Abstract
Here, we compare the regulation and localization of the Arabidopsis type III phosphatidylinositol (PtdIns) 4-kinases, AtPI4Kα1 and AtPI4Kβ1, in Spodoptera frugiperda (Sf9) insect cells. We also explore the role of the pleckstrin homology (PH) domain in regulating AtPI4Kα1. Recombinant kinase activity was found to be differentially sensitive to PtdIns-4-phosphate (PtdIns4P), the product of the reaction. The specific activity of AtPI4Kα1 was inhibited 70% by 0.5 mm PtdIns4P. The effect of PtdIns4P was not simply due to charge because AtPI4Kα1 activity was stimulated approximately 50% by equal concentrations of the other negatively charged lipids, PtdIns3P, phosphatidic acid, and phosphatidyl-serine. Furthermore, inhibition of AtPI4Kα1 by PtdIns4P could be alleviated by adding recombinant AtPI4Kα1 PH domain, which selectively binds to PtdIns4P (Stevenson et al., 1998). In contrast, the specific activity of AtPI4Kβ1, which does not have a PH domain, was stimulated 2-fold by PtdIns4P but not other negatively charged lipids. Visualization of green fluorescent protein fusion proteins in insect cells revealed that AtPI4Kα1 was associated primarily with membranes in the perinuclear region, whereas AtPI4Kβ1 was in the cytosol and associated with small vesicles throughout the cytoplasm. Expression of AtPI4Kα1 without the PH domain in the insect cells compromised PtdIns 4-kinase activity and caused mislocalization of the kinase. The green fluorescent protein-PH domain alone was associated with intracellular membranes and the plasma membrane. In vitro, the PH domain appeared to be necessary for association of AtPI4Kα1 with fine actin filaments. These studies support the idea that the Arabidopsis type III PtdIns 4-kinases are responsible for distinct phosphoinositide pools.
The negatively charged polyphosphorylated inositol lipids, phosphatidylinositol (PtdIns)-4-phosphate (PtdIns4P) and PtdIns(4,5)P2, are important biochemical cues for regulating vesicle formation and trafficking, cytoskeletal dynamics, membrane protein binding, and enzyme activity (Munnik et al., 1998; Stevenson et al., 2000; Wang, 2001; De Matteis et al., 2002) in addition to being precursors of the second messenger inositol(1,4,5)trisphosphate (Berridge and Irvine, 1989). Because of the multiple roles of the phosphoinositides (PIs) in cellular metabolism, regulation of PI biosynthesis is integral to both the response to and recovery from external stimuli. PtdIns 4-kinase, the enzyme that synthesizes PtdIns4P, is the first committed step in PtdIns(4,5)P2 biosynthesis; therefore, regulation of PtdIns 4-kinase activity and/or its subcellular distribution could affect not only PtdIns4P biosynthesis but also the biosynthesis and distribution of PtdIns(4,5)P2.
In plants, PtdIns 4-kinase activity has been reported to be associated with many cellular compartments such as the plasma membrane (Sommarin, 1988; Gross et al., 1992; Cho et al., 1993), cytosol (Okpodu et al., 1995), cytoskeleton (Tan and Boss, 1992; Xu et al., 1992), and the nucleus (Hendrix et al., 1989; Bunney et al., 2000). This wide distribution of PtdIns 4-kinase activity throughout the cell implies that there are distinct isoforms that are targeted to the various compartments where they perform distinct functions. Two functional PtdIns 4-kinases have been cloned from Arabidopsis cDNA. These are designated AtPI4Kα1 (formerly AtPI4Kα; Stevenson et al., 1998) and AtPI4Kβ1 (formerly AtPI4Kβ; Xue et al., 1999), which were renamed in a recent description of the genes involved in the PI pathway based on database predictions (Mueller-Roeber and Pical, 2002).
PtdIns 4-kinases historically have been subgrouped as type II and type III based on biochemical studies of partially purified native protein that revealed higher substrate Km values for the type III enzymes and a decreased sensitivity to adenosine. Subsequently, amino acid sequence comparison has provided a more reliable classification system for the type II and III PtdIns 4-kinases. AtPI4Kα1 and AtPI4Kβ1 are orthologous to the human type III PI4Kα and -β isoforms and the yeast (Saccharomyces cerevisiae) PtdIns 4-kinases, STT4 and PIK1, respectively. In addition to these two type III PtdIns 4-kinases, there appears to be at least one other smaller type III isoform based on immunological data (Westergren et al., 1999). The smaller isoform has been partially purified from spinach (Spinacia oleracea) plasma membranes but has yet to be cloned from any plant. Other putative isoforms of the PtdIns 4-kinase have been predicted based on genomic database searches (Mueller-Roeber and Pical, 2002); however, none have been shown to be functional enzymes.
In this work, we have compared the two major isoforms of Arabidopsis type III PtdIns 4-kinases, AtPI4Kα1 (AC007504) and AtPI4Kβ1 (AB008266). AtPI4Kα1 and AtPI4Kβ1 share significant sequence similarity within selected domains. Both isoforms contain a well-conserved catalytic lipid kinase domain and a lipid kinase unique domain. AtPI4Kα1 also contains a PI-binding domain known as a pleckstrin homology (PH) domain. Although PH domains can function to recruit proteins to membranes through binding to PIs, the finding that the AtPI4Kα1 PH domain binds the product of the reaction, PtdIns4P, and is located adjacent to the catalytic domain suggests that this PH domain may have additional functions in regulating enzyme activity. AtPI4Kβ1 does not contain a PH domain. It contains a unique domain of approximately 300 amino acids in length that is enriched in charged amino acids. Interestingly, this domain appears to be specific to the plant PI4Kβ1 isoform (Xue et al., 1999), but its function in regulating the enzyme is not known.
Studies of the yeast orthologs (PIK1 and STT4) indicate that the two isoforms of PtdIns 4-kinase have distinct functions. Specifically, neither PIK1 nor STT4 complement each other in PtdIns 4-kinase mutant strains (Audhya et al., 2000). Low concentrations of the fungal (Penicillium fumiculosum) toxin wortmannin inhibit Stt4 but not Pik1 in vitro, and the wortmannin toxicity of cells can be suppressed by overexpression of STT4 but not PIK1 (Cutler et al., 1997). Stt4 function is essential for generating the PtdIns(4,5)P2 pool involved in the organization of the plasma membrane-associated actin cytoskeleton, whereas Pik1 activity is essential for Golgi vesicle trafficking (Audhya et al., 2000; Audhya and Emr, 2002).
The activity of the PtdIns 4-kinases can be affected by association with other proteins such as eukaryotic elongation factor 1A, Arf1, frequenin (Yang and Boss, 1994; Hendricks et al., 1999; Jones et al., 2000; Audhya and Emr, 2002), and the soybean (Glycine max) PI transfer protein Ssh1p (Monks et al., 2001). To our knowledge, the sensitivity of the PtdIns 4-kinases to non-proteinaceous factors such as membrane lipids has not been explored. The influence of the membrane lipid environment on enzyme activity is suggested from yeast data where overexpression of PIK1 and not STT4 somewhat alleviates the defects of a SEC14 mutation compromised in its PI/phosphatidyl-choline (PtdCho) lipid transfer function (Hama et al., 1999). In addition, the binding of the PH domain of AtPI4Kα1 to the product of the reaction implies that either PtdIns4P regulates the kinase or that PtdIns4Kα1 regulates PtdIns4P localization or availability for further metabolism. To test these hypotheses, we have expressed full-length AtPI4Kα1 and AtPI4Kβ1 and mutated versions of AtPI4Kα1 in Spodoptera frugiperda (Sf9) insect cells to compare their biochemistry and localization. The recombinant enzymes show distinct differences in activity in response to added phospholipids. PtdIns4P inhibits AtPI4Kα1 and enhances the activity of AtPI4Kβ1. In addition, we show differential subcellular localization of the Arabidopsis PtdIns 4-kinase isoforms and demonstrate that the PH domain of the AtPI4Kα1 isoform is, in part, responsible for the differences in both distribution and the lipid sensitivity of enzyme activity.
