Abstract
In plants, Na+/H+ exchangers in the plasma membrane are critical for growth in high levels of salt, removing toxic Na+ from the cytoplasm by transport out of the cell. The molecular identity of a plasma membrane Na+/H+ exchanger in Arabidopsis (SOS1) has recently been determined. In this study, immunological analysis provided evidence that SOS1 localizes to the plasma membrane of leaves and roots. To characterize the transport activity of this protein, purified plasma membrane vesicles were isolated from leaves of Arabidopsis. Na+/H+ exchange activity, monitored as the ability of Na to dissipate an established pH gradient, was absent in plants grown without salt. However, exchange activity was induced when plants were grown in 250 mm NaCl and increased with prolonged salt exposure up to 8 d. H+-coupled exchange was specific for Na, because chloride salts of other monovalent cations did not dissipate the pH gradient. Na+/H+ exchange activity was dependent on Na (substrate) concentration, and kinetic analysis indicated that the affinity (apparent Km) of the transporter for Na+ is 22.8 mm. Data from two experimental approaches supports electroneutral exchange (one Na+ exchanged for one proton): (a) no change in membrane potential was measured during the exchange reaction, and (b) Na+/H+ exchange was unaffected by the presence or absence of a membrane potential. Results from this research provide a framework for future studies into the regulation of the plant plasma membrane Na+/H+ exchanger and its relative contribution to the maintenance of cellular Na+ homeostasis during plant growth in salt.
Na+/H+ exchangers or antiporters are transport proteins that move Na+ either out of cells or into organelles in exchange for H+ (Blumwald et al., 2000; Padan et al., 2001). These transporters play a major role in cellular pH and Na+ homeostasis (Padan et al., 2001; Wiebe et al., 2001). The exchangers were first described by West and Mitchell (1974) and have since been shown to exist widely in both prokaryotic and eukaryotic cells (Blumwald et al., 2000; Padan et al., 2001).
In recent years, significant advances have been made toward understanding the structure and function of Na+/H+ exchangers as well as the mechanisms underlying the exchange reaction and its regulation (Schachtman and Liu, 1999; Blumwald, 2000; Blumwald et al., 2000; Hasegawa et al., 2000a, 2000b; Padan et al., 2001). Genes encoding Na+/H+ exchangers have been cloned from bacterial, yeast, animal, and plant cells (Gaxiola et al., 1999; Counillon and Pouyssegur, 2000; Padan et al., 2001). These transporters are composed of a single polypeptide chain with a molecular mass ranging from 56 to 100 kD. They typically have 12 transmembrane segments with the C- and N-terminal ends of the protein predicted to be toward the cytoplasm. Depending on the organism, the exchange reactions have been found to be electroneutral or electrogenic, exchanging one or more H+ for one Na+ (Wiebe et al., 2001). Recent studies of the molecular mechanisms of Na+/H+ exchange have identified specific amino acid residues that play critical roles in the pH sensitivity of the Na+/H+ exchangers and are important for the transport of Na+ and H+ (Wiebe et al., 2001). Genetic and biochemical studies have provided evidence that Na+/H+ exchangers are regulated by phosphorylation, Ca2+/calmodulin, calcineurin, HSP70, G-proteins, and lipids (Orlowski and Grinstein, 1997; Counillon and Pouyssegur, 2000).
In plants, Na+/H+ exchangers have been characterized at the vacuolar and plasma membranes where they function to remove Na+ from the cytoplasm by transport into the vacuole or out of the cell to prevent toxic cellular accumulations of Na+ (Blumwald, 2000; Blumwald et al., 2000; Hasegawa et al., 2000a). These transport proteins are believed to be critical components of plant salt tolerance; evidence to support this comes from overexpression studies of the vacuolar exchanger AtNHX1 in yeast and plants (Apse et al., 1999; Darley et al., 2000; Quintero et al., 2000; Zhang and Blumwald, 2001; Zhang et al., 2001; Venema et al., 2002).
Although transport studies provided evidence for Na+/H+ exchangers in the plasma membrane of numerous plants (Ratner and Jacoby, 1976; Watad et al., 1986; Allen et al., 1995), the proteins involved had not been identified at the molecular level until recently. In a genetic screen designed to identify components of the cellular machinery that contribute to salt tolerance in Arabidopsis, three salt-overly sensitive genes (SOS1, SOS2, and SOS3) were found to function in a common pathway (Wu et al., 1996; Liu and Zhu, 1997; Zhu et al., 1998; Zhu, 2000). One of these genes, SOS1, was shown to encode a 127-kD membrane protein with 12 putative membrane-spanning domains and a long hydrophilic tail at the C-terminal end of the protein (Shi et al., 2000). The predicted membrane-spanning domains in the SOS1 protein display significant similarity to domains of the plasma membrane-localized Na+/H+ exchangers from animal, bacterial, and fungal cells. Recent studies have demonstrated that Na+/H+ exchange activity was significantly reduced in plasma membrane vesicles isolated from sos1 plants relative to activity in wild-type plants providing direct evidence that SOS1 encodes a plant plasma membrane Na+/H+ exchanger (Qiu et al., 2002). In addition, increases in plasma membrane Na+/H+ exchange activity in vesicles isolated from sos2 and sos3 plants in the presence of constitutively active recombinant (activated) SOS2 protein added in vitro have shown that SOS2 (a protein kinase; Halfter et al., 2000) and SOS3 (a myristoylated calcium-binding protein; Liu and Zhu, 1998) regulate SOS1 transport activity (Qiu et al., 2002).
In the present study, we have used plasma membrane-enriched vesicles from Arabidopsis leaves to monitor Na+-induced dissipation of an ATP-dependent pH gradient to characterize plasma membrane Na+/H+ exchange (SOS1) activity. Initial rates of Na+/H+ exchange as a function of substrate concentration were determined and compared in vesicles isolated from wild-type and mutant plants in the absence and presence of activated SOS2 protein added in vitro. Ion specificity and the effect of membrane potential on exchange activity were investigated and the membrane localization of the exchanger was confirmed.
RESULTS
SOS1 Localizes to the Plasma Membrane in Arabidopsis
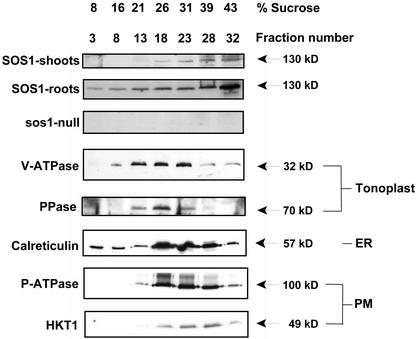
SOS1 has been shown to localize to the plasma membrane in green fluorescent protein-SOS1 transformed Arabidopsis (Shi et al., 2002). To confirm the subcellular localization of the Na+/H+ exchanger, microsomal membranes from wild-type and sos1 Arabidopsis plants were fractionated. Centrifugation through a continuous Suc gradient was first used to compare the distribution of the exchanger (SOS1) with that of markers for the vacuolar and plasma membranes and the endoplasmic reticulum (Fig. 1). When membrane fractions of increasing density were assayed for SOS1 accumulation, a protein of approximately 127 kD increased in both shoot and root tissue as the density increased (most abundant in fractions 28 and 32 corresponding to 39% and 43% [w/v] Suc, respectively). SOS1 was more abundant in roots than shoots as previously observed for SOS1 mRNA (Shi et al., 2000); however, the pattern of localization was the same in both tissues. As expected, no protein was detected in the corresponding fractions from sos1 plants. SOS1 distribution did not correspond to the peak fractions for any of the vacuolar membrane markers (vacuolar H+-ATPase and vacuolar H+-pyrophosphatase). Rather, SOS1 localized to higher density fractions. To determine whether these fractions contained mitochondrial membranes, cytochrome c oxidase activity was measured throughout the gradient, mitochondria were purified, and the membrane proteins were screened with the SOS1 antibody. Cytochrome c oxidase activity was not detected in the gradient fractions, and no SOS1 protein was present in the purified mitochondrial membrane protein preparations (data not shown) demonstrating that SOS1 does not localize to the mitochondrion. SOS1 localized to fractions enriched in plasma membranes and rough endoplasmic reticulum as indicated by sedimentation profiles overlapping with resident proteins from these membranes (P-ATPase; HKT1, plasma membrane K+/Na+ cotransporter; Calreticulin, rough endoplasmic reticulum). The fact that SOS1 peaked toward the upper end of the plasma membrane-enriched fractions suggests that this novel transporter may be associated with a more dense population of plasma membrane vesicles. The SOS1 antibody also recognized a protein of about 127 kD in the upper phase of aqueous two phase preparations from leaves of plants treated with NaCl (data not shown).
