35S-Luc plasmids. After bombardment, leaves were examined by fluorescent microscopy. Leaf sections displaying a large number of GFP-expressing cells (>60) were selected for assay of luciferase activity in leaf homogenates, which was then used to normalize GUS activity measurements for the same sample. Results (Fig. 2) showed that constructs P3 and Ea did not differ significantly in expression, but displayed approximately 2-fold higher expression than constructs Eb, Ec, and P5 (P > 0.01). This suggested that the putative enhancer element of the PDF1.2 promoter is located downstream of -465. Because no significant difference was observed among Eb, Ec, and P5, these results indicate that the 68-nucleotide region of the PDF1.2 promoter lying between -465 and -398, or spanning the -398 region, is critical for high-level expression.
Figure 2.
Delineation of enhancer element using quantitative transient expression analysis of the PDF1.2-promoter deletion constructs in Arabidopsis leaves. Relative expression data for each construct represent the mean of GUS/luciferase ratios for at least six separate leaf samples. Bars indicate se. The data shown here are compiled from several independent bombardment experiments that produced similar results.
Jasmonate Responsiveness Conferred by the PDF1.2 Promoter Deletion Constructs
We next examined transgenic plants carrying the PDF1.2 promoter deletion constructs to identify regions of the promoter that may regulate the response to jasmonate. In both sterile- and soil-grown plants, although GUS activity levels were higher after MeJA treatment, differences between constructs appeared to mirror the results from untreated plants in that specific GUS activities were considerably higher in lines carrying constructs P1, P2, and P3 than lines carrying constructs P4, P5, and P6 (data not shown). However, a different pattern emerged when -fold induction ratios were compared (Fig. 1C). In sterile-grown plants, constructs P3 and P4 did not differ significantly from one another in -fold induction ratios and showed significantly higher -fold induction by MeJA than P1 and P2 (P < 0.01). Similarly, in soil-grown plants, lines carrying the P3 construct showed a higher level of induction than lines carrying P1 and P2 constructs.
Using sterile-grown transgenic plants, the deletion of the 15-bp region between constructs P4 and P5 resulted in a significant decrease in induction by MeJA (P < 0.01; Fig. 1C). A further decrease in induction was observed in P6 with the deletion of an additional 7 bp comprising the core GCC motif of AGCCGCC. In contrast to sterile-grown plants, the stably transformed soil-grown plants carrying the P5 construct gave a similar -fold induction of the reporter gene to plants carrying the P3 or P4. However, similar to sterile-grown plants, deletion of the core GCC-box (P6 construct) significantly reduced the level of MeJA-mediated induction of GUS activity. Although these results indicate the importance of this GCC-box motif for induction of the PDF1.2 gene by jasmonate, deletion of this box did not completely abolish jasmonate responsiveness, indicating that other elements downstream of -255 may also be involved in the regulation of this response. The biological significance of the difference between sterile- and soil-grown plants carrying construct P5 is not known but if confirmed, could be related to additional positive signaling pathways initiated by biotic or abiotic factors.
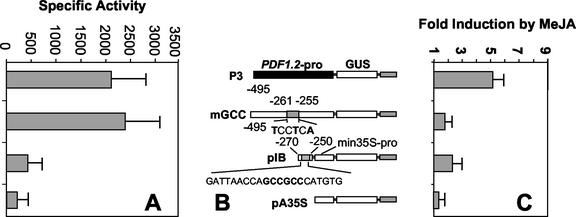
In an effort to confirm the role of the GCC-box in mediating jasmonate responsiveness, we generated two additional PDF1.2 promoter constructs, mGCC and pIB (Fig. 3B). The mGCC consisted of the -495 PDF1.2 promoter fragment with substitution of the core GCC-box sequence (GCCGCC) with TCCTCA. The pIB construct was aimed at determining whether the GCC region alone was sufficient to confer jasmonate-responsive gene expression. This construct was a fusion of the extended GCC-box region (-250 to -270) upstream of a truncated 35S (-46) promoter derived from CaMV.
Figure 3.
A through C, Further confirmation that the GCC-box confers jasmonate responsiveness to the PDF1.2 promoter using quantitative analysis of stably transformed plants. C, Results are presented as average -fold induction values from two to five transgenic lines harboring each PDF1.2 promoter deletion construct. A, The basal expression of GUS from each construct is also given. The mGCC construct is the same as construct P3 but otherwise contains three substitutions in the first (G–T), fourth (G–T), and sixth (C–A) nucleotides (shown in bold) to convert the core GCCGCC box sequence to TCCTCA. B, Construct pIB contains a 20-nucleotide-long sequence including the GCC-box and the surrounding nucleotides from the -270 to -250 of the PDF1.2 promoter linked to a truncated 35S cauliflower mosaic virus (CaMV) promoter. se bars are also shown for each construct.
Analysis of the mean GUS expression in MeJA-treated sterile-grown whole seedlings stably transformed with the mGCC construct indicated that there was no significant induction by MeJA compared with mock-treated controls (Fig. 3C). This experiment included plants stably transformed with construct P3 as control, which showed the same highly significant increase in MeJA-responsive induction (P < 0.01) as the previous experiment described in Figure 1. This result directly indicates that the GCC-box sequence confers jasmonate responsiveness to the PDF1.2 promoter.
Next, we examined the jasmonate responsiveness conferred by the pIB construct linking the GCC-box to a minimal 35S promoter in stably transformed sterile-grown plants. After MeJA treatment, these plants showed some jasmonate response, but the level of induction was significantly less than that measured for construct P3 (P < 0.05). Plants transformed with a 35S minimal promoter construct alone did not show any significant induction of GUS activity after MeJA treatment (Fig. 3C).
Identification of MeJA-Responsive Arabidopsis ERFs
Jasmonate-responsive expression of GUS activity in plants stably transformed with PDF1.2 promoter constructs including the GCC-box suggested that this conserved sequence element might act as binding site for jasmonate-responsive ERF transcription factors. To identify gene family members encoding putative Arabidopsis ERFs, we used the consensus amino acid sequence of the DNA-binding domain of all known ERF members that bind to the GCC-box domain as a query sequence for BLASTP searches of Arabidopsis sequences available on GenBank. The searches returned 145 sequences representing unique proteins (see also Sakuma et al., 2002), of which 131 contained a single AP2/ERF-type DNA-binding domain. Because of the relatively large size of this transcription factor family, we selected representatives from sequence clusters for expression profiling. With the exception of APETALA2, all of the genes were selected from the subfamily that contains only a single ERF domain. The selected sequences included ERF1 (Solano et al., 1998), AtERF2, AtERF3, AtERF4, AtERF5 (Fujimoto et al., 2000), and 12 other ERFs (see “Materials and Methods” for locus numbers) for which no expression data were available in the literature. Of the latter, AP6 was selected because it had the highest homology of any Arabidopsis ERF to ORCA2. DNA sequences for each of these 18 ERFs were generated by PCR amplification from divergent regions outside the conserved DNA-binding domain, to avoid cross-hybridization among the different members of the gene family.