RESULTS
Sequence Comparisons of Arabidopsis Type III PtdIns 4-Kinases
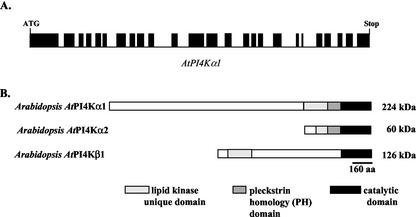
The full-length AtPI4Kα1 cDNA was cloned by designing primers for PCR based on the AtPI4Kα1 genomic sequence identified from the Arabidopsis genome sequencing initiative on chromosome 1 (GenBank accession no. AC007504/At1g49340). The resulting cDNA contained an open reading frame of 6,066 nucleotides and encodes a protein with a predicted molecular mass of 224 kD and a pI of 6.06. When the cDNA sequence was compared with the genomic sequence, it was apparent that there were 25 exons. This agreed with the putative splicing sites from the computer prediction performed by the Arabidopsis sequencing initiative (Fig. 1A).
Figure 1.
Arabidopsis Type III PtdIns 4-kinases. A, Linear representation of the introns and exons (solid bars) of the genomic sequence of AtPI4Kα1. The open reading frame of AtPI4Kα1 is 6,066 nucleotides long. B, Amino acid sequence homology among Arabidopsis PtdIns 4-kinases. AtPI4Kα1, AtPI4Kα2, and AtPI4Kβ1 homologies in the lipid kinase unique domain, PH domain, and the catalytic domain are indicated.
Comparison of the amino acid sequence of AtPI4Kα1 and AtPI4Kβ1 revealed that the two PtdIns 4-kinase isoforms contained significant similarity in the catalytic lipid kinase domain (48% identity, 51% similarity) and the lipid kinase unique domain (33% similar) but otherwise were dissimilar (Fig. 1B). A recent submission of genomic sequence from chromosome 1 into GenBank (accession no. AAG50530) revealed a third putative PtdIns 4-kinase isoform designated AtPI4Kα2 because it has significant predicted amino acid sequence identity to the AtPI4Kα1 isoform (Mueller-Roeber and Pical, 2002). It is predicted to encode 525 amino acids with a molecular mass of approximately 60 kD. The putative catalytic domain, PH domain, and the C-terminal one-half of the AtPI4Kα2 lipid kinase unique domain are 88% identical and 93% similar to the same domains in AtPI4Kα1. The first one-half of the N-terminal putative lipid kinase unique domain of AtPI4Kα2 contains 114 amino acids that have only 14% identity and 33% similarity to AtPI4Kα1 and no significant similarity to any other protein (Fig. 1B).
Biochemical Comparison of PtdIns 4-Kinase Isoforms
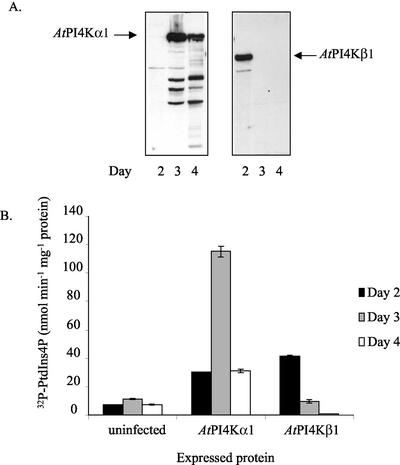
We previously isolated endogenous AtPI4Kα1 from solubilized Arabidopsis microsomes by immunoaffinity purification (Stevenson et al., 1998). This technique gave low yields of unstable enzyme and the antiserum bound to PtdIns4Kβ1 and an unidentified 60-kD polypeptide (which may be the abovedescribed AtPI4Kα2); therefore, to compare the biochemical properties of AtPI4Kα1 and AtPI4Kβ1, we chose to express the full-length Arabidopsis PtdIns4-kinases in baculovirus-infected Sf9 insect cells. The Sf9 cells provide a robust eukaryotic recombinant protein synthesis system whereby recombinant polypeptides are properly processed and targeted within the cells (Luckow, 1995). Sf9 cells were infected with recombinant baculovirus particles harboring the desired cDNAs for the indicated periods of time, and production of the recombinant proteins was monitored by immunoblotting with anti-AtPI4Kα1 antiserum (Fig. 2A). Under the assay conditions used, at 3 d after infection the AtPI4Kα1-specific activity was consistently 12- to 24-fold higher than that of uninfected cells and 2- to 5-fold higher than that of cells expressing AtPI4Kβ1 at 2 d after infection (Fig. 2B). By 4 d after infection, the AtPI4Kα1 protein was still being produced; however, the amount of functional enzyme decreased (Fig. 2B). In 3-d cells expressing AtPI4Kα1, the recombinant enzyme accounted for greater than 90% of the PtdIns 4-kinase-specific activity and in 2-d cells expressing AtPI4Kβ1, the recombinant enzyme accounted for greater than 75% of the specific activity. These respective lysates were used for the biochemical studies.
Figure 2.
Expression and PtdIns 4-kinase activity of AtPI4Kα1 and AtPI4Kβ1 in Sf9 insect cells. A, Immunoblots of the lysates of insect cells expressing either AtPI4Kα1 or AtPI4Kβ1 on indicated days after infection. Thirty micrograms of the cleared lysates was separated by SDS-PAGE, blotted onto polyvinylidene difluoride (PVDF) membrane, and probed with anti-AtPI4Kα1 antisera. Similar results were obtained from at least three independent recombinant lysates. B, PtdIns 4-kinase-specific activity of the lysates of insect cells expressing PtdIns 4-kinases. The cells were lysed and sonicated, and the cleared lysates were assayed for PtdIns 4-kinase activity on indicated days after infection. The averages of duplicate samples of a representative experiment are shown. Similar results were obtained from at least three independent recombinant lysates.
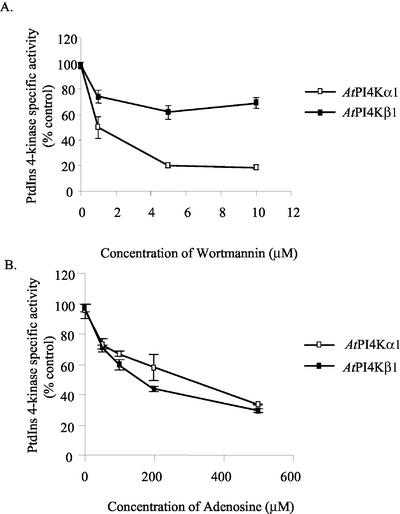
Classically, adenosine and wortmannin sensitivity have been used as tools to characterize the PtdIns 4-kinases (Pike, 1992; Drøbak et al., 1999; Westergren et al., 1999; Balla et al., 2002; Mueller-Roeber and Pical, 2002). We compared the effect of wortmannin and adenosine on the recombinant α- and β-isoforms from Arabidopsis (Fig. 3A). AtPI4Kα1 activity was inhibited 50% in the presence of 1 μm wortmannin and 80% by 5 μm wortmannin. In contrast, AtPI4Kβ1 was only inhibited 25% by wortmannin at concentrations up to 10 μm. Adenosine has been shown in other systems to inhibit PtdIns 4-kinases at concentrations ranging from 20 μm for the type II PtdIns 4-kinases to 1.5 mm for the type III PtdIns 4-kinases (Carpenter and Cantley, 1990; Pike, 1992; Gehrmann and Heilmeyer, 1998; Minogue et al., 2001; Balla et al., 2002). Both AtPI4Kα1 and AtPI4Kβ1 retained 40% of their activity at 500 μm adenosine (Fig. 3B). In contrast to type II PtdIns 4-kinases, which retain negligible activity at 500 μm adenosine (Carpenter and Cantley, 1990; Minogue et al., 2001), the plant PtdIns 4-kinase activities, described thus far, appear to be relatively insensitive to high concentrations of adenosine (Yang and Boss, 1994; Okpodu et al., 1995; Westergren et al., 1999). This is typical of the type III kinases.
Figure 3.