Figure 1.
Membrane localization of the Arabidopsis SOS1 protein. Microsomal membranes from plants grown in the absence of salt were fractionated over 5% to 45% (w/v) Suc gradients, and 1-mL fractions were collected. Membrane protein (18 μg) from the indicated fractions was separated by 10% (w/v) SDS-PAGE and transferred to nitrocellulose membranes. Unless indicated, immunological detection was carried out on fractions isolated from shoots of wild-type plants. Antibodies used from top to bottom: SOS1, SOS1 (membrane protein isolated from roots), SOS1 (membrane protein isolated from shoots of sos1 mutant plants), V-ATPase (VMA-E, vacuolar H+-ATPase E-subunit; Dietz and Arbinger, 1996), PPase (vacuolar H+-pyrophosphatase; PAB-HK antibody; Kim et al., 1994), Calreticulin (Nelson et al., 1997), P-ATPase (plasma membrane H+-ATPase; PMA1 isoform; Pardo and Serrano, 1989), and HKT1 (plasma membrane K+/Na+ cotransporter; Su et al., 2003). Bands were detected by chemiluminescence. Molecular masses of the bands are indicated.
Transport-Competent Plasma Membrane Vesicles Can Be Isolated from Arabidopsis
For transport studies, plasma membrane vesicles were isolated and purified from leaves of Arabidopsis using aqueous two-phase partitioning. The purity of these membrane preparations was examined by assaying ATPase activity (substrate hydrolysis) in the absence and presence of a number of inhibitors. Using plants that had been treated with 250 mm NaCl for 3 d, ATPase activity was measured in microsomes and in membranes collected from the lower and upper phases after partitioning. As shown in Table I, vanadate (an inhibitor of the plasma membrane H+-ATPase) reduced ATPase activity 45% in upper phase membranes. However, azide and nitrate (inhibitors of mitochondrial and vacuolar ATPases, respectively) and molybdate (an inhibitor of nonspecific phosphatases) had little effect on ATPase activity in this fraction. High levels of sensitivity to all inhibitors in both the microsomal and lower phases suggested that a mixture of membranes was present in these fractions. A characteristic feature of the plant plasma membrane H+-ATPase is the in vitro stimulation of its hydrolytic activity by K+ at acidic pH (Vara and Serrano, 1982; Sze, 1985). This cation stimulation provides an additional method to distinguish the activity of the plasma membrane H+-ATPase from that of the vacuolar H+-ATPase (stimulated by anions) and was used as another indicator of the contribution of the plasma membrane to the upper phase membranes used in this study. When K+ was added to the assay, activity was stimulated more than 8-fold (0.28 versus 2.48 μmol Pi mg-1 protein min-1 in the absence and presence of K+, respectively). Taken together, these results demonstrate that upper phase preparations isolated from Arabidopsis are enriched in plasma membranes without any significant contamination from other cellular membranes.
Table I.
Determination of vesicle purity and sidedness
Membrane vesicles were isolated by two-phase partitioning from leaves of wild-type Arabidopsis treated with 250 mm NaCl for 3 d. ATPase activity was assayed as described in “Materials and Methods.” Numbers in parentheses represent percentage of activity relative to the value with KCl. ATPase activity in upper phase vesicles in the absence of KCl is 0.28 ± 0.01 μmol Pi mg-1 protein min-1. Data represent means ± se of three replicate experiments. Each replicate experiment was performed using independent membrane preparations. nd, Not determined.
| Addition
|
ATPase Activity
|
||
|---|---|---|---|
| Upper Phase | Microsomes | Lower Phase | |
| μmol Pimg-1protein min-1 | |||
| KCl (50 mm) | 2.48 ± 0.01 (100) | 0.62 ± 0.03 (100) | 0.55 ± 0.01 (100) |
| Na3VO4 (0.1 mm) | 1.36 ± 0.07 (45.2)a | 0.38 ± 0.02 (38.7)a | 0.38 ± 0.02 (30.9)a |
| NaN3 (1 mm) | 2.34 ± 0.08 (5.6)a | 0.53 ± 0.03 (14.5)a | 0.46 ± 0.00 (16.4)a |
| Na2MoO4 (0.1 mm) | 2.39 ± 0.09 (3.6)a | 0.43 ± 0.02 (30.6)a | 0.39 ± 0.02 (29.1)a |
| NaNO3 (50 mm) | 2.45 ± 0.06 (1.2)a | 0.51 ± 0.02 (17.7)a | 0.46 ± 0.00 (16.4)a |
| Triton X-100 (0.02%) | 5.81 ± 0.05 (134.3)b | nd | nd |
a and b represent inhibition and stimulation relative to K+ control, respectively
The sidedness of the membrane preparations was determined as described (Qiu and Su, 1998). When 0.02% (w/v) Triton X-100 was added to the assays, activity increased 57%, indicating that approximately one-half of the vesicles were oriented right side out (relative to the orientation of the plasma membrane in vivo) and one-half inside out (Table I). On the basis of substrate accessibility, measurements of H+ transport activity and Na+/H+ exchange likely took place in inside out vesicles.
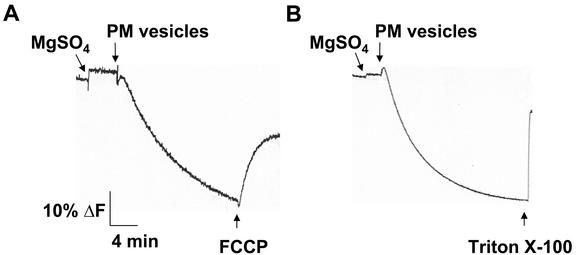
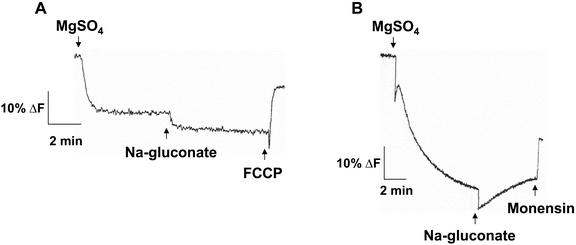
The H+ transport activity of these membrane vesicles was also characterized to demonstrate their transport competence. Formation of a pH gradient (ΔpH; measured as a quench in the fluorescence of the pH-sensitive probe quinacrine; Bennett and Spanswick, 1983) was dependent on the presence of plasma membrane protein because, in the absence of vesicles, no quench was observed (Fig. 2). Once formed, the ΔpH was abolished with a H+-ionophore (protonophore; FCCP; Fig. 2A) or low levels of the detergent Triton X-100 (Fig. 2B), which, at the concentration used, has been shown to effectively produce holes in membrane vesicles (Sandelius and Morre, 1990).