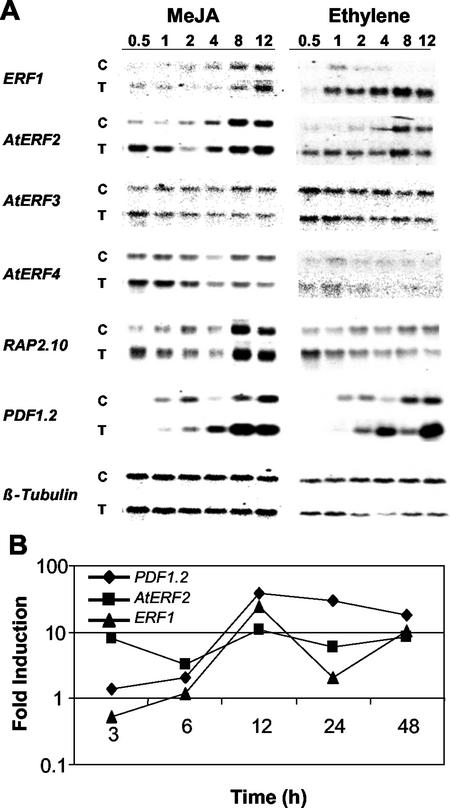
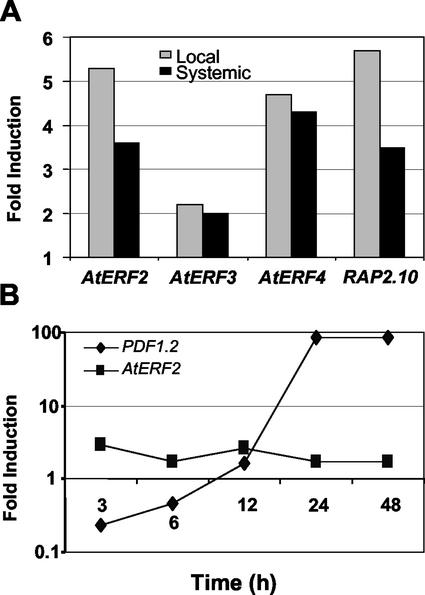
To identify jasmonate-responsive Arabidopsis ERFs, we first examined the expression patterns of the 18 selected Arabidopsis ERFs by reverse northern hybridization analysis of a macroarray of the variable sequence from each gene. Expression patterns of ERFs showing induction after MeJA treatment were then confirmed by northern-blot analysis. Only five (ERF1, AtERF2, AtERF3, AtERF4, and RAP2.10) of the 18 AP2/ERF gene family members present on the macroarray, hybridized with cDNA prepared from MeJA-treated plants and their associated controls. All of these five genes showed induction by MeJA at 0.5 h, with a peak in induction after 1 h of MeJA exposure (data not shown). With the exception of AtERF4, which remained induced until 4 h, the transcription levels of all genes had returned to those observed in control plants by 2 h (data not shown). Examination of expression patterns by northern blotting confirmed the results obtained from macroarray analysis (Fig. 4A, left panel). Induction of PDF1.2 was observed after 4 h of exposure to MeJA in these experiments (Fig. 4A, left panel).
Figure 4.
A, Expression profiling of selected ERF genes by northern blot in a time-course analysis. Left panel, Expression in response to MeJA; right panel, expression in response to ethylene for each gene. Top and bottom panels represent RNA from control (C) and treated (T) plants, respectively. B, Induction ratios of the Arabidopsis ERF1, AtERF2, and PDF1.2 genes obtained by real-time quantitative reverse transcriptase (RT)-PCR after MeJA treatment.
We further quantified the induction ratios of ERF1, AtERF2, and PDF1.2 after MeJA treatment by quantitative real-time RT-PCR, which is particularly suitable for specific detection of transcripts from the members of multigene families (Charrier et al., 2002). In these time-course experiments, we used RNA isolated from plants grown and treated at separate occasions. Results from these experiments which included early as well as late time-points (e.g. 24 and 48 h) were in agreement with the results from northern-blot and macroarray analyses as both ERF1 and AtERF2 displayed distinct yet overlapping expression patterns (Fig. 4B). AtERF2 was substantially more responsive to MeJA treatment than ERF1, especially at earlier time points (e.g. 3 and 6 h), whereas a sustained level of expression was observed for both genes at late time points. Consistent with known expression pattern of PDF1.2, we detected a strong induction of the PDF1.2 transcripts starting at 6 to 12 h after treatment.
We also tested the ethylene response profiles of those ERFs that were induced by MeJA by northern-blot analysis. These analyses showed that AtERF2, AtERF3, AtERF4, and RAP2.10 responded in a similar manner to ethylene as observed for MeJA (Fig. 4A, right panel). They displayed rapid induction on ethylene exposure, with expression returning to control levels after one to 2 h. ERF1 expression after ethylene treatment was more sustained and continued to increase over the time points tested.
We next used macroarray analysis to investigate the effect of inoculation by fungal pathogen A. brassicicola on the expression profiles of selected ERF genes in Arabidopsis plants 5 h after inoculation. Expression patterns were assessed in local, inoculated tissue and systemic noninoculated tissue of inoculated plants and were compared with equivalent tissues of control, uninoculated plants. Expression of AtERF2, AtERF3, AtERF4, and RAP2.10 was induced in both local and systemic tissue 5 h after A. brassicicola inoculation (Fig. 5A). For AtERF2 and RAP2.10, induction was substantially higher in local compared with systemic tissue at this early time point. No significant induction was observed from any of the other ERFs tested or the known defense genes PDF1.2 and PR-1 (data not shown). Inoculation with A. brassicicola has been previously demonstrated to induce expression of these defense genes (Penninckx et al., 1996; Thomma et al., 1998), but at later time periods after inoculation (48–72 h). We further quantified the induction ratios of AtERF2 and PDF1.2 by real-time quantitative RT-PCR after inoculation with A. brassicicola. Analysis of RNA samples obtained from an independent (i.e. RNA isolated from plants grown and inoculated in a different occasion) time-course experiment showed induction of both ERF1 and AtERF2 by this incompatible pathogen at all time points examined although ERF1 induction was substantially higher than that observed for AtERF2 (Fig. 5B).
Figure 5.
Arabidopsis ERF genes respond to fungal inoculation. A, Macroarray analysis showing -fold induction of four Arabidopsis ERFs after challenge by incompatible fungal pathogen A. brassicicola in inoculated (local) leaves and uninoculated (systemic) leaves of the same plant. Samples were collected 5 h after inoculation. Bars show -fold induction of each gene compared with the equivalent tissue from inoculated plants. Expression was determined using ImageQuant software, and values were adjusted against β-tubulin, as a loading control. B, Induction ratios (local) of the Arabidopsis AtERF2 and PDF1.2 genes obtained by real-time quantitative RT-PCR after inoculation with A. brassicicola.
Overexpression of AtERF2 in Arabidopsis Activates PDF1.2, Thi2.1, and PR4 Expression
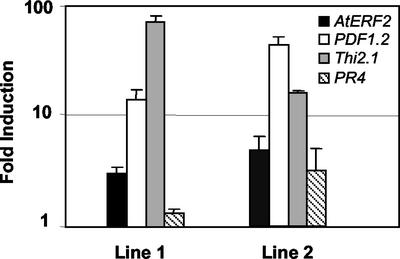
As described above, transcripts of some AtERF2 genes substantially increased at earlier time points after MeJA treatment than the first detection of elevated levels of transcripts of the PDF1.2 gene. To examine whether there was causal relationship between elevated levels of ERF transcripts and subsequent induction of the PDF1.2 gene, we stably transformed Arabidopsis plants with the AtERF2 gene driven by the constitutive 35S promoter. Examination of the transcript levels T2 plants in four stably transformed lines by real-time quantitative RT-PCR revealed that two of the lines were overexpressing AtERF2 3- to 5-fold over untransformed control plants (Fig. 6). Expression levels of the AtERF2 gene in two other stably transformed lines were similar to that observed in untransformed control plants (data not shown). We also detected a 16- and 51-fold increase in PDF1.2, an 80- and 50-fold increase in Thi2.1, and 2- and 3-fold increase in PR4 (basic chitinase) transcript levels, respectively, in these two AtERF2 overexpressing lines (Fig. 6), whereas no such increase in the expression of these genes was evident in untransformed control plants and in two other transgenic lines with wild-type levels of AtERF2 transcripts. Overall, these results suggest that the AtERF2 protein is capable of activating the PDF1.2, Thi2.1, and PR4 genes when overexpressed in transgenic plants.
Figure 6.