Wortmannin and adenosine sensitivity of AtPI4Kα1 and AtPI4Kβ1. Sf9 insect cells expressing AtPI4Kα1 and AtPI4Kβ1 were harvested 2 (AtPI4Kβ1) and 3 (AtPI4Kα1) d after infection with recombinant baculoviruses. Eight micrograms of the cleared lysates was assayed for PtdIns 4-kinase activity in the presence of 0, 1, 5, and 10 μm of wortmannin (A) or 50, 100, 200, and 500 μm of adenosine (B). Wortmannin was dissolved in dimethyl sulfoxide; hence, the PtdIns 4-kinase assay control was determined in the presence of an equal volume (1 μL) of dimethyl sulfoxide alone. Values are the average of duplicate samples of a representative experiment. Experiments from two independent recombinant baculovirus infections gave similar results.
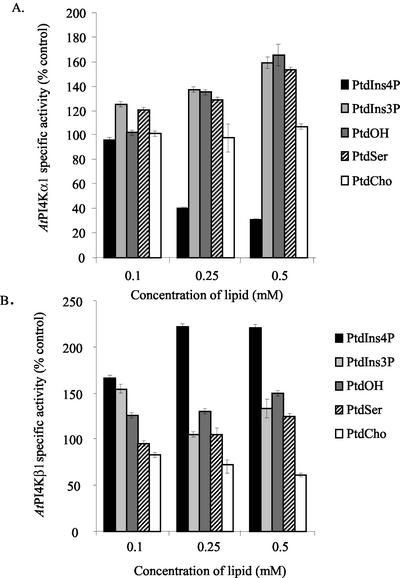
Because the PtdIns 4-kinases are lipid kinases and must associate with phospholipids for activity and because the PH domain of AtPI4Kα1 was previously shown to bind specifically to PtdIns4P (Stevenson et al., 1998), we explored the possibility that various phospholipids, including PtdIns4P, might influence the activity of the PtdIns 4-kinases. Equal amounts of insect cell lysates from cells overexpressing either AtPI4Kα1 or AtPI4Kβ1 were pre-incubated with 0.1, 0.25, and 0.5 mm PtdIns3P, PtdIns4P, PtdOH, PtdSer, or PtdCho delivered as Triton X-100 detergent lipid micelles and then assayed for PtdIns 4-kinase activity. The final concentrations of Triton X-100 and PtdIns were kept constant (0.2% [v/v] and 5.8 mm, respectively) for all reactions. Control reactions were pre-incubated with Triton X-100 alone instead of the detergent lipid micelles. As shown in Figure 4, the PtdIns 4-kinase isoforms were affected differently by the presence of certain phospholipids. The most striking differences occurred when PtdIns4P, the product of the reaction, was added. AtPI4Kα1 was inhibited 60% and 70% by the presence of 0.25 and 0.5 mm PtdIns4P, respectively. The effect was specific for PtdIns4P because 0.5 mm PtdIns3P resulted in a 50% increase in the specific activity of AtPI4Kα1. In comparison, both PtdIns4P and PtdIns3P increased AtPI4Kβ1-specific activity 50% at low concentrations (0.1 mm), and PtdIns4P increased the specific activity of AtPI4Kβ1 more than 2-fold at higher concentrations (0.25 or 0.5 mm PtdIns4P). The differential effects of the PtdInsP isomers, PtdIns3P and PtdIns4P, offer a potentially distinctive mechanism of regulation for each of these type III PtdIns 4-kinase isoforms.
Figure 4.
Phospholipids affect type III PtdIns 4-kinase activities. Sf9 insect cells expressing AtPI4Kα1 (A) and AtPI4Kβ1 (B) were harvested 2 (AtPI4Kβ1) and 3 (AtPI4Kα1) d after infection with recombinant baculovirus. Eight micrograms of the cleared lysates was assayed for PtdIns 4-kinase activity in the presence of increasing concentrations of PtdIns4P, PtdIns3P, PtdOH, PtdSer, and PtdCho. The lipids were prepared in 1% (v/v) Triton X-100, and the final concentrations of Triton X-100 and PtdIns in the PtdIns 4-kinase assay were kept constant at 0.2% (v/v) and 5.8 mm, respectively. Values are the average of duplicate samples of a representative experiment. Experiments from two independent recombinant baculovirus infections gave similar results.
A comparison of other negatively charged lipids (PtdOH and PtdSer) and a zwitterionic lipid (PtdCho) indicated that, in contrast to the effect of PtdIns4P, PtdOH and PtdSer increased the specific activity of AtPI4Kα1. AtPI4Kβ1 was affected by the negatively charged lipids to a lesser extent. Specifically, AtPI4Kα1-specific activity increased up to 70% when either 0.5 mm PtdOH or PtdSer were added, whereas at the same concentrations, PtdOH and PtdSer increased the specific activity of AtPI4Kβ1 only 25% to 50% (Fig. 4). PtdCho had no effect on AtPI4Kα1-specific activity; however, 0.5 mm PtdCho inhibited AtPI4Kβ1-specific activity by 50%.
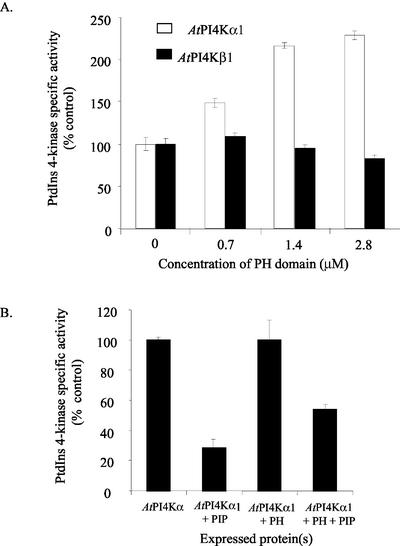
To test the ability of the AtPI4Kα1 PH domain to affect PtdIns 4-kinase activity, we added purified recombinant PH domain to the lysates of insect cells overexpressing AtPI4Kα1 or AtPI4Kβ1. The addition of less than 3 μm PH domain enhanced AtPI4Kα1 activity over 2-fold, whereas the addition of the same amount of PH domain slightly decreased AtPI4Kβ1 activity (Fig. 5A). These results are consistent with the finding that AtPI4Kα1 is inhibited by PtdIns4P, whereas AtPI4Kβ1 is not. Therefore, when the PH domain is added to the AtPI4Kα1 reaction mixture, the polypeptide should bind to PtdIns4P as it is synthesized and relieve product inhibition. The decrease in activity of the AtPI4Kβ1 isoform when high concentrations of PH domain peptide are added suggests that the production of PtdIns4P normally enhances enzyme activity and is consistent with the data in Figure 4B.
Figure 5.
The AtPI4Kα1 PH domain affects PtdIns 4-kinase activity. A, Sf9 insect cells expressing AtPI4Kα1 and AtPI4Kβ1 were harvested 2 (AtPI4Kβ1) and 3 (AtPI4Kα1) d after infection with recombinant baculovirus. Eight micrograms of the cleared lysates was assayed for PtdIns 4-kinase activity in the presence of increasing concentrations of affinity-purified recombinant AtPI4Kα1 PH domain. Values are the average of duplicates from a representative experiment. Similar results were seen in two independent recombinant baculovirus infections. B, Effect of co-expression of the PH domain with AtPI4Kα1 on PtdIns4P inhibition of PtdIns 4-kinase activity. Sf9 insect cells expressing AtPI4Kα1 alone or cells co-expressing AtPI4Kα1 and the cDNA encoding the PH domain were lysed and analyzed for PtdIns 4-kinase activity in the presence or absence of 0.5 mm PtdIns4P. The PtdIns 4-kinase-specific activity of each recombinant baculovirus infection in the absence of PtdIns4P was normalized to 100%. For each infection, the effect of the addition of 0.5 mm PtdIns4P was compared with the same infection without PtdIns4P. Values shown are the average of duplicates in a representative experiment. Similar results were seen with three independent recombinant baculovirus infections.