Figure 2.
Characterization of ΔpH formation in aqueous two-phase membrane vesicles isolated from salt-treated Arabidopsis. Plasma membrane vesicles were isolated by two-phase partitioning from leaves of wild-type plants treated with 250 mm NaCl for 3 d. ΔpH was established by the activity of the plasma membrane H+-ATPase and was measured as a decrease (quench) in the fluorescence of the pH-sensitive fluorescent probe quinacrine. Assays (1 mL) contained: 5 μm quinacrine, 3 mm ATP, 100 mm KCl, 25 mm 1,3-bis[tris(hydroxylmethyl) methylamino]propane (BTP)-HEPES (pH 6.5), 250 mm mannitol, and 50 μg of plasma membrane protein. Reactions were illuminated at 430 nm and quinacrine fluorescence was monitored at 500 nm. The protonophore, 5 μm carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP; A) or the detergent 0.02% (w/v) Triton X-100 (B) was added after the ΔpH had reached steady. One representative experiment of three replicates is shown. Each replicate experiment was performed using independent membrane preparations.
Plasma Membrane Na+/H+ Exchange Activity Is Present in Arabidopsis
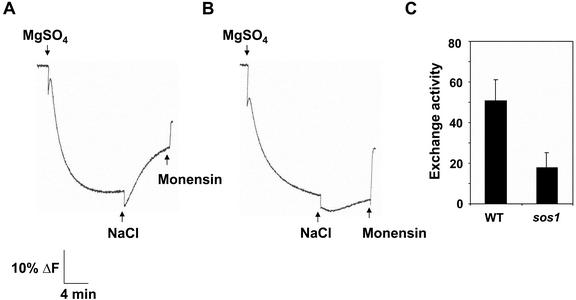
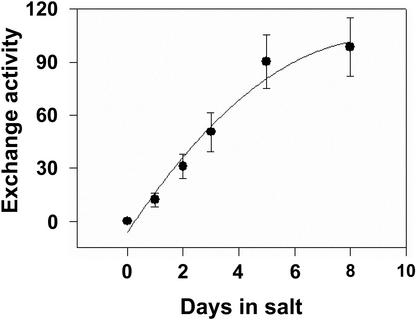
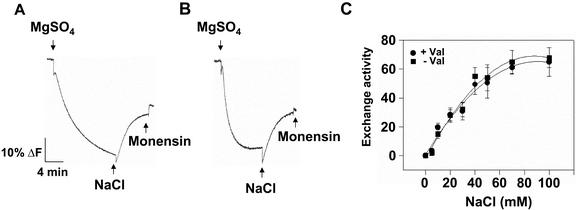
Once ΔpH reached steady state, addition of Na+ led to a dissipation of the ΔpH in plasma membrane vesicles isolated from wild-type Arabidopsis grown in 250 mm NaCl for 3 d (Fig. 3A). Treatment of plants with 250 mm NaCl for at least 1 d was required to measure Na+/H+ exchange, and initial rates of exchange activity in salt-treated plants increased with growth in salt up to 8 d (Fig. 4). In contrast, little Na+-induced dissipation of ΔpH could be measured in vesicles isolated from sos1 plants grown in the presence (Fig. 3B) or absence of NaCl (data not shown). Unless indicated, all subsequent assays were done with vesicles isolated from plants grown in 250 mm NaCl for 3 d.
Figure 3.
Plasma membrane Na+/H+ exchange activity in salt-treated wild-type and sos1 Arabidopsis plants. Plasma membrane vesicles were isolated by two-phase partitioning from the leaves of wild-type and sos1 plants treated with 250 mm NaCl for 3 d. Reaction mixes and assay conditions were as described in the legend to Figure 2. After formation of ΔpH, NaCl (50 mm) was added to initiate Na+/H+ exchange activity (dissipation of ΔpH). Na+/H+ exchange in plasma vesicles isolated from wild-type (A) or sos1 (B) plants. The electroneutral Na+/H+ exchanger Monensin (150 μm) was added at the end of the assay to determine whether a ΔpH was present. For A and B, one representative experiment of three replicates is shown. Each replicate experiment was performed using independent membrane preparations. C, Initial rates of Na+/H+ exchange (means ± se) of the three experiments; units are Δ%F mg-1 protein min-1.
Figure 4.
Plasma membrane Na+/H+ exchange activity in Arabidopsis as a function of plant growth in NaCl. Plasma membrane protein (50 μg) isolated from the leaves of wild-type Arabidopsis treated with 250 mm NaCl for 0 to 8 d was used in the assays. Reaction mixes and assay conditions were as described in the legend to Figure 2. After formation of ΔpH, 50 mm NaCl was added, and initial rates of Na+/H+ exchange activity (dissipation of ΔpH) were calculated. Units of Na+/H+ exchange are Δ%F mg-1 protein min-1. All data represent means ± se of at least three replicate experiments. Each replicate experiment was performed using independent membrane preparations.
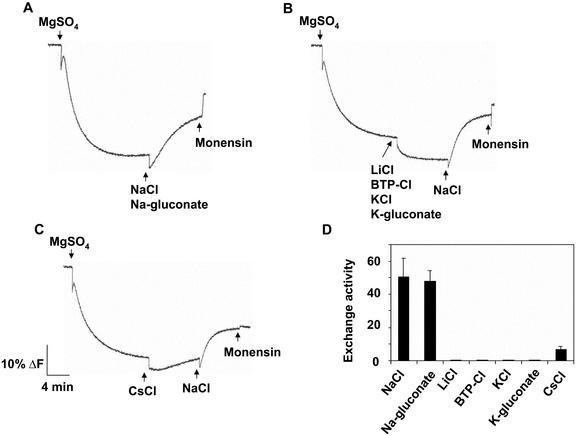
To determine the ion specificity of the exchange reaction, the ability of various salts to dissipate ΔpH was monitored. When either NaCl or sodium-gluconate was added to initiate the dissipation of ΔpH, similar rates of exchange activity were observed (Fig. 5), suggesting that Na+ and not the anion is responsible for the dissipation of ΔpH. Addition of the chloride salt of Cs+ produced a small but reproducible dissipation of ΔpH. When chloride salts of K+, Li+, and BTP were used instead of Na+, no dissipation was observed (Fig. 5), demonstrating Na+ specificity.
Figure 5.
Ion specificity of plasma membrane H+-coupled exchange activity in salt-treated Arabidopsis. Plasma membrane vesicles were isolated from leaves of wild-type Arabidopsis treated with 250 mm NaCl for 3 d. Reaction mixes and assay conditions were as described in the legend to Figure 2. After formation of ΔpH, various chloride or gluconate salts (50 mm) were added to initiate H+-coupled exchange activity (dissipation of ΔpH). Monensin (150 μm) was added at the end of the assay to determine whether a ΔpH was present. For A through C, one representative experiment of three replicates (for each ion) is shown. Each replicate experiment was performed using independent membrane preparations. D, Initial rates of H+-coupled exchange (means ± se) of the three experiments for each ion assayed; units are Δ%F mg-1 protein min-1.