Overexpression of AtERF2 activates defense gene expression in Arabidopsis. Shown here are -fold expression values of the AtERF2, PDF1.2, Thi2.1, and PR4 genes in two independent stably transformed transgenic T2 lines containing the 35S-AtERF2 transgene over those observed in untransformed control plants. Error bars are also shown for each line.
AtERF2 Transcript Level Remains Unchanged in Arabidopsis Mutants Constitutively Expressing the PDF1.2 Gene
To determine whether enhanced levels of the AtERF2 transcript can be found in Arabidopsis defense-signaling mutants that constitutively express the PDF1.2 and Thi2.1 genes, we examined the transcript levels of AtERF2 by real-time quantitative RT-PCR analysis in Arabidopsis defense-signaling mutants cev-1 and cpr5 (Bowling et al., 1997; Ellis and Turner, 2001). These analyses showed no significant change in the transcript level of AtERF2 in these mutants (data not shown). However, consistent with previous reports, we detected enhanced transcript levels of PDF1.2 in both cev-1 and cpr5 mutants (data not shown).
DISCUSSION
The GCC-Box Is a Major Element Conferring the Jasmonate Responsiveness of the PDF1.2 Promoter
In this report, we have identified the -255 to -277 promoter region as being critical for regulation of the PDF1.2 gene by jasmonate. Plant lines carrying constructs with deletions in this region (P5 and particularly P6) showed significantly lower induction by MeJA in comparison with all other deletion constructs. Penninckx et al. (1998) and Manners et al. (1998) have reported that the level induction of PDF1.2 by both MeJA and ethylene in tissue culture-grown plants was increased by growth of the plants under non-sterile conditions. Therefore, we used both types of growth conditions in assessing the expression from the promoter-deletion constructs. In general, results reported in this manuscript supported this observation for the induction of PDF1.2 promoter-driven reporter gene expression by MeJA in soil-grown plants. Augmented induction in soil-grown plants may be a consequence of physiological differences between plants grown in soil and in vitro or as a result of potentiation by soil-associated biological factors.
The -255 to -277 region contains the GCC-box, which is a common motif in the promoters of various genes encoding defense-related proteins, and has been previously identified as the specific binding site for members of the ERF subfamily of AP2/ERF transcription factors (Ohme-Takagi and Shinshi, 1995; Buttner and Singh, 1997; Zhou et al., 1997). Analysis of Arabidopsis lines stably transformed with a PDF1.2 promoter-GUS reporter transgene revealed a stepwise reduction in induction by jasmonate as the sequences immediately preceding the GCC-box (P4 and P5) were deleted, with a further reduction of induction after deletion of the core GCC motif itself (P6). In other studies, a 21-nucleotide sequence containing the GCC-box in the promoter of the PDF1.2 promoter provided jasmonate responsiveness when linked to a minimal 35S CaMV promoter, and mutations introduced into the GCC-box significantly reduced the jasmonate responsiveness of the plants carrying P3 construct. Taken together, these results indicate that the GCC-box in the promoter of the PDF1.2 gene modulates the jasmonate responsiveness of the PDF1.2 gene. Similar to our findings reported here, Menke et al. (1999a) have recently shown that jasmonate- and elicitor-responsive elements found in the promoter of the secondary metabolite biosynthetic gene, Str, of Madagascar periwinkle, contains a GCC-like box that is both necessary and sufficient for jasmonate- and elicitor-responsive expression.
It has been well established that members of ERF transcription factor family interact with the GCC-box sequence. Detailed analysis of the in vitro binding requirements of three ERF transcription factors from tobacco indicated that whereas the contact of specific amino acid residues of the ERFs are confined to the 6-bp core GCCGCC region of the GCC-box, surrounding nucleotides are necessary for high-affinity binding (Hao et al., 1998). These sequences may also modulate differential binding by different ERFs. This may explain the observation that plants carrying the P5 construct, with a deletion of the 5′-flanking sequence to the core GCC motif, showed decreased responsiveness to jasmonate but significantly higher basal expression levels compared with the P4 plants carrying an extended GCC motif. The removal of the nucleotides specifying recruitment of the specific ERFs that up-regulate PDF1.2 transcription in response to jasmonate could allow competitive binding of other ERF subfamily members, which although unresponsive to jasmonate, may increase basal gene expression.
Although the results presented here identify the -255 to -277 region, containing the core GCC-box, and flanking sequences as critical for gene regulation, they do not exclude the additional significance of cis-elements located further downstream in the PDF1.2 promoter. In fact, other cis-elements also appear to contribute to jasmonate responsiveness, as indicated by residual induction displayed by lines carrying the P6 construct. Interestingly, in P6, a partial GCC-box sequence (AGCCGC, -219 to -214) occurs in the PDF1.2 promoter downstream from the consensus GCC-box at -255 to -277. It is possible that some transcription factors can bind to this GCC-box like region and activate gene expression through interactions with ERFs. Coregulation of gene expression by the GCC-box and other promoter elements has also been observed previously. For example, the GCC-box is required but not sufficient for high-level induction of the tobacco osmotin gene by ethylene (Raghothama et al., 1997). Also, a G-box is associated with jasmonate responsiveness in defense-associated genes in various plant species (Kim et al., 1992; Mason et al., 1993). In the PDF1.2 promoter, the GCC-box also directly follows a putative G-box motif (-255 to -250) with the sequence CATGTG (Manners et al., 1998). Close proximity of these G- and GCC-box elements is a common feature of tobacco PR genes and is postulated to allow cross-linking of ERF and bZIP transcription factors for regulation of gene expression during plant defense responses (Sessa et al., 1995; Buttner and Singh, 1997). Although the G-box has been shown to be important in regulating the expression of certain jasmonate-responsive genes (Kim et al., 1992; Mason et al., 1993), the putative G-box in the PDF1.2 promoter lacks the exact ACGT core sequence characteristic of bZIP transcription factor binding sites (Izawa et al., 1993) and thus may not represent a functional G box. Further work would be needed to determine the role of these additional elements in the regulation of the PDF1.2 gene.
AtERF2, a Jasmonate/Ethylene- and Pathogen-Inducible ERF, Constitutively Activates Transcription from the GCC-Box Containing Defense Genes
The deletion analysis of the PDF1.2 promoter suggested that the GCC-box is critical for jasmonate-mediated regulation of the PDF1.2 gene expression. It thus seemed a reasonable hypothesis that one or more members of the Arabidopsis ERF transcription factor family homologous to the periwinkle ORCA2 and ORCA3 transcription factors (Menke et al., 1999a; van der Fits and Memelink, 2000) might act to regulate the expression of PDF1.2 in response to jasmonates. Therefore, we examined the mRNA levels of selected ERFs in Arabidopsis plants after treatment with inducers of PDF1.2 expression and identified ERFs that show distinct induction profiles. ERF1, AtERF2, AtERF3, AtERF4, and RAP2.10 all displayed immediate early induction, with mRNA accumulating after only 0.5 h of exposure to MeJA. MeJA-induced expression of each of these genes occurred earlier than that observed for PDF1.2, suggesting that one or more of these ERFs may regulate PDF1.2 expression. Rapid and transient induction of AtERF2 by MeJA mimicked the expression patterns of the Orca2 and Orca3 transcripts, which also showed a strong induction 0.5 h after exposure to MeJA (Menke et al., 1999a, 1999b; van der Fits and Memelink, 2000, 2001). In addition, the deduced amino acid sequence of AtERF2 is highly similar to that of the ORCA3, particularly around the Ser-rich region, a putative phosphorylation site located at the C terminus of ORCA3 protein (van der Fits and Memelink, 2000). These similarities between AtERF2 and ORCA2/3 suggested that AtERF2 might play a role in MeJA-mediated induction of the PDF1.2 gene. We specifically quantified expression from the PDF1.2 gene by quantitative real-time RT-PCR to distinguish the expression of PDF1.2 from three other PDF1.2-like expressed sequences that exist in Arabidopsis. The predicted mature peptides encoded by genes PDF1.2a (same as PDF1.2), PDF1.2b, and PDF1.2c are identical (R.L. Brown and K. Karzan, unpublished data; Thomma et al., 2002). Moreover, the upstream regulatory sequences of the latter two genes contain regions with substantial homology to the GCC-box and neighboring sequences found in the promoter of the PDF1.2 gene (R.L. Brown and K. Karzan, unpublished data). The identification of multiple family members homologous to PDF1.2 means that studies of the expression of this gene based solely on hybridization signals may be misleading. This was especially critical to rule out the possibility that in AtERF2-overexpressing plants, expression of PDF1.2 but not any other PDF1.2-like genes is detected because it is possible that other PDF1.2-like genes might also be activated due to having GCC-box like sequences in their promoters. Our analysis showed that overexpression of AtERF2 activated transcription of defense genes such as PDF1.2, Thi2.1, and PR4. All of these genes contain GCC-boxes in their promoters (Epple et al., 1995; Zhou et al., 1997; Manners et al., 1998), and their activation by AtERF2 probably occurs via the GCC-box. Fujimoto et al. (2000) have demonstrated the ability of AtERF2 to interact with the GCC-box for the activation of reporter gene expression in transient expression assays.