Feedback inhibition of AtPI4Kα1 could also be alleviated if the recombinant PH domain was coexpressed in the same cells with AtPI4Kα1. The PH domain cDNA was co-expressed with AtPI4Kα1 in the insect cells, and the lysates were assayed for PtdIns 4-kinase activity with and without the addition of 0.5 mm PtdIns4P. As can be seen in Figure 5B, when the cells expressed only AtPI4Kα1, the PtdIns 4-kinase-specific activity was reduced by approximately 70% in the presence of 0.5 mm PtdIns4P, but when the cells produced the PH domain and the AtPI4Kα1 polypeptides together, the PtdIns 4-kinasespecific activity was inhibited less (approximately 50%) in the presence of 0.5 mm PtdIns4P. Together, these data and previous binding studies (Stevenson et al., 1998) indicate that the AtPI4Kα1 PH domain alleviates feedback inhibition of AtPI4Kα1 activity by binding PtdIns4P.
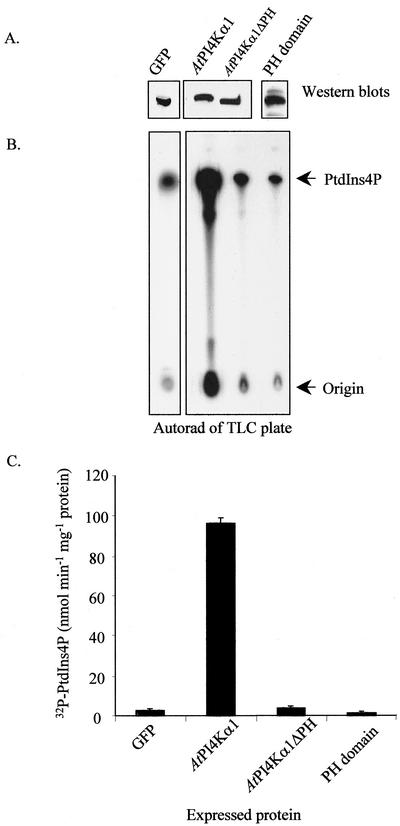
To gain insight as to whether the PH domain was essential for AtPI4Kα1 activity, we constructed a deletion mutation in the AtPI4Kα1 cDNA, designated AtPI4Kα1ΔPH, and expressed the mutant and the PH domain alone in insect cells. An identical time course of expression was performed as described for the expression of recombinant AtPI4Kα1 and AtPI4Kβ1. Optimal production for both the AtPI4Kα1ΔPH and the PH domain polypeptides was found to be on the 3rd d after infection; therefore, cells producing these polypeptides were harvested after 3 d and used in the subsequent studies. Figure 6 shows a comparison of the PtdIns 4-kinase activity of insect cell extracts that expressed the recombinant forms of AtPI4Kα1 cDNA with and without its PH domain. Figure 6A is a western blot using the anti-AtPI4Kα antibody to identify the recombinant AtPI4Kα1, AtPI4Kα1ΔPH, and PH domain polypeptides and to show that they were produced at approximately equally amounts. Insect cells expressing GFP were used as an infection control for comparison of PtdIns 4-kinase activity. Cells overproducing GFP consistently had less PtdIns 4-kinase activity than uninfected cells. Figure 6B shows the autoradiogram of the TLC plate that separated the radioactive products of the PtdIns 4-kinase assay, and Figure 6C shows the quantification of the 32P-labeled PtdIns4P. Insect cells overexpressing the full-length AtPI4Kα1 had 32-fold higher PtdIns 4-kinase-specific activity than cells overexpressing the GFP cDNA. Cells that were expressing AtPI4Kα1ΔPH had greatly reduced PtdIns 4-kinase activity compared with those expressing full-length AtPI4Kα1. AtPI4Kα1 was 22-fold more active than AtPI4Kα1ΔPH. Interestingly, cells overproducing the PH domain itself had lowered endogenous PtdIns 4-kinase-specific activity compared with cells overproducing GFP, which suggested that the PH domain was interfering with endogenous PtdIns 4-kinases of the insect cells or that overproduction of the PH domain was toxic to the cells.
Figure 6.
The PH domain is essential for efficient PtdIns 4-kinase activity. Sf9 insect cells were infected with recombinant baculoviruses harboring the cDNAs for cytosolic green fluorescent protein (GFP), AtPI4Kα1, AtPI4Kα1ΔPH, and the AtPI4Kα1 PH domain alone. Three days after infection, the cells were harvested, lysed, and 8 μg of the cleared lysates was assayed for PtdIns 4-kinase activity in the presence of added substrate, PtdIns, and [γ-32P]ATP. A western blot probed with anti-AtPI4Kα1 or anti-GFP identifying the expressed recombinant polypeptides is shown at the top of A. Below the western is the resulting autoradiograph of the thin-layer chromatography (TLC) plate used to separate the 32P-labeled lipid products of the PtdIns 4-kinase assay. B, Quantification of the TLC plate shown in A using a Bioscan Imaging Scanner. Shown are the averages of duplicate samples of a representative experiment. Similar results were obtained from at least four independent recombinant baculovirus infections.
Table I shows the specific activity of the affinity-purified AtPI4Kα1 and AtPI4Kα1ΔPH polypeptides. When the specific activity of AtPI4Kα1 in insect cell lysates (90–150 nmol min mg protein-1 where over 90% of PtdIns 4-kinase activity was from the overexpressed polypeptide) was compared with that of purified AtPI4Kα1 (2.27 nmol min mg protein-1), it was evident that the enzyme lost activity during purification, implying that there might be some cellular cofactors that enhanced or stabilized the PtdIns 4-kinase activity of AtPI4Kα1. This appears to be a common problem among PtdIns 4-kinases because the activity of recombinant bovine brain PI4Kβ1 was also compromised as it was purified (Zhao et al., 2000). Purified AtPI4Kα1ΔPH exhibited a 94% reduction in activity compared with the full-length AtPI4Kα1. These data show that when the PH domain was deleted from the PtdIns 4-kinase, the activity was greatly compromised, and the PH domain was necessary for optimal PtdIns 4-kinase activity and/or protein folding. When 0.6 μm purified PH domain was added back to the purified AtPI4Kα1ΔPH polypeptide, the PtdIns 4-kinase activity was enhanced almost 2-fold but did not recover to the specific activity of the full-length AtPI4Kα1. These data suggested that at least some of the effect of the PH domain deletion in the purified protein could be rescued by the addition of the peptide domain.
Table I.