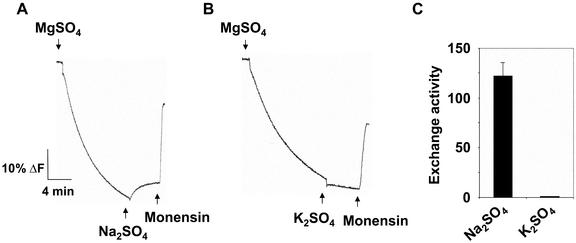
In the experiments described in Figure 5, KCl was included in all assays to generate the ΔpH (the H+ transport activity of the plasma membrane ATPase does not require K+, but this electrogenic H+-pump requires a permeant anion to dissipate the electrical or membrane potential (ΔΨ) for maximal ΔpH formation; Vara and Serrano, 1982). Under these conditions, measurements of K+-coupled H+ transport might be masked by the high concentration of KCl present when ΔpH was formed. Therefore, K+-coupled H+ transport was also measured in plasma membrane vesicles in which the ΔpH was generated in the absence of K+, using BTP-Cl (Fig. 6). Once the ΔpH reached steady state, 50 mm Na2SO4 (Fig. 6A) or 50 mm K2SO4 (Fig. 6B) was added. Although a Na+-induced dissipation of the gradient (Na+/H+ exchange) could be measured (Fig. 6, A and C), no K+-induced dissipation (K+/H+ exchange) was observed (Fig. 6, B and C).
Figure 6.
Analysis of plasma membrane H+-coupled K+ exchange activity in salt-treated Arabidopsis. Plasma membrane vesicles were isolated from leaves of wild-type Arabidopsis treated with 250 mm NaCl for 3 d. Assays (1 mL) contained 5 μm quinacrine, 3 mm ATP, 50 mm BTP-Cl, 25 mm BTP-HEPES (pH 6.5), 250 mm mannitol, and 50 μg of plasma membrane protein. Assay conditions were as described in the legend to Figure 2. Na2SO4 (50 mm; A) or K2SO4 (50 mm; B) was added after the ΔpH had reached steady state. Monensin (150 μm), was added at the end of the assay to determine whether a ΔpH was present. For A and B, one representative experiment of three replicates is shown. Each replicate experiment was performed using independent membrane preparations. C, Initial rates of H+-coupled exchange (means ± se) of the three experiments for each ion assayed; units are Δ%F mg-1 protein min-1.
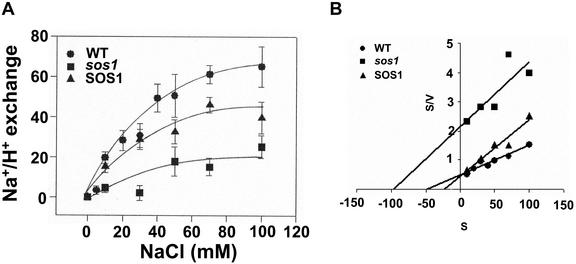
Na+-induced dissipation of ΔpH was measured as a function of Na+ (substrate) concentration in vesicles isolated from wild-type and sos1 plants (Fig. 7). Na+/H+ exchange was dependent on Na+ (substrate) concentration as initial rates of exchange increased with increasing NaCl up to approximately 80 mm (Fig. 7). We have previously reported that the low level of transport activity seen in sos1 vesicles when higher levels of NaCl were added to assays is independent of the electrical potential and likely represents Na+/H+ exchange originating from another as-yet-unidentified exchanger (Qiu et al., 2002). Kinetic analysis of the data showed that SOS1 (calculated as the difference in activity in vesicles isolated from wild-type and sos1 plants) has an apparent Km for Na+ of 22.8 mm and a Vmax of 51.5 change in % fluorescence (Δ%F) mg-1 protein min-1 (Fig. 7B; Table II). We have previously shown that in vitro addition of activated SOS2 protein increased Na+/H+ exchange activity in salt-treated wild-type, sos2, and sos3 plants (Qiu et al., 2002). A comparison of the apparent Km and Vmax values for exchange activity in wild-type and sos plants in the absence and presence of activated SOS2 protein added in vitro is shown in Table II. In the absence of activated SOS2 protein, the affinity of plasma membrane Na+/H+ exchange for substrate was highest in vesicles where SOS1 activity is present (wild-type plants). This is in contrast to the maximum velocities of the exchange reaction, which were similar in plasma membrane vesicles isolated from wild-type and mutant plants. In the presence of activated SOS2 protein, the affinity of the exchanger for substrate was unchanged in sos1 plants but increased in vesicles isolated from wild-type, sos2, and sos3 plants, respectively. Under these conditions, the maximum velocities increased in vesicles isolated from wild-type, sos2, and sos3 plants, respectively, and decreased in vesicles isolated from sos1 plants.
Figure 7.
Initial rates of plasma membrane Na+/H+ exchange activity in salt-treated Arabidopsis as a function of substrate (Na+) concentration. Plasma membrane vesicles were isolated from wild-type and sos1 plants grown in 250 mm NaCl for 3 d. Reaction mixes and assay conditions were as described in the legend to Figure 2. A, Initial rates of Na+-induced dissipation of ΔpH in vesicles isolated from wild-type (•) and sos1 (▪) plants were calculated over a range of Na+ concentrations from 0 to 100 mm. Units of Na+/H+ exchange are Δ%F mg-1 protein min-1. SOS1 (▴) represents the values calculated as the difference in Na+/H+ exchange activity in vesicles isolated from wild-type and sos1 plants. All data represent means ± se of at least three replicate experiments. Each replicate experiment was performed using independent membrane preparations. B, Data shown in A were transformed using a Hanes-Woolf plot to determine kinetic parameters (-X intercept = Km, Km/Y intercept = Vmax).
Table II.
The influence of constitutively active SOS2 protein on kinetic parameters of the plasma membrane Na+/H+ exchanger from wild-type and sos Arabidopsis plants
Membrane vesicles were isolated from leaves of salt-treated (250 mm NaCl for 3 d) plants using aqueous two-phase partitioning. Initial rates of Na+-induced dissipation of quinacrine fluorescence were calculated over a range of Na+ concentrations from 0 to 100 mm (Qiu et al., 2002). The data were transformed using a Hanes-Woolf plot to determine kinetic parameters. SOS1 represents the values calculated as the difference in Na+/H+ exchange activity in vesicles isolated from wild-type and sos1 plants. Data represent apparent Km (mm) and Vmax (Δ% F mg-1 protein min-1) values ± se of three replicate experiments.