The AtERF2 protein also shares high amino acid sequence identity with the tomato ERF, Pti4, whose overexpression in Arabidopsis also results in elevated expression of PDF1.2, Thi2.1, and PR4 and enhanced resistance against the fungal pathogen Erysiphe orontii and the bacterial pathogen P. syringae pv. tomato (Gu et al., 2002). Defense gene activation and an enhanced disease resistance phenotype were also reported for Arabidopsis plants overexpressing ERF1 (Berrocal-Lobo et al., 2002; Lorenzo et al., 2003). This may indicate that ERF1 and AtERF2 may have overlapping or redundant functions. Although we have not tested the level of disease resistance in plants overexpressing AtERF2, we predict that these plants may also show elevated levels of disease resistance due to constitutive overexpression of a number of defense genes which may be further induced during infection due to availability of additional positive regulators of defense genes, which may require induction by a pathogen.
Convergence of Multiple Signaling Pathways on ERFs
Recent studies (Fujimoto et al., 2000; Ohta et al., 2000; Park et al., 2001; Cheong et al., 2002; Gu et al., 2002; Mazarei et al., 2002; Nishiuchi et al., 2002; Oñate-Sánchez and Singh, 2002; Wu et al., 2002) suggest that ERFs are regulated differentially at both the transcriptional and posttranscriptional levels and that different ERF proteins may regulate various subsets of GCC-box-containing genes. For instance, the ERF genes studied here including AtERF2 were responsive to the known inducers of the PDF1.2 gene, namely MeJA and ethylene (Fujimoto et al., 2000; Lorenzo et al., 2003; this study), and inoculation with fungal pathogen (this study). Identification of jasmonate-, ethylene-, pathogen-, wounding-, nematode-, and stress-inducible ERF transcription factors suggest that ERFs can spatially and temporally regulate gene expression in response to biotic and abiotic cues. As recently suggested by Fujimoto et al. (2000), both ethylene and stress-related signaling pathways seem to converge on a subset of different ERFs, which subsequently regulate a subset of GCC-box containing genes. Results reported here, together with studies on ORCA2/3 proteins, and ERF1 (Lorenzo et al., 2003) suggest that components of jasmonate-signaling pathways converge with those of other pathways (e.g. abiotic stress, wounding, ethylene, and pathogen/elicitor) on ERF transcription factors in regulating gene expression.
Interestingly, so far a number of ERF proteins have been shown to be able to trans-activate the PDF1.2 expression when expressed stably or transiently in Arabidopsis (Fujimoto et al., 2000; Ohta et al., 2001; Berrocal-Lobo et al., 2002; Gu et al., 2002; Wu et al., 2002; this study). However, it is not clear which ERFs might be responsible for activating the PDF1.2 expression in planta in response to jasmonate or pathogen signals. More recently, Lorenzo et al. (2003) have suggested that ERF1 and AtERF2 may have redundant functions. It is also possible that the activation of the PDF1.2 gene in transgenic plants constitutively expressing a GCC-box binding protein occurs somewhat nonspecifically. In this study, AtERF2, a jasmonate-, pathogen-, and ethylene-inducible ERF, was able to activate the expression from the PDF1.2 gene when overexpressed in transgenic plants. However, examination of the constitutive PDF1.2-expressing Arabidopsis mutants, cev1 and cpr5, did not reveal any increase in the level of AtERF2 transcript. These results are consistent with those of Oñate-Sánchez and Singh (2002), who have recently reported that the expression of a number of jasmonate-, pathogen-, and SA-inducible ERFs, including AtERF2 have not changed in cep1 and cpr5-2 mutants. Taken together, these data suggest that PDF1.2 overexpression observed in these mutants may be mediated by other ERFs. Alternatively, as suggested by Gu et al. (2002), relatively low levels of AtERF2 may be activated posttranslationally by a constitutively expressed putative kinase without any detectable increase at the transcript level. Such a phosphorylation event could influence the localization of AtERF2 to nucleus and modulate its DNA-binding ability or its interaction with other transcription factors. Phosphorylation by Pto kinase enhances binding of the tomato ERF Pti4 to the GCC-box (Gu et al., 2000). Several other AP2/ERFs, such as CBF1, CBF2, CBF3 ORCA2, ORCA3, and AtERF5, contain potential recognition sites for protein kinases, which may play important roles in modulating the function of these proteins (Medina et al., 1999; Yamamoto et al., 1999; Fujimoto et al., 2000).
Alternatively to ERF activation, inactivation of a putative repressor protein, perhaps also an ERF, might be responsible for constitutive expression of the PDF1.2 gene in the cev1 and cpr5 mutants noted above. Recent studies have indicated that some ERFs may act as repressors rather than activators of transcription (Fujimoto et al., 2000; Ohta et al., 2001). A conserved sequence motif called ethylene-associated amphiphilic repression within the C-terminal region of these transcription factors seems to be essential for repression by such repressor ERFs (Ohta et al., 2001). For example, the AtERF3, AtERF4, and RAP2.10 proteins all contain the ethylene-associated amphiphilic repression motif, and AtERF3 and AtERF4 have been shown to function as repressors in in vitro assays (Fujimoto et al., 2000). Thus, even though the MeJA-, ethylene-, and A. brassicicola-induced expression of AtERF3, AtERF4, and RAP2.10 precedes induction of PDF1.2 in the research reported herein, it cannot be concluded that these genes are involved in the specific activation of PDF1.2.
Enhancer Region in the PDF1.2 Promoter
In this paper, we have also reported on promoter elements other than the GCC region, that appear to play roles in regulating the overall levels of expression of the PDF1.2 gene of Arabidopsis. Analysis of reporter gene expression in multiple transgenic lines for each promoter-reporter deletion line showed that a strong enhancer element is located within the -277 to -495 region of the PDF1.2 promoter. Further deletion analysis of the PDF1.2 promoter using the transient expression system allowed delineation of this element to a 68-nucleotide region located between -398 and -465. This region of the PDF1.2 promoter contains several motifs with similarity to regulatory elements of other genes, and/or known recognition sites for transcription factors. These included the sequences TTCGAC (-398 to -404), involved in the SA responsiveness to the PR-2d gene of tobacco (Shah and Klessig, 1996), and CGACG (-399 to -403), necessary for high-level expression of the rice (Oryza sativa) Amy3D gene under conditions of sugar starvation (Hwang et al., 1998). In addition, this 68-nucleotide region contains the sequence CAACA (-454 to -458), which was revealed by in vitro binding experiments as the DNA recognition sequence for the RAV1 transcription factor of Arabidopsis (Kagaya et al., 1999). The RAV1 protein binds to this motif through a protein domain that strongly resembles the AP2 domain found in AP2/ERF proteins (Kagaya et al., 1999). Also located within the 68-nucleotide putative enhancer region of the PDF1.2 promoter are sequences with homology to the GT- and GATA (I box) elements, which mediate light-responsive expression of many genes (Zhou, 1999). GT-elements also exist in many light-unresponsive genes, indicating a diverse range of functions and, depending on promoter structure, may have a positive or negative effect on transcription (Zhou, 1999). Further experimentation is necessary to determine whether one or more of the elements described above modulate high-level expression on the PDF1.2 promoter, or whether the putative enhancer contained within this region of the PDF1.2 promoter represents a novel element.