A comparison of the specific activities of purified recombinant AtPI4Kα1 and AtPI4Kα1ΔPH
| Sample | Specific Activity | Total Activity |
|---|---|---|
| nmol C min-1mg-1 ± sd(n = 4) | % | |
| AtPI4Kα1 | 2.27+0.16 | 100 |
| AtPI4Kα1ΔPH | 0.14+0.04 | 6 |
| AtPI4Kα1ΔPH + 0.65 | 0.25+0.01 | 11 |
| μm PH domain | ||
| PH domain | 0 | 0 |
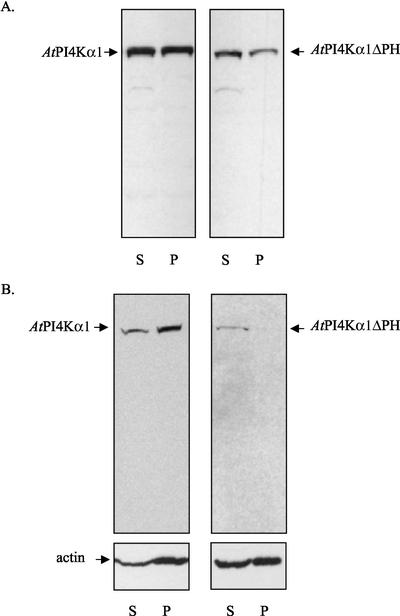
An additional function proposed for some PH domains aside from binding polyphosphoinositides is binding and modulating F actin (Yao et al., 1999). F-actin-binding regions of the PH domains of oxysterol-binding protein and Btk were mapped to the amino-terminal ends that were rich in basic amino acids (Yao et al., 1999). The PH domain of AtPI4Kα has a similar region of basic amino acids, and an F-actin fraction had been shown previously to be enriched in AtPI4Kα1 (Stevenson et al., 1998); therefore, we considered the hypothesis that the PH domain of AtPI4Kα1 might play a role in the association of AtPI4Kα1 with the F-actin enriched fraction of the cell. To test this hypothesis, we assessed the ability of the recombinant AtPI4Kα1 and AtPI4Kα1ΔPH polypeptides to associate with polymerized F-actin in vitro. Extracts from Sf9 cells that were overexpressing AtPI4Kα1 and AtPI4Kα1ΔPH were solubilized in 0.5% (v/v) Triton X-100 and then centrifuged at 20,000g or 100,000g to obtain a supernatant devoid of insoluble proteins, aggregates, and polymerized F actin. The supernatants were then treated to enhance the polymerization of the remaining endogenous G actin by the addition of KCl and MgCl2 as described in “Materials and Methods” (Carraway and Carraway, 1992). After polymerization had occurred, the samples were again centrifuged at 20,000g or 100,000g, respectively. Figure 7 reveals that the distribution of AtPI4Kα1 was equal between the supernatant and the pellet at 20,000g and that more of AtPI4Kα1 resided in the F-actin 100,000g pellet. In contrast, AtPI4Kα1ΔPH was more prevalent in the 20,000g supernatant than the pellet and was detected only in the supernatant in 100,000g fraction. These data imply that although both AtPI4Kα1 and to a lesser extent, AtPI4Kα1ΔPH, may associate with the larger F-actin filaments that were sedimented at 20,000g, only AtPI4Kα1 preferentially associated with the fine filaments that were sedimented at 100,000g. The western blot of actin showing an increased distribution of actin in the 100,000g pellet when the full-length AtPI4Kα1 was present suggested that in the presence of the full-length PtdIns 4-kinase, proportionally more of the fine filaments were recovered. These data suggest that the PH domain of AtPI4Kα1 may play a role in associating AtPI4Kα1 with actin filaments and that this interaction may enhance fine filament formation.
Figure 7.
The AtPI4Kα1 PH domain binds F actin. Sf9 insect cells expressing either AtPI4Kα1 or AtPI4Kα1ΔPH were harvested 3 d after infection with recombinant baculoviruses. The cells were lysed and solubilized as described in “Materials and Methods.” The 20,000g (A) and 100,000g (B) supernatants were treated with actin polymerizing conditions and then centrifuged again at the same speed. The resulting supernatants (S) and pellets (P) were separated by SDSPAGE, electroblotted onto PVDF membrane, and probed with antiAtPI4Kα1 antisera (A and B) and anti-actin (B). The experiment was repeated at least three times from two independent baculovirus infections with similar results.
GFP Localization of Arabidopsis PtdIns 4-Kinases in Sf9 Cells
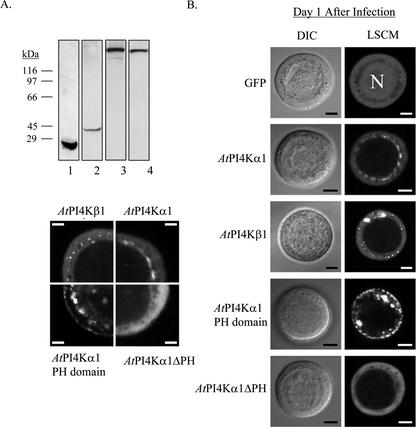
To determine whether there were differences in the subcellular localization of the different isoforms of the Arabidopsis PtdIns 4-kinases in Sf9 cells and whether the PH domain played a role in the localization of AtPI4Kα1, we constructed GFP fusion proteins for baculovirus expression and visualized the localization of the fusion proteins with using laser scanning confocal microscopy (LSCM). To avoid potential problems or artifacts from overproduction of recombinant polypeptides and to avoid the effects of late-stage viral infection on the cell morphology, we visualized cells 24 h after infection. Figure 8A shows a western blot of the GFP fusion proteins present in the insect cells after 1 d of infection probed with anti-AtPI4Kα1 and anti-GFP antibodies. The immunoblots show that the fusion proteins are correctly synthesized intact with no breakdown products (Persson et al., 2002). The polypeptide representing GFP-AtPI4Kβ1 could not be identified by immunoblotting, presumably because of low expression levels after only 1 d. However, immunoblots of extracts from identical cells overexpressing AtPI4Kβ1 for 2 d revealed intact GFP fusion proteins (not shown).
Figure 8.
A. western blot of GFP-fusion proteins produced in Sf9 cells 1 d after infection by the recombinant baculoviruses. Insect cells expressing GFP (lane 1), GFP-PH domain (lane 2), GFPAtPI4Kα1 (lane 3), and GFP-AtPI4Kα1ΔPH (lane 4) were harvested 24 h postinfection. The cells were lysed by sonication in lysis buffer. Thirty micrograms of the cleared lysate was loaded onto a 10% (w/v) acrylamide SDS-PAGE gel. The gel was electroblotted onto PVDF, and the blot was probed with anti-GFP (lane 1) or antiAtPI4Kα1 antisera (lanes 2–4). B, AtPI4Kα1 and AtPI4Kβ1 are differentially localized in Sf9 cells. Sf9 insect cells infected with recombinant baculoviruses harboring the specified cDNAs fused to GFP were visualized by differential interference contrast and LSCM 24 h (d 1) after infection as described in “Materials and Methods.”C, An enlarged image showing one-quarter of a representative cell from each recombinant baculovirus infection, as in B. N, Nucleus. Scale bars = 5 μm. Cells shown are representative of two independent expression experiments.
As seen in Figure 8, the GFP fusion proteins are readily detected and show differences in their distribution within the transgenic cells. GFP alone expressed in the insect cells was widely distributed throughout the cell, both in the cytoplasm and the nucleus. GFP-AtPI4Kα1 was excluded from the nucleus and appeared to be mostly associated with perinuclear membranes. In contrast, GFP-AtPI4Kα1ΔPH appeared to be more evenly dispersed throughout the cytosol and not obviously associated with any particular membrane component, indicating that the PH domain was necessary for the association of GFP-AtPI4Kα1 to perinuclear membranes. The fluorescence from the GFP-PH domain had a very different appearance, with a striking punctate fluorescence associated with what appeared to be membranous components throughout the cytosol and the plasma membrane. Finally, the fluorescence associated with the GFP-AtPI4Kβ1 was primarily located in the cytoplasm with increased fluorescence around dispersed, cytoplasmic vesicles.
DISCUSSION
The plant PtdIns 4-kinases historically have been difficult to purify to homogeneity. For this reason, the comparative characterization of each isoform has been lacking. In this study, the baculovirus expression system was used to overexpress functional PtdIns 4-kinases for comparing their regulation and biochemistry. It is noteworthy that the cells overexpressing AtPI4Kβ1 consistently grew at approximately one-half the rate of cells overexpressing AtPI4Kα1, and expression of AtPI4Kβ1 was virtually halted after the 2nd d of infection. This indicated that overexpression of AtPI4Kβ1 was toxic, whereas overexpression of the AtPI4Kα1 isoform was much more tolerable to the cells.
In addition to our findings described in the results, other insights were gained from the heterologous expression system. The large-molecular mass AtPI4Kα1 isoform has been difficult to detect on immunoblots of plant extracts unless an F-actin-enriched fraction is used (Stevenson et al., 1998). When overproduced in insect cells, AtPI4Kα1 was readily detectable by immunblotting, indicating that the lack of detection in plant extracts was not the result of difficulties with blotting or the immunological reaction but rather suggesting that AtPI4Kα1 is present in very low abundance in planta. Neither cold nor osmotic stress seemed to increase the presence of the AtPI4Kα1 polypeptide on immunoblots of isolated plant membranes (data not shown).