| - SOS2 Protein
|
+ SOS2 Protein
|
|||
|---|---|---|---|---|
| Km | Vmax | Km | Vmax | |
| Wild type | 45.8±5 | 97.1±6 | 37.1±6 | 192.3±11 |
| sos1 | 96.7±11 | 41.0±12 | 97.1±8 | 56.8±8 |
| sos2 | 102.3±18 | 45.3±11 | 65.6±8 | 125.0±16 |
| sos3 | 158.8±21 | 48.7±12 | 99.0±11 | 157.3±20 |
| SOS1 | 22.8±5 | 51.5±5 | 18.2±4 | 138.9±10 |
Plasma Membrane Na+/H+ Exchange Activity in Arabidopsis Is Electroneutral
Two approaches were used to provide information about the relative number of ions (Na+ and H+) being transported during the plasma membrane Na+/H+ exchange reaction. If one Na+ is exchanged for one H+, the reaction is electroneutral (involves no net charge transfer across the membrane and does not generate ΔΨ). If the exchange of Na+ for H+ is more than one for one (or vice versa), the reaction is electrogenic (generating a ΔΨ). The stoichiometry of Na+/H+ exchange activity was first examined using Oxonol V, a fluorescent probe that accumulates inside membrane vesicles in response to an increase in positive charge (an inside-positive ΔΨ; Bennett and Spanswick, 1983). Oxonol V accumulated inside the vesicles when H+ transport was established by the plasma membrane H+-ATPase (shown as a quench of Oxonol fluorescence; Fig. 8A). In these experiments, potassium iminodiacetic acid and sodium-gluconate rather than KCl and NaCl were used to stimulate the ATPase and dissipate the ΔpH, respectively, to limit the concentrations of permeant anions while maintaining the required concentrations of cations. Once a steady-state ΔpH was formed, Na+ was added to initiate Na+/H+ exchange. Under conditions of optimum Na+/H+ exchange, no change in Oxonol V fluorescence was observed (Fig. 8A), demonstrating that Na+/H+ exchange did not alter the ΔΨ and indicating that the exchange reaction is electroneutral. To demonstrate that a ΔpH was present under these assay conditions and that it could be used to drive Na+/H+ exchange, ΔpH formation and Na+-induced dissipation of the gradient were measured (Fig. 8B). These results show that a sizeable ΔpH was formed and could be used for Na+/H+ exchange, supporting the results from the Oxonol V experiments that Na+/H+ exchange does not alter the ΔΨ and is electroneutral. Additional evidence for the electroneutral nature of Na+/H+ exchange was supported by experiments in which generation of ΔΨ was eliminated by the K+-ionophore valinomycin in the presence of K+. If the Na+/H+ exchange reaction is electrogenic, elimination of the ΔΨ would lead to an increase in Na+/H+ exchange. If the exchange reaction is electroneutral, it would be unaffected by either the presence or absence of any ΔΨ. Similar rates of Na+/H+ exchange in the presence of K+ with or without valinomycin demonstrate that the exchange reaction is independent of ΔΨ (Fig. 9) and is, therefore, electroneutral.
Figure 8.
Influence of plasma membrane Na+/H+ exchange on formation of a membrane potential in salt-treated Arabidopsis. Plasma membrane vesicles were isolated from the leaves of wild-type plants treated with 250 mm NaCl for 3 d. A, Oxonol V fluorescence. Assays (1 mL) contained 3 μm Oxonol V (Sigma-Aldrich), 3 mm ATP, 4 mm MgSO4,20mm potassium iminodiacetic acid, 25 mm BTP-HEPES (pH 6.5), 250 mm mannitol, and 50 μg of plasma membrane protein. Reactions were illuminated at 580 nm, and Oxonol V fluorescence was monitored at 650 nm. An inside-acid positive ΔΨ was formed when plasma membrane protein (50 μg) was added to the reaction mix. Once ΔΨ reached steady state, sodium-gluconate (50 mm) was added to initiate Na+/H+ exchange. At the end of the experiment, the protonophore FCCP (5 μm) was added to determine whether a ΔΨ was present. B, Na+-induced dissipation of ΔpH. Assays (1 mL) contained 5 μm quinacrine, 3 mm ATP, 4 mm MgSO4, 20 mm potassium iminodiacetic acid, 25 mm BTP-HEPES (pH 6.5), 250 mm mannitol, and 50 μg of plasma membrane protein. Assay conditions were as described in the legend to Figure 2. Once ΔpH reached steady state, sodium-gluconate (50 mm) was added to initiate Na+/H+ exchange. Monensin (150 μm), was added at the end of the assay to determine whether a ΔpH was present. For A and B, one representative experiment of three replicates is shown. Each replicate experiment was performed using independent membrane preparations.
Figure 9.
Influence of the membrane potential on plasma membrane Na+/H+ exchange in salt-treated Arabidopsis. Plasma membrane vesicles were isolated from the leaves of wild-type plants treated with 250 mm NaCl for 3 d. ΔpH was established as described in the legend to Figure 2 in the absence (A) or presence (B) of the K+-ionophore, valinomycin (1 μm). When ΔpH reached steady state, 100 mm NaCl was added to initiate Na+/H+ exchange activity (dissipation of ΔpH). C, Na+/H+ exchange was measured over a range of final Na+ concentrations (0–100 mm) in the absence (▪) and presence (•) of valinomycin (1 μm) and initial rates of dissipation of ΔpH as a function of Na+ concentration are shown. Units of Na+/H+ exchange are Δ%F mg-1 protein min-1. For A and B, one representative experiment of three replicates is shown. Data in C represent means ± se of at least three replicate experiments. Each replicate experiment was performed using independent membrane preparations.
Measurements of transport in the presence and absence of valinomycin also provide evidence that the exchange reaction represents H+-dependent Na+ transport and not Na+-Na+ equilibration. Valinomycin also moves Na+ (with much lesser affinity), so if H+ were moving out of the vesicles in response to an electrical gradient while Na+ equilibrates across the membrane, the dissipation would be expected to be greater in the presence of valinomycin (faster equilibration). This was not the case because no difference was found when dissipation was compared with and without valinomycin (data not shown).
DISCUSSION
SOS1 is a major determinant of salt tolerance in Arabidopsis, and sequence analysis indicated that it encodes a Na+/H+ exchanger (Shi et al., 2000). In recent studies, significantly reduced Na+/H+ exchange activity in sos1 plants relative to activity in wild-type plants demonstrated that SOS1 encodes a Na+/H+ exchanger and that this protein contributes the major portion of the plasma membrane Na+/H+ exchange activity measured in leaves of Arabidopsis (Qiu et al., 2002; Fig. 3). Localization studies also support the sequence and transport data that SOS1 is a plasma membrane Na+/H+ exchanger: (a) Shi et al. (2002) found that the fluorescence associated with an SOS1-green fluorescent protein fusion protein localized to the plasma membranes in young Arabidopsis root cells, and (b) in the present study, SOS1 protein localized to fractions enriched in plasma membranes isolated using either continuous Suc gradients (Fig. 1) or two-phase preparations (data not shown).
Plasma membrane Na+/H+ exchange activity (SOS1 activity) in leaves of Arabidopsis was induced as a result of salt stress; no Na+/H+ exchange activity could be measured in vesicles isolated from plants grown in the absence of NaCl (Fig. 4). Transport activity was induced by plant growth in salt for as little as 1 d and continued to increase with prolonged exposure to salt up to 8 d (Fig. 4). Shi et al. (2000) previously found that SOS1 mRNA was expressed in shoots of wild-type plants grown in the absence of salt and that exposure to salt resulted in an induction of mRNA expression. In the present study, immunoblot analysis indicated that the SOS1 protein was also present in the plasma membranes of plants grown in the absence of salt (Fig. 1). Taken together, these results suggest that Na+/H+ exchange activity is likely to be regulated at the posttranslational level. Possible candidates for regulation include other components of the SOS pathway. SOS2 (a Ser/Thr protein kinase) has been shown to directly stimulate Na+/H+ exchange activity in vitro, and reduced activity has been detected in plasma membranes isolated from sos2 and sos3 (encoding a myristoylated calcium binding protein) plants (Qiu et al., 2002). Salt induction of plasma membrane Na+/H+ exchange activity is in contrast to activity of the vacuolar transporter in Arabidopsis culture cells (data not shown) and shoots (Apse et al., 1999) where Na+/H+ exchange activity is constitutive. This differential salt induction implies diverse roles for the vacuolar and plasma membrane transporters in ion homeostasis and salt tolerance. It is possible that the vacuolar membrane plays a major role in maintaining Na+ homeostasis during normal growth and when the plant first experiences increasing levels of Na+. With prolonged exposure to high levels of Na+, the transporter on the plasma membrane may begin to play a greater role in reducing cellular Na+ levels and ultimately in the plant's ability to continue to grow during salt stress.