In conclusion, our results have demonstrated a role for the GCC-box in conferring jasmonate responsiveness to the PDF1.2 promoter. This motif and its surrounding bases most probably interact with several members of the ERF family of transcription factors, some of which may be also induced by jasmonates. Analysis of the PDF1.2 promoter has also indicated the existence of other, probably interactive, cis elements necessary for high-level jasmonate-mediated regulation of PDF1.2 expression during plant defense responses.
MATERIALS AND METHODS
Production of the PDF1.2 Promoter Constructs
The cloned promoter of the PDF1.2 gene described in Manners et al. (1998) was used as template for the production of all PDF1.2-promoter deletions. Promoter fragments of varying lengths were amplified by PCR using the following forward primers for constructs P2, P3, P4, P5, and P6, respectively (Fig. 1B): 5′-ATCAAAGCT TTGAGCCTCTCACTTGCGGTC-3′, 5′-ATCAAA GCTTTGCTGCTCTTGAGATCAACC-3′, 5′-GCCGAAGCTTCCATTCAGATTAACCAGCC-3′, 5′-ATCAAAGCTTAGCCGCCCATGTGAACGATG-3′, 5′-ATCAAAGCTTCATGTGAACGATGTAGCATTAGC-3′, and a common reverse primer 5′-GAGAGAGGATCCTGATGGAAGCAAACTTAGCCATG-3′ for all constructs. The PDF1.2-promoter deletions were cloned into the HindIII-BamHI site of the binary vector pBI101.3 (CLONTECH, Hampshire, UK), in frame with the UidA gene, using standard molecular biology techniques (Sambrook et al., 1989). The pA35S construct was made by amplifying the -46 to +10 region of the CaMV 35S promoter using the primer pair 5′-CACTATGTCGACCAAGACCTTTCCTCTATATAAG-3′ and 5′-CCCGGGGATCCGTCCTCTCAAAATGAATGGAAC-3′. Amplification product was digested with SalI and BamHI and inserted in front of the UidA gene in pBI101.3 also digested with SalI and BamHI. This construct is termed pB35S. The transgene cassette in pB35S was then removed by digesting HindIII and EcoRI and inserted into HindIII/EcoRI digested binary vector pAOV, which carries a gene for Basta resistance (Mylne and Botella, 1998). The pIB construct was made by amplifying the -270 to -250 region of the PDF1.2 promoter using primers 5′-TTAATCAAGCTTGATTAACCAGCCGCC-3′ and 5′-TGCTACGTCGACCACATGGGCGGCTGG-3′. Amplification product was then digested with SalI and HindIII and ligated onto SalI/HindIII-digested pB35S, which was called pB6. The transgene cassette in pB6 was then removed by digesting with HindIII/EcoRI and cloned into HindIII/EcoRI-digested pAOV. The mGCC construct was generated via a two-step PCR process using primer pairs: (a) 5′-ATCAAAGCTTTGCTGCTCTTGAGATCAACC-3′ and 5′-CGTTCACATGTGAGGATGGTTAATC-3′ and (b) 5′-GATTAACCATCCTCACATGTGAACG-3′ and 5′-GAGAGAGGATCCTGATGGAAGCAAACTTAGCCATG-3′. The amplification product was digested with BamHI and HindIII and was cloned into BamHI/HindIII-digested binary vector PBI101.3. The transgene cassette was then removed from this plasmid by HindIII/EcoRI digest and cloned into HindIII/EcoRI digested pAOV. Plasmids were introduced into Escherichia coli DH5-α cells via heat shock transformation (Sambrook et al., 1989). The fusion constructs were verified by sequencing using the antisense primer (5′-CAGTTTTCGCGATCCAGACTG-3′) for the UidA gene. Binary vectors were transferred to Agrobacterium tumefaciens via triparental mating. The procedure outlined by Valvekens et al. (1988) was used for the production of transgenic Arabidopsis plants of C24 ecotype, and transformants were selected based on kanamycin resistance. The mGCC, pIB, pA35, and an additional P3 construct were introduced into Col-0 plants by using the flower-dip transformation procedure (Clough and Bent, 1998).
Plant Treatments for Promoter Activity Assays
For sterile-grown plants, Arabidopsis seeds were surface sterilized and evenly positioned across petri dishes containing germination medium (Valvekens et al., 1988) at a density of 70 seeds per plate. Seeds were stratified at 4°C for 3 d, and the petri dishes were transferred to growth cabinets with a 12-h photoperiod, at 24°C. Induction experiments were carried out on 16-d-old seedlings. For soil-grown plants, seeds were germinated on germination medium in petri dishes, and 10-d-old seedlings were transferred to soil, with five individuals planted in each 4- × 4-cm pot for induction experiments using exogenous chemicals. For ease of inoculation, only four individuals were transplanted per pot for experiments when plants were challenged by the fungal pathogen Alternaria brassicicola. Plants were grown under short days (8-h photoperiod) to delay bolting. Daytime temperature was 21°C with 70% humidity, and nighttime temperature was 24°C, with 60% humidity. Induction experiments were performed on 5- to 6-week-old plants.
For plant treatments, the plants pots or tissue culture plates were placed into 1-L phytocon vessels (Sigma-Aldrich, St. Louis), which were sealed around the rim with plasticine. Ethylene was applied at a concentration of 200 μL L-1 by injection of gaseous ethylene through a rubber septum. MeJA was applied at a concentration of 0.1 μmol L-1 air, by inoculation of 5 μL of a 21.8 mm solution of MeJA (in ethanol) to a ball of cotton wool taped to the inside of the container. Control plants were placed in identical chambers, but without the addition of inducing agents. Samples were collected after 24 h of incubation in continuous light. For the seedling-based assay, 20 individual whole transgenic plants (T2 generation) were bulked for each replicate sample. For each treatment, three replicate sets of samples were taken from each of two independent experiments. Five plants were bulked for each replicate in the mature plant assays. MeJA treatment of plants used in additional real-time quantitative RT-PCR time-course experiments was done according to Schenk et al. (2000).
Fungal Inoculation
Leaves of soil-grown plants were inoculated with 5-μL drops of a freshly harvested conidial spore suspension (5 × 105 spores mL-1 in water) of A. brassicicola (isolate UQ4273). The plants were placed in a propagator flat under a clear polystyrene lid to maintain high humidity. The treated leaves were collected separately from the untreated leaves of the same plant, 5 h after inoculation.
Quantitative GUS Assays for Promoter Activity
Transgenic plants at T2 generation (20 individuals) stably transformed with each promoter reporter construct were used. Promoter activity was assessed by quantitative GUS assays of the UidA gene, using 4-methyl-umbelliferyl-β-d-glucuronide (Sigma-Aldrich) as a substrate. The assay was carried out according to the procedure of Jefferson (1987), adapted for use with microtitre plates and the Fluoroskan Ascent fluorometer (Labsystems, Helsinki). The total protein content of samples was assessed using the dye-binding protein assay adapted for microtitre plates (Bio-Rad Laboratories, Hercules, CA).