The antisera used in this study were generated against the C-terminal third region of AtPI4Kα1 and recognized both recombinant AtPI4Kα1 and AtPI4Kβ1 in addition to a smaller polypeptide in both Arabidopsis and maize (Zea mays; J. Stevenson-Paulik and W.F. Boss, unpublished data; Shank et al., 2001). The putative smaller isoform of AtPI4Kα1, AtPI4Kα2, has significant sequence similarity to AtPI4Kα1. Its deduced amino acid sequence shares 93% similarity with AtPI4Kα1 beyond the first 100 N-terminal amino acids. Interestingly, when the C-terminal 537 amino acids of AtPI4Kα1, which encompassed the same domains and size as AtPI4Kα2, was expressed as a polypeptide in Sf9 cells, it did not possess any PtdIns 4-kinase activity (data not shown). That the truncated polypeptide of AtPI4Kα1 did not have activity and yet is very similar to AtPI4Kα2 implies that either the N-terminal 114 amino acids of AtPI4Kα2 confer a unique and necessary function that regulates the catalytic domain or that AtPI4Kα2 is not active and may be a pseudogene that was duplicated and truncated from AtPI4Kα1. The suggestion that AtPI4Kα2 is an unexpressed pseudogene in the Arabidopsis genome seems unlikely because a prominent 60-kD polypeptide that would represent AtPI4Kα2 was recognized on western blots of fractionated Arabidopsis total microsomes (data not shown). Furthermore, the 60-kD putative isoform identified here may represent the previously observed smaller isoform partially purified, type III PtdIns 4-kinase from spinach plasma membranes (Westergren et al., 1999) and a putative 65-kD PtdIns 4-kinase identified in the maize mutant, floury-2 and in soybean cells, where it was found to increase during an endoplasmic reticulum stress response (Shank et al., 2001).
For biochemical comparisons of AtPI4Kα1 and AtPI4Kβ1, we used proteins of similar purity assayed under the same conditions. The fungal metabolite wortmannin is established as a potent inhibitor of PtdIns 3-kinases at nanomolar concentrations and was believed to be a specific inhibitor of this enzyme. Recently, wortmannin, at higher (micromolar) concentrations, also has been shown to be an effective inhibitor of PtdIns 4-kinase activity (Balla et al., 1997; Cutler et al., 1997; Meyers and Cantley, 1997; Xue et al., 1999; Jung et al., 2002; Mueller-Roeber and Pical, 2002). There are some discrepancies as to what extent the α- and β-isoforms from different systems are inhibited by wortmannin. These discrepancies in the effective concentrations of wortmannin may arise from interactions with other potentially wortmanninor PtdIns 4-kinase-interacting proteins present. For example, the decrease in wortmannin sensitivity for AtPI4Kβ1 reported here compared with the previous study (Xue et al., 1999) may be because we did not assay purified protein, whereas the other study did. Although the majority of the PtdIns 4-kinase activity in cells overexpressing AtPI4Kβ1 was due to the recombinant protein, there may have been other wortmannin-binding or AtPI4Kβ1-interacting proteins or other factors in the cells that reduced the sensitivity of AtPI4Kβ1 to wortmannin. Most importantly, using the same heterologous system and assay conditions, we show here that AtPI4Kβ1 was less sensitive to wortmannin (inhibited only 25% by 10 μm wortmannin) than AtPI4Kα1 (inhibited 80% by 5 μm and 50% by 1 μm wortmannin).
A comparison of phospholipid sensitivities proved to be enlightening. AtPI4Kα1 is significantly inhibited by PtdIns4P, whereas AtPI4Kβ1 is activated by its product under conditions where the substrate was in 20-fold excess. Selective end product inhibition of AtPI4Kα1 suggests that once formed, PtdIns4P can bind the AtPI4Kα1 PH domain, which is in close proximity to the catalytic site, and block further enzyme activity. Stoichiometric analysis indicates that a 1:1 ratio of added PH domain (1.4 μm) and to the predicted product formed (0.16 mm PtdIns4P) is not required to relieve inhibition. These data suggest that the PH domain acts allosterically, perhaps binding to the enzyme-product complex, and enabling maximal enzyme activity. It is possible that in vivo other PtdIns 4-kinase- or PtdIns4P-binding proteins, such as PtdInsP kinase, PtdIns4P phosphatase, or PtdIns4P lipid transfer protein, profilin (Drøbak et al., 1994), actin depolymerizing factor (Gungabissoon et al., 1998), and/or dynamins (Kim et al., 2001) could also selectively increase AtPI4Kα1 activity by relieving feedback inhibition. The need for the involvement of a lipid transfer protein to pass PtdIn4P from enzyme to enzyme has been proposed by Cockcroft (1997, 1998). That the β-isoform is product stimulated may suggest that PtdIns4P recruits an activating protein; for instance, Arf1, which has been shown to activate the β isoform of PtdIns 4-kinase (Audhya et al., 2000; Jones et al., 2000). The prevalence of the β-isoform relative to the α-isoform in studies of whole-plant extracts is consistent with the relatively high ratio of PtdIns4P over other polyphosphoinositides in plants and the apparent lack of feedback regulation of PtdIns4P biosynthesis.
Furthermore, the sensitivity of the β-isoform to PtdCho may contribute to the selective effect of PIK1 (the AtPI4Kβ1 ortholog), but not STT4 (the AtPI4Kα1 ortholog), on the sec14 mutant, which lacks a PtdIns/PtdCho transfer protein (Hama et al., 1999). In either event, our data are consistent with a model of interfacial binding of PtdIns 4-kinases (Carman et al., 1995; Gelb et al., 2000), which means that the membrane lipid composition should affect the subcellular localization and activity of the PtdIns 4-kinase isoforms, thereby contributing to their distinct roles in regulating membrane trafficking, cytoskeletal dynamics, and signal transduction.
We have shown here that the PH domain may be involved in the binding of AtPI4Kα1 to F actin and that AtPI4Kα1 sediments with and may facilitate the formation of fine F-actin filaments. If this occurs in vivo, then one scenario would be that as PtdIns4P is formed and subsequently removed from the PH domain by cytoskeleton-binding proteins such as gelsolin and profilin, actin polymerization would be enhanced. The PH domain would then facilitate the translocation of the AtPI4Kα1 to an F-actin-enriched region of fine filaments closely associated with membranes. Alternatively, the PH domain of the PtdIns 4-kinase may bind PtdIns4P and F-actin, simultaneously enhancing actin filament formation at actin nucleation sites along membrane surfaces. Of course, these ideas need to be experimentally tested, but this model agrees with the recent finding that yeast cells harboring temperature sensitive stt4 consistently had disorganized actin at restrictive temperatures when the function of the α-isoform of the PtdIns 4-kinase was compromised (Audhya et al., 2000).
Expression of the GFP fusions with the various PtdIns 4-kinases proved to be useful in visualizing the differences in localization of the plant PtdIns 4-kinase α- and β-isoforms. Based on the data generated by confocal microscopy of the Sf9 cells expressing the GFP fusions, it appears that AtPI4Kα1 associates mostly with internal membranes that surround the nucleus and that the PH domain is necessary for this localization. Furthermore, the PH domain alone appears to be associated very strongly with most membranes outside the nucleus, including the plasma membrane. The punctate staining suggests that the PH domain is localized to very specific regions of the membranes. Perhaps these regions are enriched with PtdIns4P or are points of actin/membrane connections. Wong et al. (1997) showed by subcellular fractionation and immunolocalization in Chinese hamster ovary (CHO) and HeLa cells that the human PI4Kα isoform was associated with the endoplasmic reticulum around the nucleus, whereas the β-isoform of the PtdIns 4-kinase was cytosolic with some association with the Golgi. Our results are consistent with these findings and indicate that the association of the kinases to distinct membrane and cellular compartments may be dictated mainly by components that are common to all eukaryotic cells such as lipids and the actin cytoskeleton. The fact that the structures of the α- and β-isoforms of the PtdIns 4-kinases appear to be conserved across species and that they have nonoverlapping roles based on yeast genetics suggests that they have distinct roles in regulating membrane trafficking, cytoskeletal dynamics, and signal transduction in other species including plants. The challenge for future studies will be to elucidate the distinct functions of the different PtdIns 4-kinases.