Different roles for the transporters at the vacuolar and plasma membranes are also indicated by their different affinities for Na+. Kinetics studies showed that the Arabidopsis vacuolar and plasma membrane exchangers have apparent Km values for Na+ of 7 (Apse et al., 1999) and 22.8 mm (Fig. 4), respectively. Under normal growth conditions, cells in glycophytes maintain low levels of Na+ in the cytosol (from 1–10 mm; Blumwald et al., 2000), suggesting that the vacuolar exchanger may be most critical for maintaining Na+ homeostasis during normal growth and the early stages of salt stress, whereas the plasma membrane transporter may be the line of defense after acute and/or prolonged exposure to salt.
A number of properties of the plasma membrane Na+/H+ exchanger in Arabidopsis distinguish it from plasma membrane exchangers in other organisms and from the vacuolar membrane exchangers in plants. Inhibition of activity by the diuretic drug amiloride is a typical property of Na+/H+ exchangers in mammalian, fungal, and bacterial cells (Counillon and Pouyssegur, 2000; Wiebe et al., 2001). In plants, amiloride has been shown to inhibit vacuolar membrane Na+/H+ exchange activity in sugar beet (Beta vulgaris; Blumwald and Pool, 1985; Barkla and Blumwald, 1991), the euhalophyte Salicornia bigelovii (Parks et al., 2002), and Arabidopsis (Q.-S. Qiu, Y. Guo, J.-K. Zhu, K.S. Schumaker, unpublished data). In contrast, structural analysis revealed that the SOS1 does not have an amiloride-binding site (Shi et al., 2000), and results from the present study show that plasma membrane Na+/H+ exchange activity is insensitive to amiloride (data not shown). Differences in ion specificity for the plant plasma membrane exchanger relative to that of transporters in other organisms and on the plant vacuolar membrane implies that the exchangers have different mechanisms for ion transport and regulation as well as unique physiological roles in ion homeostasis. Na+/H+ exchangers in mammalian, fungal, and bacterial cells are able to transport Li+ in exchange for H+ (Wiebe et al., 2001). Low levels of H+-coupled Li+ and Cs+ transport have been demonstrated in vacuolar Na+/H+ exchangers isolated from shoots of S. bigelovii (Parks et al., 2002) and Arabidopsis culture cells (Q.-S. Qiu, Y. Guo, J.-K. Zhu, K.S. Schumaker, unpublished data) and when the vacuolar exchanger AtNHX1 from Arabidopsis was reconstituted in liposomes (Venema et al., 2002). Transport of these monovalent cations is in contrast to the plasma membrane Na+/H+ exchanger in Arabidopsis, which transported mainly Na+ and Cs+ at only very low levels (Fig. 5). These transport differences are supported by molecular evidence showing that the SOS1 protein has a unique structure that includes a long C-terminal tail (Shi et al., 2000).
In Arabidopsis, the absence of ΔΨ formation during plasma membrane Na+/H+ exchange (Fig. 8) along with the unchanged activity of the exchanger when the ΔΨ was clamped with K+ and valinomycin (Fig. 9) demonstrate that this exchange activity was electroneutral. Similar results have been found for Na+/H+ exchangers in mammalian and fungal cells, and for the vacuolar Na+/H+ exchangers in plants (Blumwald and Pool, 1985; Barkla et al., 1995; Darley et al., 2000). In contrast, the exchangers in bacteria are electrogenic transporting two or three H+ for each Na+ for the products of NhA and NhB genes, respectively (Wiebe et al., 2001). The electroneutral nature of the Arabidopsis plasma membrane Na+/H+ exchanger suggests that, whereas the transport reaction contributes to the regulation of cellular Na+ and H+ ion homeostasis, it could not contribute to generation of an electrical driving force for the transport of other ions and solutes.
Our previous studies established that there is a significant stimulation of plasma membrane Na+/H+ exchange activity in vesicles isolated from wild-type, sos2, and sos3 plants by in vitro addition of activated SOS2 protein (Qiu et al., 2002). In contrast, no stimulation of activity was observed in vesicles isolated from sos1 plants (Qiu et al., 2002). These results demonstrated that SOS1 transport activity is regulated by the SOS2 kinase. Kinetic analyses of exchange activity as a function of substrate concentration in the presence and absence of activated SOS2 protein provides additional evidence for this regulation. (a) In the absence of activated SOS2 protein, the exchanger in wild-type plants (where SOS1 activity is present) had an affinity for Na+ (apparent Km = 45.8 mm) 2.2 and 3.5 higher than exchangers active in sos2 and sos3 plants, respectively (Table II). (b) When activated SOS2 protein was added to vesicles isolated from sos2 and sos3 plants, both the exchange activity (Vmax) and the affinity (decreased apparent Km) of the exchanger for substrate increased significantly (Qiu et al., 2002; Table II). The increase in affinity of the exchangers for substrate indicates that the stimulation of transport activity by SOS2 is likely due to a change in the structure of the Na+-binding site of the SOS1 protein. Sequence analysis has shown that SOS1 has 12 putative membrane-spanning domains and, based on studies of exchangers in bacteria, yeast, and animals, the cation-binding and transport site is likely to be localized to the membrane-spanning domains. From our studies, a number of interesting questions remain to be answered including: (a) how does SOS2 regulate the Na+-binding site of the SOS1 protein? (b) What additional factors are responsible for the differences in the regulation of the exchangers in sos2 and sos3 plants? (b) What is the identity of the other plasma membrane Na+/H+ exchanger(s) in Arabidopsis?
Genetic dissection of salt tolerance in Arabidopsis has established the involvement of the SOS pathway in the plant's response to the ionic effects of salt stress (Zhu, 2001). This novel protein kinase pathway is activated by calcium signaling and regulates a Na+/H+ exchanger on the plasma membrane (Qiu et al., 2002). The studies described in this manuscript add to our understanding of the transport properties of this exchanger. It should now be possible to investigate the mechanisms underlying the regulation and function of the plasma membrane Na+/H+ exchanger by reconstitution in liposomes or heterologous expression in yeast. When plants are exposed to high levels of salt, the activities of multiple ion transporters on several membranes must be altered. Testing the involvement of the SOS pathway in regulation of these transporters as well as identification of additional regulatory components remains an important task to enable us to understand the contributions of the Na+/H+ exchangers to plant salt tolerance.
MATERIAL AND METHODS
Plant Material
Arabidopsis ecotype Columbia was used in all experiments. Plants were grown in potting soil (Metro-Mix, Grace Sierra Horticultural Products Co., Milpitas, CA) in a growth room with a cycle of 16 h of light (approximately 100 μE m-2 s-1) at 22°C and 8 h of dark at 20°C. Plants were watered with tap water three times per week. After 4 weeks, plants were treated with 250 mm NaCl for 1 to 8 d by placing the pots in a tray containing the NaCl solution. Rosette leaves were harvested and used for membrane isolation.
For linear Suc gradients, plants were sown and grown to maturity in soil in propagation trays, or for root isolation, seeds were germinated and plants were grown in liquid culture in Murashige and Skoog (1962) medium at 25°C with continuous shaking (120 rpm) in the dark.