Transient Expression Assays
The pBINm-GFP5-ER plasmid (Haseloff et al., 1997) containing a synthetic GFP gene under the control of the 35S promoter (CaMV) was used as a transformation control in all experiments. The pSP35Slucnos plasmid containing the firefly luciferase gene under the control of the 35S promoter (CaMV) was a gift from Dr. Robert Birch (Department of Botany, The University of Queensland, Australia). PCR amplified PDF1.2-promoter deletion fragments using the forward primers 5′-AACAAGCTTTTTAGTCCACAACG-3′, 5′-TGTAAGCTTATGACGAAGGTC-3′, and 5′-AAGCTTGACTATGAACTGC-3′ for constructs Ea, Eb, and Ec, respectively, and a common reverse primer 5′-GAGAGAGGATCCTGATGGAAGCAAATTAGCCATG-3′ for all constructs was cloned, in frame with the UidA gene, into the 4.87-kb vector pGEM4.GUS3 using standard molecular biology techniques (Sambrook et al., 1989). Bombardments were carried out using a custom-made helium pressure-driven particle inflow gun as previously described (Schenk et al., 1998). GFP-expressing cells in the bombarded leaves were viewed using an MZ6 stereomicroscope with fluorescence GFP Plus filter module (Leica Microscopy and Scientific Instruments, Bannockburn, IL), containing a 480/40-nm excitation filter, a 505-nm long pass dichromatic beam-splitting mirror, and a 510-nm long pass barrier filter. Luciferase activity was detected using Luciferase Assay Reagent (Promega, Madison, WI) in a Fluoroskan Ascent FL luminometer (Labsystems).
Macroarray Analysis
The consensus amino acid sequence for the DNA-binding domain of the AP2/ERF transcription factor family (YRGV RRPW GKWA AEIR DPKR VWLG TFTA EEAA RAYD AARG KALN FP; Riechmann and Meyerowitz, 1998) was used as a query sequence in a BLASTP search of the nonredundant GenBank protein database, limited to Arabidopsis. Subfamily allocation was determined by searching for the conserved linker sequence (E-V—LRR-S—F-RG; Klucher et al., 1996) occurring between the two conserved AP2 domains of AP2 subfamily members. ClustalW was used to analyze sequence similarity within the conserved domain and to produce a guide tree of relatedness. For macroarray construction, genomic DNA was extracted from Arabidopsis ecotype Col-0 plants using the CTAB method. The following forward and reverse primers sequences were used for amplification of divergent regions of AP2/ERF sequences from the Arabidopsis genome: for ERF1 (At3g23240), 5′-CGGCGGAGAGAGTTCAAGAGTC-3′ and 5′-TCCCACTATTTTCAGAAGACCCC-3′; for AtERF2 (At5g47220), 5′-TTGGGGAGGTTTGCCATTG-3′ and 5′-TCCATCGCCGTAAAGTTCTCAG-3′; for AtERF3 (At1g50640), 5′-CCGACCATCAACAACAGTTCCC-3′ and 5′-CGACGAATCGCAATCGCTG-3′; for EREBP4/RAP2.5/AtERF4 (At3g15210), 5′-TCTACTTTTTGGACCTGATGGGG-3′ and 5′-TCGCTTTGGGCACCACAAG-3′; for AtERF5 (At5g47230), 5′-ACGAGGCTTCTCCTGTGGCTAC-3′ and 5′-GCGAAATCTTCAATGGCGG-3′; for putative EREBP (At2g44970), 5′-CGATAACTGGAGCGACTTGCC-3′ and 5′-GGAGAGGTAACGGGAGGAACG-3′; for TINY isolog (At1g22810), 5′-TCCTTCACTCGTTTCCAGAACTTC-3′ and 5′-GCCGCCCAGATAATCATACACTG-3′; for gene similar to tobacco EREBP3 (At1g12980), 5′-AGAAATCTTCGCCGTCTGCTC-3′ and 5′-TCTTGATACCCCCACTCGTTTG-3′; for AP9 (At1g80580), 5′-AACCCGAAAACACGCATC-3′ and 5′-GCTTCACACCACCTTCTTTGG-3′; for RAP2.10 (At4g36900), 5′-AGGAGGAGTGAACGGTGGTG-3′ and 5′-GATGATGATGATGACGATTCCC-3′; for putative AP2 (At1g63030), 5′-ATTCTGCTTGGAGGTTGCCG-3′ and 5′-CTCCAAAGTGACAAATCTTC-3′; for putative AP2 (At2g35700), 5′-CATCTCCCACAGTTACGGAAAC-3′ and 5′-AAAAGAGTCCCAGAAGCCATC-3′; for putative AP2 (At1g63040), 5′-TCTTACCCCATTCCCCTTTCC-3′ and 5′-AGCCACCATCATCCCTTTGG-3′; for putative AP2 (At2g46310), 5′-CGAGGGAATCGCAAATCAGTC-3′ and 5′-CGAGAAACAAAGGGTCAGGGG-3′; for AP2 transcription factor (At2g40340), 5′-CAAGTTCAGGTTTTGGTCAGGTG-3′ and 5′-GCAATCTCCATAGGGTTGAGGC-3′; for putative AP2 (At2g22200), 5′-TGAATCCTCTCCCTTCCTCTGTTG-3′ and 5′-CATCGCTTCTCGGTGACTCATTAG-3′; for putative protein (At4g31060), 5′-AGGCAACCAAAAAGTGGCG-3′ and 5′-CAACCATTCCGTGTTCTCCATC-3′; and for APETALA2 (At4g36920), 5′-AACGACGCACCACACCAAAC-3′ and 5′-AAAGCCAGAAGCAACACCACC-3′. For normalization purposes, the macroarray filters also contained DNA sequences from the following genes amplified using the following primer pairs: PR-1, 5′-CGGCGACAAGACTACCTTGATG-3′ and 5′-TGGCACATCCAACCCACTCTG-3′; PDF1.2, 5′-CGCACCGGCAATGGTGGAAG-3′ and 5′-CACACGATTTAGCACCAAAG-3′; BGL-2, 5′-GTAAAACGACGGCCAGT-3′ and 5′-GGAAACAGCTATGACCATG-3′; UidA, 5′-ACCCTTACGCTGAAGAGATGC-3′, and 5′-AACGTATCCACGCCGTATTCG-3′; Actin, 5′-GGATGCTTATGTTGGCGATG-3′ and 5′-GCTGGTTTTGGCTGTCTCG-3′; and Tubulin β-8 chain, 5′-GTAAAACGACGGCCAGT-3′ and 5′-GGAAACAGCTATGACCATG-3′. Macroarray filters were prepared as follows. Probe DNA (40 ng spot-1) was denatured and transferred in duplicate to a Hybond N+ membrane (Amersham Biosciences, Piscataway, NJ) using the Bio-Dot microfiltration apparatus (Bio-Rad Laboratories), according to the manufacturer's instructions. Total cDNA synthesis was performed using 750 ng of purified poly(A+) RNA using Superscript II RT (Invitrogen, Carlsbad, CA) as recommended by the manufacturer. The total cDNA prepared was labeled either using the Megaprime DNA labeling system (Amersham Biosciences) or the Strip-EZ system (Ambion, Austin TX) as instructed by the manufacturer. Blots were hybridized as previously described (Mitter et al., 1998) and were visualized by exposure to a PhosporImaging system, and data were processed with the ImageQuant analysis software (Molecular Dynamics, Sunnyvale, CA). For each hybridization blot, a reference figure was calculated by averaging the hybridization signal from the duplicate β-tubulin spots minus the background. Data for probes from all other genes were determined similarly and then divided by the β-tubulin reference for that blot to allow comparison of relative expression levels of the different genes.