MATERIALS AND METHODS
Cloning of Full-Length Arabidopsis PtdIns 4-Kinases
The cDNA for AtPI4Kα1 was amplified from Arabidopsis RNA by RTPCR using primers that were designed based on the predicted open reading frame found on chromosome 1 (GenBank accession no. AC007504/At1g49340) and that correspond with our previously described partial cDNA for AtPI4Kα1 (Stevenson et al., 1998). The sequence of the sense primer was ATG GAG GCA CTG ACG GAG CTT, and the antisense primer sequence was TTA CTT CTC GAT GCC TTG TTG CAA ATA. A region of 6.1 kb was amplified using Platinum Taq DNA High Fidelity DNA polymerase (Life Technologies/Gibco-BRL, Cleveland) from cDNA synthesized from Arabidopsis whole-plant poly(A+) RNA. The 6.1-kb PCR product was subcloned into pBluescript. The cDNA was sequenced in its entirety by the Iowa State Sequencing Facility (Ames). Multiple PCR products were sequenced and compared with the genomic sequence to obtain a clone devoid of mutations.
The full-length cDNA for the open reading frame of AtPI4Kβ1 was amplified from Arabidopsis cDNA with primers that were designed based on the computer prediction of the open reading frame of AtPI4Kβ1 sequenced on chromosome 5 from the Arabidopsis genome sequencing initiative (accession no. AB008266/At5g64070). This sequence corresponds to that of AtPI4Kβ1 identified by Xue et al. (1999). The 3.5-kb PCR product was subcloned into pCR-Blunt and sequenced by the Iowa State Sequencing Facility. Multiple PCR products were sequenced to find an un-mutated cDNA.
To delete the PH domain sequence from the AtPI4Kα1 cDNA, a 3.1-kb partial cDNA containing the 3′ one-half of the AtPI4Kα1 cDNA was used initially to make the PH domain deletion. PCR primers were designed to amplify the cDNA on either side of the PH domain sequence. The primer sequences for cDNA amplification downstream of the PH domain were: sense, 5′CAG GTT CGG CAG GGA GAT GAT TGC-3′; and antisense, 5′- AAA AGT CGA CTT ACT TCT CGA TG-3′. The primers used for upstream amplification were: sense, 5′- GGA CCG AAT ATG CTA AA-3′; and antisense, 5′- GCA ATC ATC TCC CTG CCG AAC CTG-3′. The primers that amplified the cDNA on either side of the PH domain were designed so that their 5′ sequences were complimentary to the 12 nucleotides that flank the PH domain sequence on the opposite side of which they annealed. The resulting PCR products were gel purified and then placed into another PCR amplification reaction in a 1:1 ratio with each other and with the primers that amplify the extreme ends of either fragment. A faint band that resulted from the amplification of the two fragments together was gel purified, reamplified, and subcloned into pCR-blunt (Invitrogen, Carlsbad, CA). The newly formed cDNA of the 3′ one-half of AtPI4Kα1 with a deleted PH domain was sequenced to verify deletion of the PH domain sequence and to check for any additional mutations that may have been incorporated by the DNA polymerase in the process of making the deletion. The above cDNA fragment containing the PH domain deletion and the full-length AtPI4Kα1 cDNA in pBluescript were digested at the 3′ end SalI site and an internal BamHI site that was upstream of the PH domain sequence. The resulting fragments were gel purified and ligated together. Correct ligation was verified by sequence analysis.
The cDNA encoding the AtPI4Kα1 PH domain was amplified by PCR using Pfu DNA polymerase with primers that were complimentary to either end of the PH domain sequence. The sense primer contained a BamHI site on its 5′ end, and the antisense primer contained an EcoRI site on its 5′ end for efficient subcloning of the cDNA into expression vectors. The cDNA encoding soluble GFP (Davis and Vierstra, 1998) was amplified by several different primers that were engineered with various restriction sites on their 5′ ends so that the resulting GFP-cDNA could be efficiently ligated in frame with the PtdIns 4-kinase cDNAs. In every case, the PCR-amplified cDNA was verified for sequence integrity.
The cDNAs encoding AtPI4Kα1, AtPI4Kβ1, AtPI4Kα1ΔPH, the PH domain, GFP, GFP-AtPI4Kα1, GFP-AtPI4Kβ1, GFP-AtPI4Kα1ΔPH, and GFP-PH domain were each subcloned in frame with the N-terminal polyHis tag of the pFastBac HT plasmid. AtPI4Kα1 and AtPI4Kα1ΔPH were subcloned into the EcoRI and SalI sites of the pFastBac HT plasmid. AtPI4Kβ1 was subcloned using NotI and SalI sites. The cDNA encoding the AtPI4Kα1 PH domain was amplified by PCR with a high-fidelity DNA polymerase (Pfu) with primers that had a BamHI site on the 5′ end of the sense primer and an EcoRI site on the 5′ end of the antisense primer. The amplified PCR product was cloned into the pFastBac HT plasmid via the BamHI and EcoRI sites. Primers containing restriction sites for NotI on the sense primer and SalI on the antisense primer were used to generate GFP for ligating in frame with the 5′ end of AtPI4Kα1 and AtPI4Kα1ΔPH in the pFastBac HT plasmid. Primers containing NotI (sense) and XbaI (antisense) sites were used to amplify GFP for in-frame ligation to the 5′ end of AtPI4Kβ1. Primers containing EcoRI (sense) and SalI (antisense) were used to amplify GFP for in-frame ligation to the 3′ end of the PH domain sequence.
Insect Cell Maintenance
Serum-free Spodoptera frugiperda (Sf9) cells were obtained from Life Technologies/Gibco-BRL and maintained at 28°C at a concentration of 2.5 × 105 to 5 × 106 cells mL-1 in Sf-900 II insect cell serum-free medium (Life Technologies/Gibco-BRL). The cells were maintained as 30- to 50-mL suspension cultures in 125-mL shaker flasks with loosened caps and shaken in an air-regulated incubator at 140 rpm.
Heterologous Expression of Lipid Kinases in Sf9 Insect Cells
The expression system used was the Bac-to-Bac Baculovirus Expression System (Life Technologies/Gibco-BRL). Recombinant baculoviruses were generated from the recombinant expression vectors according to the manufacturer's recommendations (Life Technologies/Gibco-BRL). For recombinant polypeptide production, a monolayer of 4 × 106 Sf9 cells was infected at a multiplicity of infection of 9 for 2 to 3 d, depending on the recombinant virus. Optimal production of AtPI4Kβ1 and the PH domain polypeptide was on the 2nd d after infection, and optimal production of AtPI4Kα1 was on the 3rd d after infection. All assays were performed using cells optimally producing the respective polypeptide. Infected cells were then harvested by pouring off the media, rinsing with cold phosphate-buffered saline, and shaking the cells loose by hand five times in the presence of lysis buffer (50 mm Tris [pH 8.5], 5 mm 2-mercaptoethanol, 100 mm KCl, 1 mm phenylmethylsulfonyl fluoride, and 0.05% [v/v] Triton X-100, and 1 mg per 100 mL leupeptin). Cells were then sonicated four times for 15 s on ice, and cell debris was removed by centrifugation at 5,000g for 5 min. The cleared lysate was analyzed for protein concentration by the Bradford method (Bio-Rad Laboratories, Hercules, CA) and assayed for PtdIns 4-kinase activity.
Preparation of Membranes
Arabidopsis plants were coarsely macerated and ground in a Virtis homogenizer with an equal volume (1 g of tissue per 1 mL of buffer) of ice-cold buffer containing 3 mm EDTA, 2 mm EGTA, 30 mm Tris (pH 7.4), 250 mm Suc, 14 mm β-mercaptoethanol, 2 mm dithiothreitol, 0.1 mm phenylmethylsulfonyl fluoride, 1 mg per 100 mL leupeptin, and 2 mm benzamidine. The homogenate was filtered through cheesecloth and centrifuged at 2,000g for 5 min at 4°C. The supernatant was centrifuged at 40,000g for 60 min at 4°C. The microsomal pellet was resuspended with 1/50 the original grinding volume in ice-cold deionized water, and the plasma membranes were isolated by aqueous two-phase partitioning as described by Wheeler and Boss (1987). The membrane fractions were assayed for protein concentration according to the Bradford method (Bio-Rad Laboratories), and equal concentrations of proteins were separated by SDS-PAGE and electroblotted onto PVDF for immunodetection. Triton X-100 (1% [v/v]) solubilization was used to release the polypeptide from a Triton-insoluble fraction, and this enhanced the cross-reaction of the 60-kD polypeptide with the antibody over membranes that were untreated with detergent.