Membrane Isolation
Plasma membrane vesicles were isolated using two-phase partitioning as described (Qiu and Su, 1998; Qiu et al., 2002). For linear Suc gradients and immunoblots, shoots and roots were harvested, and tissue was placed directly into 200 mL of ice-cold homogenization medium (400 mm mannitol, 10% [w/v] glycerol, 5% [w/v] polyvinylpyrrolidone-10, 0.5% [w/v] bovine serum albumin, 1 mm phenylmethylsulfonyl fluoride, 30 mm Tris, 2 mm dithiothreitol, 5 mm EGTA, 5 mm MgSO4, 0.5 mm butylated hydroxytoluene, 0.25 mm dibucaine, 1 mm benzamidine, and 26 mm K+-metabisulfite, adjusted to pH 8.0 with H2SO4). Microsomal membranes were isolated as previously described (Barkla et al., 1995). The microsomal suspension was then layered onto continuous linear Suc gradients (5%–45% [w/v] Suc) in suspension medium. Suc gradients were then centrifuged at 100,000g (3 h at 4°C) using a SW28 swinging bucket rotor (Beckman Coulter, Fullerton, CA) in a Beckman Coulter L8-M ultracentrifuge. Membrane fractions (1 mL) of increasing density were collected from the continuous Suc gradients (32 fractions in total). Membranes were frozen directly in liquid N2 and stored at -80°C.
Plasma Membrane Characterization
To characterize the activity of the plasma membrane H+-ATPase, its substrate hydrolytic activity was determined by measuring the release of Pi from ATP according to previous methods (Qiu, 1999). Activities for the plasma membrane, mitochondrial and vacuolar ATPases were measured using optimal conditions for each enzyme (Gallagher and Leonard, 1982; Giannini and Briskin, 1987). The purity and latency of the membrane preparations were determined as described (Qiu and Su, 1998).
The formation of ΔpH was established by the activity of the plasma membrane H+-ATPase and was measured as a decrease (quench) in the fluorescence of the pH-sensitive fluorescent probe quinacrine (Bennett and Spanswick, 1983; Qiu, 1999; Qiu et al., 2002). Assays (1 mL) contained 5 μm quinacrine, 3 mm ATP, 100 mm KCl, 25 mm BTP-HEPES (pH 6.5), 250 mm mannitol, and 50 μg of plasma membrane protein. Reactions were mixed by inversion several times and then placed in a dark chamber in a luminescence spectrophotometer (model LS-5B, Perkin-Elmer, Shelton, CT). Reactions were equilibrated in the dark with stirring for 5 min before beginning fluorescence readings, and all additions to reactions were made in a darkened room. Assays were initiated with the addition of 4 mm MgSO4, and formation of ΔpH was measured at excitation and emission wavelengths of 430 and 500 nm, respectively.
Control experiments were conducted to monitor potential effects of organic solvents used with inhibitors and ionophores in H+ transport assays; no solvent effects were observed (data not shown).
Immunoblot Analysis
For SDS-PAGE and immunoblotting, protein samples were prepared according to the method of Parry et al. (1989). Protein was precipitated by 50-fold dilution in ice-cold 1:1 (v/v) ethanol:acetone and incubated overnight at -30°C. Samples were then centrifuged at 13,000g (20 min at 4°C) using an F2402 rotor in a GS-15R table-top centrifuge (Beckman Coulter). Pellets were air-dried, resuspended with Laemmli (1970) sample buffer (2.5% [w/v] SDS final concentration), and heated at 60°C for 2 min before loading onto a 10% (w/v) acrylamide gel. Eighteen micrograms of protein was loaded per lane. After electrophoresis, proteins were electrophoretically transferred onto nitrocellulose membranes (ECL, Amersham-Biosciences, Piscataway, NJ) using a Mini-Transblot Transfer Cell (Bio-Rad Laboratories, Hercules, CA) for 1 h at 100 mV. Membranes were blocked with Tris-buffered saline (10 mm Tris-HCl, pH 7.5, and 150 mm NaCl) containing 0.02% (w/v) sodium-azide and 5% (w/v) fat-free powdered milk for 2 h at room temperature. Membranes were then incubated for 12 h at room temperature with the appropriate antibody. Immunocomplexes were detected using a 1:15,000 dilution of peroxidase-labeled anti-rabbit IgG secondary antibody (Sigma-Aldrich, St. Louis) and were developed using the chemiluminescent ECL detection substrate (Amersham).
A gel purified fusion of glutathione S-transferase with the C-terminal domain (2.1 kb) of the SOS1 protein was used to generate specific polyclonal anti-SOS1 antibody used in this study. Sequence comparisons of this domain with the Arabidopsis database indicate that this region is specific for the SOS1 protein (data not shown).
Na+/H+ Exchange Assays
Na+/H+ exchange activity was measured as a Na+-induced dissipation of ΔpH (i.e. a Na+-induced increase in quinacrine fluorescence; Barkla et al., 1995; Qiu et al., 2002). The reaction medium was the same as that used for H+ transport assays. When ΔpH reached steady state, NaCl or sodium-gluconate were added to initiate Na+ transport. To determine initial rates of Na+/H+ exchange (Δ%F min-1), changes in relative fluorescence were measured during the first 15 s after addition of Na+. Specific activity was calculated by dividing the initial rate by the mass of plasma membrane protein in the reaction (Δ%F mg-1 protein min-1). Measurements of Na+-induced dissipation of ΔpH were made without inhibiting the H+-ATPase; however, initial rates of Na+/H+ exchange should be unaffected by the presence of an active H+-pump (B.J. Barkla, R. Vera-Estrella, and O. Pantoja, unpublished data).
Membrane Potential Assays
Oxonol V, an inside-positive ΔΨ-sensitive fluorescent probe, was used to measure the membrane potential (Bennett and Spanswick, 1983; Kaestner and Sze, 1987). Assays (1 mL) contained 3 μm Oxonol V (Sigma-Aldrich), 3 mm ATP, 4 mm MgSO4, 20 mm potassium iminodiacetic acid, 25 mm BTP-HEPES (pH 6.5), 250 mm mannitol, and 50 μg of plasma membrane protein. Assays were carried out as described for measurements of ΔpH and formation of ΔΨ was measured at excitation and emission wavelengths of 580 and 650 nm, respectively.
Protein Determination
The protein content of membrane vesicles isolated by aqueous two-phase partitioning or linear Suc gradients was determined by the dye-binding method of Bradford (1976) with bovine serum albumin as a standard.
Acknowledgments
We gratefully acknowledge the following individuals for supplying antibodies: Dr. Ramon Serrano (Universidad Politecnica de Valencia, Spain) for P-ATPase; Dr. Phil Rea (University of Pennsylvania, Philadelphia) for vacuolar H+-pyrophosphatase; Dr. Karl-Josef Dietz (Bielefeld Universität, Bielefeld) for V-ATPase; and Dr. Hans J. Bohnert (University of Illinois, Urbana) Calreticulin.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.010421.
This work was supported by the National Institutes of Health (grant no. R01GM59138 to J.-K.Z.) and by the Southwest Consortium on Plant Genetics and Water Resources (to J.-K.Z. and K.S.S.). B.J.B. and R.V.-E. were supported by Dirección General de Asuntas para el Personal Académico (grant no. IN#230998) and Consejo Nacional de Ciencia y Tecnológia (grant no. 33054–N). Q.-S.Q. was supported in part by the Major State Basic Research and Development Plan of the People's Republic of China (grant no. G1999011705).