Northern-Blot Analysis
Arabidopsis ecotype Col-0 plants were grown under short-day conditions (8 of h light, 16 h of dark) for 3 weeks and treated with MeJA, ethylene, or nothing (controls), as described above for GUS activity assays. Total cellular RNA was extracted from the aerial portion of the plants using the guanidine thiocyanate method and a 20-μg sample of total RNA was run on the gel and blotted onto a Hybond N+ membrane (Amersham Biosciences). Probes were amplified by PCR and purified using the Qiaquick PCR purification kit (Qiagen USA, Valencia, CA). The ERF probes were amplified using the primer sequences given above. The PDF1.2 probe was amplified from Clone 10a (Manners et al., 1998) using the primers 5′-CGCACCGGCAATGGTGGAAG-3′ and 5′-CACACGATTTAGCACCAAAG-3′. The tubulin β-8 chain probe was amplified from EST clone number 108K21T7 (GenBank accession no. T41808; Arabidopsis Biological Resource Center, Columbus, OH) using the M13 forward and M13 reverse primers. The hybridizations and analysis of hybridization signals were done as described for macroarray hybridizations.
Real-Time Quantitative RT-PCR Analysis
Total cellular RNA was extracted from the aerial portion of the plants using Plant RNA extraction kit (Bio-Rad Laboratories). First-strand cDNA was synthesized using 2 μg of total RNA, Superscript II RT (Invitrogen), and random hexamer primers to a total volume of 10 μL. The reaction mix was incubated at 42°C for 50 min, before heat inactivation of the enzyme at 70°C for 15 min. The cDNA template was then diluted by adding 240 μL of milli-Q water, to give a final concentration of 8 ng μL-1 for use in real-time PCR assays. A second 2-μg aliquot of each RNA sample was subjected to the same conditions, without the addition of RT (the minus RT controls). Real-time PCR amplification and detection was carried out in an ABI model 7700 Sequence Detection System (Applied Biosystems, Foster City, CA). Reactions were in final volumes of 25 μL and contained 40 ng of cDNA template and 5 μL of primer mix containing a 1 μm solution of each of the AtERF2 primers, 5′-TGAGGTTAATTCCGGTGAACC-3′ and 5′-TCAACTTCCCGTTTTCAGACGA-3′; ERF1 primers, 5′-CGAGAAGCTCGGGTGGTAGT-3′ and 5′-GCCGTGCATCCTTTTCC-3′; PDF1.2 (At5g44420) primers, 5′-TTGCTGCTTTCGACGCA-3′ and 5′-TGTCCCACTTGGCTTCTCG-3′; Thi2.1 (L41244.1) primers (Epple et al., 1995), 5′-CTCAGCTGATGCTACCAATGAGC-3′ and 5′-GCTCCATTCACAATTTCACTTGC-3′; or PR4 (At3g12500) primers, 5′-ATCAGCGCTGCAAAGTCCTTC-3′ and 5′-GTGCTGTAGCCCATCCACCTG-3′; and 12.5 μL of SYBR Green PCR master mix (Applied Biosystems). The thermal cycling conditions were: 10 min at 95°C, 45 cycles of 15 s at 95°C and 1 min at 60°C. Duplicate SYBR Green assays for each gene target were performed on both plus and minus RT samples. No-template controls for each primer pair were included in each run. Expression detected from three β-actin genes of Arabidopsis, β-actin-2 (At3g18780), β-actin-7 (At5g09810), and β-actin-8 (At1g49240) used the following mixture of primers (R. Simpson, personal communication; a universal actin forward primer 5′-AGTGGTCGTACAACCGGTATTGT-3′ and specific reverse primers 5′-GATGGCATGGAGGAAGAGAGAAAC-3′, 5′-GAGGAAGAGCATTCCCCTCGTA-3′, and 5′-GAGGATAGCATGTGGAACTGAGAA-3′, respectively) were used as combined internal standards to normalize small differences in template amounts. Real-time PCR results were captured and analyzed using the Sequence Detector Software (SDS v1.7, Applied Biosystems) and data analysis procedures described in the User Bulletin 2 for the ABI model 7700 Sequence Detection System.
Construction of the 35S-AtERF2 Binary Vector and Plant Transformation
The AtERF2 (AT5g47220) gene of Arabidopsis was amplified from the Col-0 ecotype using the primers 5′-CTGAAAATGTACGGACAGTGC-3′ and 5′-AACTTATGAACCAATAACTC-3′. The amplification product was treated with 1 unit of polynucleotide kinase (Roche Diagnostics, Mannheim, Germany), ligated into the binary vector pKEN cut by HindIII, filled in by Klenow DNA polymerase (Roche Diagnostics), and dephosphorylated. pKEN was constructed by cloning a SacI/XhoI fragment containing the double 35S promoter/terminator cassette from the plasmid pJIT163 (Hellens et al., 2000) into SacI/XhoI-digested pGreen229 (Hellens et al., 2000). The recombinant plasmid was electroporated into competent A. tumefaciens (strain AGL-1) cells previously transformed with pSoup plasmid (Hellens et al., 2000). Arabidopsis Col-0 plants were then transformed using the floral dip transformation procedure (Clough and Bent, 1998), and the transformants were selected based on their resistance to Basta.
Statistical Analysis of Data
The JMP IN Statistics package v3.2.6 (SAS Institute Inc., Cary, NC) was used for all statistical analyses. The significance of differences between transgenic plants carrying the various promoter deletion constructs and independent transgenic lines within these constructs was determined by nested ANOVA. The non-parametrical Wilcoxen-Mann-Whitney test was used to determine statistical significance of differences between constructs in bombardment experiments. Biological significance was assigned at P < 0.05.
Acknowledgments
We thank Dr. John Turner and the Arabidopsis Stock Center at Ohio State University for cev1 and cpr5 seeds, respectively; Jonathan Anderson for cpr5 cDNA samples; Dr. Peer Schenk for advice in plasmid construction and some of the cDNA samples; Anca Rusu for GUS activity analyses of some of the promoter-reporter lines; Dr. Philip Mullineaux for the pGreen0229, pSoup, and pJIT163 plasmids; Dr. Robert Birch for the pSP35Slucnos plasmid; Dr. Jimmy Botella for the pAOV plasmid; Dr. Jim Haseloff for the pBINm-GFP5-ER plasmid; and Dr. Willem Broekaert for his advice on plant treatments and collaboration.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.017814.
This work was supported by the Grains Research and Development Corporation (postgraduate fellowship to R.L.B.).