References
- Allen GJ, WYN Jones RG, Leigh RA (1995) Sodium transport measured in plasma membrane vesicles isolated from wheat genotypes with differing K+/Na+ discrimination traits. Plant Cell Environ 18: 105-115 [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256-1258 [DOI] [PubMed] [Google Scholar]
- Barkla BJ, Blumwald E (1991) Identification of a 170-kDa protein associated with the vacuolar Na+/H+ antiport of Beta vulgaris. Proc Natl Acad Sci USA 88: 11177-11181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkla BJ, Zingarelli L, Blumwald E, Smith JAC (1995) Tonoplast Na+/H+ antiport activity and its energization by the vacuolar H+-ATPase in the halophyte Mesembryanthemum crystallinum. Plant Physiol 109: 549-556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett AB, Spanswick RM (1983) Optical measures of ΔpH and ΔΨ in corn root membrane vesicles: kinetic analysis of Cl- effects on a proton-translocating ATPase. J Membr Biol 71: 95-107 [Google Scholar]
- Blumwald E (2000) Sodium transport and salt tolerance in plants. Curr Opin Cell Biol 12: 431-434 [DOI] [PubMed] [Google Scholar]
- Blumwald E, Aharon GS, Apse MP (2000) Sodium transport in plant cells. Biochim Biophys Acta 1465: 140-151 [DOI] [PubMed] [Google Scholar]
- Blumwald E, Pool R (1985) Na+/H+ antiport in isolated tonoplast vesicles from storage tissue of Beta vulgaris. Plant Physiol 78: 163-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254 [DOI] [PubMed] [Google Scholar]
- Counillon L, Pouyssegur J (2000) The expanding family of eucaryotic Na+/H+ exchangers. J Biol Chem 275: 1-4 [DOI] [PubMed] [Google Scholar]
- Darley CP, van Wuytswinkel OCM, van de Woude K, Mager WH (2000) Arabidopsis thaliana and Saccharomyces cerevisiae genes encode amiloride sensitive electroneutral Na+/H+ exchanger. Biochem J 351: 241-249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K-J, Arbinger B (1996) cDNA sequence and expression of subunit E of the vacuolar H+-ATPase in the inducible Crassulacean acid metabolism plant Mesembryanthemum crystallinum. Biochim Biophys Acta 1281: 134-138 [DOI] [PubMed] [Google Scholar]
- Gallagher SR, Leonard RT (1982) Effect of vanadate, molybdate, and azide on membrane-associated ATPase and soluble phosphatase activities of corn roots. Plant Physiol 70: 1335-1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, Fink GR (1999) The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc Natl Acad Sci USA 96: 1480-1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannini JL, Briskin DP (1987) Proton transport in plasma membrane and tonoplast vesicles from red beet (Beta vulgaris L.) storage tissue. Plant Physiol 84: 613-618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu JK (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97: 3735-3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Pardo JM (2000b) The dawn of plant salt tolerance genetics. Trends Plant Sci 5: 317-319 [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000a) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51: 463-499 [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Sze H (1987) Potential-dependent anion transport in tonoplast vesicles from oat roots. Plant Physiol 83: 483-489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Zhen R-G, Rea PA (1994) Heterologous expression of plant vacuolar pyrophosphatase in yeast demonstrates sufficiency of the substrate-binding subunit for proton transport. Proc Natl Acad Sci USA 91: 6128-6132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680-685 [DOI] [PubMed] [Google Scholar]
- Liu J, Zhu JK (1997) An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proc Natl Acad Sci USA 94: 14960-14964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu JK (1998) A calcium sensor homolog required for plant salt tolerance. Science 280: 1943-1945 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol Plant 15: 473-497 [Google Scholar]
- Nelson DE, Glaunsinger B, Bohnert HJ (1997) Abundant accumulation of the calcium-binding molecular chaperone calreticulin in specific floral tissues of Arabidopsis thaliana. Plant Physiol 114: 29-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski J, Grinstein S (1997) Na+/H+ exchangers of mammalian cells. J Biol Chem 272: 22373-22376 [DOI] [PubMed] [Google Scholar]
- Padan E, Venturi M, Gerchman Y, Dover N (2001) Na+/H+ antiporters. Biochim Biophys Acta 1505: 144-157 [DOI] [PubMed] [Google Scholar]
- Pardo JM, Serrano R (1989) Structure of a plasma membrane H+-ATPase gene from the plant Arabidopsis thaliana. J Biol Chem 264: 8557-8562 [PubMed] [Google Scholar]
- Parks GE, Dietrich MA, Schumaker KS (2002) Increased vacuolar Na+/H+ exchange activity in Salicornia bigelovii Torr. in response to NaCl. J Exp Bot 53: 1055-1065 [DOI] [PubMed] [Google Scholar]
- Parry RV, Turner JC, Rea PA (1989) High purity preparations of higher plant vacuolar H+-ATPase reveal additional subunits: revised subunit composition. J Biol Chem 264: 20025-20032 [PubMed] [Google Scholar]
- Qiu QS (1999) Characterization of PNPP hydrolysis by the plasma membrane H+-ATPase from soybean hypocotyls. J Plant Physiol 154: 628-633 [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99: 8436-8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS, Su XF (1998) The influence of extracellular Ca2+on the activity of the plasma membrane H+-ATPase from soybean hypocotyls. Aust J Plant Physiol 25: 923-928 [Google Scholar]
- Quintero FJ, Blatt MR, Pardo JM (2000) Functional conservation between yeast and plant endosomal Na+/H+ antiporters. FEBS Lett 471: 224-228 [DOI] [PubMed] [Google Scholar]
- Ratner A, Jacoby B (1976) Effect of K+, its counter anion, and pH on sodium efflux from barley root tips. J Exp Bot 27: 843-852 [Google Scholar]
- Sandelius AS, Morre DJ (1990) Plasma membrane isolation. In C Larsson, IM Moller, eds, The Plant Plasma Membrane. Spring-Verlag, Berlin, pp 44-75
- Schachtman D, Liu W (1999) Molecular pieces to the puzzle of the interaction between potassium and sodium uptake in plants. Trends Plant Sci 4: 281-286 [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896-6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14: 465-477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Balderas E, Vera-Estrella R, Golldack D, Quigley F, Zhao C, Pantoja O, Bohnert HJ (2003) Expression of the cation transporter McHKT1 in a halophyte. Plant Mol Biol (in press) [DOI] [PubMed]
- Sze H (1985) H+-translocating ATPases: advances using membrane vesicles. Annu Rev Plant Physiol Plant Mol Biol 36: 175-208 [Google Scholar]
- Vara F, Serrano R (1982) Partial purification and properties of the proton-translocating ATPase of plant plasma membranes. J Biol Chem 257: 12826-12830 [PubMed] [Google Scholar]
- Venema K, Quintero FJ, Pardo JM, Donaire JP (2002) The Arabidopsis Na+/H+ exchanger AtNHX1 catalyzes low affinity Na+ and K+ transport in reconstituted liposomes. J Biol Chem 277: 2413-2418 [DOI] [PubMed] [Google Scholar]
- Watad A-EA, Pesci P-A, Reinhold L, Lerner HR (1986) Proton fluxes as a response to external salinity in wild type and NaCl-adapted Nicotiana cell lines. Plant Physiol 81: 454-459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West IC, Mitchell P (1974) Sodium/proton ions antiport in Escherichia coli. Biochem J 144: 87-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebe CA, Dibattista ER, Fliegel L (2001) Functional role of polar amino acid residues in Na+/H+ exchangers. Biochem J 357: 1-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJ, Lei D, Zhu JK (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8: 617-627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HX, Blumwald E (2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19: 765-768 [DOI] [PubMed] [Google Scholar]
- Zhang HX, Hodson JN, Williams JP, Blumwald E (2001) Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc Natl Acad Sci USA 98: 12832-12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2000) Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol 124: 941-948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2001) Cell signaling under salt, water and cold stresses. Curr Opin Plant Biol 4: 401-406 [DOI] [PubMed] [Google Scholar]
- Zhu JK, Liu J, Xiong L (1998) Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role or potassium nutrition. Plant Cell 10: 1181-1191 [DOI] [PMC free article] [PubMed] [Google Scholar]