References
- Berrocal-Lobo M, Molina A, Solano R (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29: 23-32 [DOI] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu YD, Klessig DF, Dong XN (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573-1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner M, Singh KB (1997) Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC-box DNA-binding protein interacts with an ocs element binding protein. Proc Natl Acad Sci USA 94: 5961-5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier B, Champion A, Henry Y, Kreis M (2002) Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol 130: 577-590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Chang H-S, Gupta R, Wang X, Tong Z, Luan S (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol 129: 661-667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation. Plant J 16: 735-743 [DOI] [PubMed] [Google Scholar]
- Dong X (1998) SA, JA, ethylene, and disease resistance in plants. Curr Opin Plant Biol 1: 316-323 [DOI] [PubMed] [Google Scholar]
- Ellis C, Turner JG (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13: 1025-1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple P, Apel K, Bohlmann H (1995) An Arabidopsis thaliana thionin gene is inducible via a signal transduction pathway different from that for pathogenesis-related proteins. Plant Physiol 109: 813-820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC-box-mediated gene expression. Plant Cell 12: 393-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmig B, Gonzalez-Perez MN, Leubner-Metzger G, Vogeli-Lange R, Meins F, Hain R, Penuelas J, Heidenreich B, Langebartels C, Ernst D et al. (2003) Ozone-induced gene expression occurs via ethylene-dependent and -independent signalling. Plant Mol Biol 51: 599-607 [DOI] [PubMed] [Google Scholar]
- Gu YQ, Wildermuth MC, Chakravarthy S, Loh YT, Yang C, He X, Han Y, Martin GB (2002) Tomato transcription factors Pti4, Pti5, and Pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14: 817-831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YQ, Yang CM, Thara VK, Zhou JM, Martin GB (2000) Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12: 771-785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao D, Ohme-Takagi M, Sarai A (1998) Unique mode of GCC-box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J Biol Chem 273: 26857-26861 [DOI] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94: 2122-2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacteriummediated plant transformation. Plant Mol Biol 42: 819-832 [DOI] [PubMed] [Google Scholar]
- Hwang YS, Karrer EE, Thomas BR, Chen L, Rodriguez RL (1998) Three cis-elements required for rice alpha-amylase Amy3D expression during sugar starvation. Plant Mol Biol 36: 331-341 [DOI] [PubMed] [Google Scholar]
- Izawa T, Foster R, Chua NH (1993) Plant bZIP protein DNA binding specificity. J Mol Biol 230: 1131-1144 [DOI] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene-fusion system. Plant Mol Biol Rep 5: 387-405 [Google Scholar]
- Kagaya Y, Ohmiya K, Hattori T (1999) RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res 27: 470-478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SR, Choi JL, Costa MA, An GH (1992) Identification of G-box sequence as an essential element for methyl jasmonate response of potato proteinase inhibitor II promoter. Plant Physiol 99: 627-631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8: 137-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sanchez-Serrano JJ, Solano R (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manners JM, Penninckx I, Vermaere K, Kazan K, Brown RL, Morgan A, Maclean DJ, Curtis MD, Cammue BPA, Broekaert WF (1998) The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol Biol 38: 1071-1080 [DOI] [PubMed] [Google Scholar]
- Mason HS, Dewald DB, Mullet JE (1993) Identification of a methyl jasmonate-responsive domain in the soybean vspB promoter. Plant Cell 5: 241-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazarei M, Puthoff DP, Hart JK, Rodermel SR, Baum TJ (2002) Identification and characterization of a soybean ethylene-responsive element-binding protein gene whose mRNA expression changes during soybean cyst nematode infection. Mol Plant-Microbe Interact 15: 577-586 [DOI] [PubMed] [Google Scholar]
- Medina J, Bargues M, Terol J, Perez-Alonso M, Salinas J (1999) The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol 119: 463-469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memelink J, Verpoorte R, Kijne JW (2001) ORCAnization of jasmonate-responsive gene expression in alkaloid metabolism. Trends Plant Sci 6: 212-219 [DOI] [PubMed] [Google Scholar]
- Menke FLH, Champion A, Kijne JW, Memelink J (1999a) A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J 18: 4455-4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke FLH, Parchmann S, Mueller MJ, Kijne JW, Memelink J (1999b) Involvement of the octadecanoid pathway and protein phosphorylation in fungal elicitor-induced expression of terpenoid indole alkaloid biosynthetic genes in Catharanthus roseus. Plant Physiol 119: 1289-1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter N, Kazan K, Way HM, Broekaert WF, Manners JM (1998) Systemic induction of an Arabidopsis plant defensin gene promoter by tobacco mosaic virus and jasmonic acid in transgenic tobacco. Plant Sci 136: 169-180 [Google Scholar]
- Mylne J, Botella J (1998) Binary vectors for sense and antisense expression of Arabidopsis ESTs. Plant Mol Biol Rep 16: 257-262 [Google Scholar]
- Nishiuchi T, Suzuki K, Kitajima S, Sato F, Shinshi H (2002) Wounding activates immediate early transcription of genes for ERFs in tobacco plants. Plant Mol Biol 49: 473-482 [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of Class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959-1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Ohme-Takagi M, Shinshi H (2000) Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J 22: 29-38 [DOI] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Singh KB (2002) Identification of Arabidopsis ethylene-responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiol 128: 1313-1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH (2001) Overexpression of the Tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13: 1035-1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IAMA, Eggermont K, Terras FRG, Thomma BPHJ, De Samblanx GW, Buchala A, Metraux JP, Manners JM, Broekaert WF (1996) Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8: 2309-2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IAMA, Thomma BPHJ, Buchala A, Metraux JP, Broekaert WF (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10: 2103-2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG, Maggio A, Narasimhan ML, Kononowicz AK, Wang G, D'Urzo MP, Hasegawa PM, Bressan RA (1997) Tissue-specific activation of the osmotin gene by ABA, C2H4 and NaCl involves the same promoter region. Plant Mol Biol 34: 393-402 [DOI] [PubMed] [Google Scholar]
- Reymond P, Farmer EE (1998) Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol 1: 404-411 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM (1998) The AP2/EREBP family of plant transcription factors. Biol Chem 379: 633-646 [DOI] [PubMed] [Google Scholar]
- Rouster J, Leah R, Mundy J, Mills CV (1997) Identification of a methyl jasmonate-responsive region in the promoter of a lipoxygenase 1 gene expressed in barley grain. Plant J 11: 513-523 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi- Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998-1009 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis TA (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schenk PM, Elliott AR, Manners JM (1998) Assessment of transient gene expression in plant tissues using the green fluorescent protein as a reference. Plant Mol Biol Rep 16: 313-322 [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97: 11655-11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G, Meller Y, Fluhr R (1995) A GCC element and a G-box motif participate in ethylene-induced expression of the PRB-1b gene. Plant Mol Biol 28: 145-153 [DOI] [PubMed] [Google Scholar]
- Shah J, Klessig DF (1996) Identification of a salicyclic acid-responsive element in the promoter of the tobacco pathogenesis-related beta-1,3-glucanase gene, PR-2d. Plant J 10: 1089-1101 [DOI] [PubMed] [Google Scholar]
- Shinshi H, Usami S, Ohme-Takagi M (1995) Identification of an ethylene-responsive region in the promoter of a tobacco class I chitinase gene. Plant Mol Biol 27: 923-932 [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: A transcriptional cascade mediated by ethylene-insensitive3 and ethylene-response-factor1. Genes Dev 12: 3703-3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Yuen GY, Lehman CC (1998) Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J 15: 747-754 [DOI] [PubMed] [Google Scholar]
- Thara VK, Tang X, Gu YQ, Martin GB, Zhou JM (1999) Pseudomonas syringae pv tomato induces the expression of tomato EREBP-like genes Pti4 and Pti5 independent of ethylene, salicylate and jasmonate. Plant J 20: 475-483 [DOI] [PubMed] [Google Scholar]
- Thomma BP, Eggermont K, Tierens KF, Broekaert WF (1999) Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol 121: 1093-1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Penninckx IA, Broekaert WF, Cammue BP (2001) The complexity of disease signaling in Arabidopsis. Curr Opin Immunol 13: 63-68 [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Cammue BPA, Thevissen K (2002) Plant defensins. Planta 216: 193-202 [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95: 15107-15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M (1988) Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85: 5536-5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L, Memelink J (2000) ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289: 295-297 [DOI] [PubMed] [Google Scholar]
- van der Fits L, Memelink J (2001) The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J 25: 43-53 [DOI] [PubMed] [Google Scholar]
- Vijayan P, Shockey J, Levesque CA, Cook RJ, Browse J (1998) A role for jasmonate in pathogen defense of Arabidopsis. Proc Natl Acad Sci USA 95: 7209-7214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Tian L, Hollingworth J, Brown DCW, Miki B (2002) Functional analysis of tomato Pti4 in Arabidopsis. Plant Physiol 128: 30-37 [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Suzuki K, Shinshi H (1999) Elicitor-responsive, ethylene-independent activation of GCC-box-mediated transcription that is regulated by both protein phosphorylation and dephosphorylation in cultured tobacco cells. Plant J 20: 571-579 [DOI] [PubMed] [Google Scholar]
- Zhou D (1999) Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci 4: 210-214 [DOI] [PubMed] [Google Scholar]
- Zhou JM, Tang XY, Martin GB (1997) The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J 16: 3207-3218 [DOI] [PMC free article] [PubMed] [Google Scholar]