PtdIns 4-Kinase Assays
Five to eight micrograms of the lysate of insect cells expressing recombinant polypeptides were assayed in duplicate. The reaction mixture contained final concentrations of 7.5 mm MgCl2, 1 mm sodium molybdate, 0.25 mg mL-1 PtdIns, 0.2% (v/v) Triton X-100, 0.5 mm ATP, 30 mm Tris (pH 7.2), and 10 μCi [γ-32P]ATP (7,000 Ci mmol-1) in a total volume of 50 μL. Stock PtdIns (5 mg mL-1) was solubilized in 1% (v/v) Triton X-100. The reactions were mixed by vortexing and incubated at 25°C for 10 min with intermittent shaking. The reactions were stopped by adding 1.5 mL of ice-cold CHCl3: MeOH (1:2 [v/v]) and kept at 4°C until the lipids were extracted. Lipids were extracted, dried, separated by TLC, and analyzed with a Bioscan System 500 Imaging Scanner as previously described (Cho et al., 1993; Stevenson et al., 1998). Lipids were prepared as detergent micelles by first solubilizing the lipids in CHCl3:MeOH (2:1 [v/v]) and then dried under a constant stream of N2 gas to form a uniform layer of lipid along the glass test tube. Once dried, the lipids were solubilized with vigorous vortexing to a final stock concentration of 5 mg mL-1 in 1% (v/v) Triton X-100 in 30 mm Tris (pH 7.4). Lipids that were added to the PtdIns 4-kinase assay were incubated with the sample for 5 min before the addition of substrate and ATP.
F-Actin Sedimentation Assay
Insect cell lysates expressing recombinant AtPI4Kα1 or AtPI4Kα1ΔPH were lysed as described above, solubilized in 0.5% (v/v) Triton X-100, and centrifuged at 20,000g at 4°C for 1 h or 100,000g for 30 min at 4°C. One hundred micromolar KCl and 2 mm MgCl2 (final concentrations) were added to the resulting supernatants to enhance actin polymerization (Carraway and Carraway, 1992). The solutions were incubated for 1 h at 25°C. The samples were then centrifuged again at either 20,000g or 100,000g for 1 h or 30 min at 4°C, respectively. The entire resulting pellet and supernatant was loaded onto an SDS-PAGE gel and subjected to immunoblot analysis with either the polyclonal anti-PtdIns 4-kinase antibody (Stevenson et al., 1998) or a monoclonal anti-actin antibody (ICN Pharmaceuticals, Costa Mesa, CA).
GFP Localization by Confocal Microscopy
Sf9 cells grown in suspension and expressing the recombinant GFP alone or as fusion proteins to AtPI4Kα1, AtPI4Kα1ΔPH, AtPI4Kβ1, and the PH domain of AtPI4Kα1 were visualized by LSCM 1 d postinfection. The confocal images were obtained using a Leica RXA fluorescence microscope equipped with a Leica PL APO 100× NA 1.25 oil immersion lens (Leica, Wetzlar, Germany). The microscope was attached to a Leica TCS-SP laser scanning head, and the images were acquired using Leica TCS-NT software. Light paths and wavelengths were controlled using a 488/568/633 triple dichroic filter. GFP was excited at 488 nm using an argon laser, and GFP fluorescence emission was recorded at 525 to 550 nm. Identical electronic parameters were used for all images. Simultaneous differential interference contrast images were recorded using a transmitted light detector.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available without restrictions for noncommercial research purposes.
Acknowledgments
We would like to acknowledge the use of the laser confocal microscope in the North Carolina State University Cell and Molecular Imaging Center under the direction of Dr. N. Strömgren Allen.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.021758.
This work was supported in part by the National Science Foundation (grant no. MCD 0091090) and in part by the North Carolina Agricultural Research Service.
References
- Audhya A, Emr SD (2002) Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev Cell 2: 593-605 [DOI] [PubMed] [Google Scholar]
- Audhya A, Foti M, Emr SD (2000) Distinct roles for the yeast phosphatidylinositol 4-kinases, stt4p and pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell 11: 2673-2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A, Tuymetova G, Barshishat M, Geiszt M, Balla T (2002) Characterization of type II phosphatidylinositol 4-kinase isoforms reveals association of the enzymes with endosomal vesicular compartments. J Biol Chem 277: 20041-20050 [DOI] [PubMed] [Google Scholar]
- Balla T, Downing GJ, Jaffe H, Kim S, Zolyomi A, Catt KJ (1997) Isolation and molecular cloning of wortmannin-sensitive bovine type III phosphatidylinositol 4-kinases. J Biol Chem 272: 18358-18366 [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Irvine RF (1989) Inositol phosphates and cell signaling. Nature 341: 197-205 [DOI] [PubMed] [Google Scholar]
- Bunney TD, Watkins PA, Beven AF, Shaw PJ, Hernandez LE, Lomonossoff GP, Shanks M, Peart J, Drobak BK (2000) Association of phosphatidylinositol 3-kinase with nuclear transcription sites in higher plants. Plant Cell 12: 1679-1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Deems RA, Dennis EA (1995) Lipid signaling enzymes and surface dilution kinetics. J Biol Chem 270: 18711-18714 [DOI] [PubMed] [Google Scholar]
- Carpenter CL, Cantley LC (1990) Phosphoinositide kinases. Biochemistry 29: 11147-11156 [DOI] [PubMed] [Google Scholar]
- Carraway KL, Carraway CAC (1992) The Cytoskeleton: A Practical Approach. Oxford University Press, New York
- Cho MH, Shears SB, Boss WF (1993) Changes in phosphatidylinositol metabolism in response to hyperosmotic stress in Daucus carota L. cells grown in suspension culture. Plant Physiol 103: 637-647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft S (1997) Phosphatidylinositol transfer proteins: requirements in phospholipase C signaling and in regulated exocytosis. FEBS Lett 410: 44-48 [DOI] [PubMed] [Google Scholar]
- Cockcroft S (1998) Phosphatidylinositol transfer proteins: a requirement in signal transduction and vesicle traffic. BioEssays 20: 423-432 [DOI] [PubMed] [Google Scholar]
- Cutler NS, Heitman J, Cardenas ME (1997) STT4 is an essential phosphatidylinositol 4-kinase that is a target of wortmannin in Saccharomyces cerevisiae. J Biol Chem 272: 27671-27677 [DOI] [PubMed] [Google Scholar]
- Davis SJ, Vierstra RD (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol 36: 521-528 [DOI] [PubMed] [Google Scholar]
- De Matteis MA, Godi A, Corda D (2002) Phosphoinositides and the Golgi complex. Curr Opin Cell Biol 14: 434-447 [DOI] [PubMed] [Google Scholar]
- Drøbak BK, Dewey RE, Boss WF (1999) Phosphoinositide kinases and the synthesis of polyphosphoinositides in higher plant cells. In KW Jeon, ed, International Review of Cytology. Academic Press, New York, pp 95-130 [DOI] [PubMed]
- Drøbak BK, Watkins PAC, Valenta R, Dove SK, Lloyd CW, Staiger CJ (1994) Inhibition of plant plasma membrane phosphoinositide phospholipase C by the actin-binding protein, profilin. Plant J 6: 389-400 [Google Scholar]
- Gehrmann T, Heilmeyer LM (1998) Phosphatidylinositol 4-kinases. Eur J Biochem 253: 357-370 [DOI] [PubMed] [Google Scholar]
- Gelb MH, Min JH, Jain MK (2000) Do membrane-bound enzymes access their substrates from the membrane or aqueous phase: interfacial versus non-interfacial enzymes. Biochim Biophys Acta 1488: 20-27 [DOI] [PubMed] [Google Scholar]
- Gross W, Yang W, Boss WF (1992) Release of carrot plasma membraneassociated phosphatidylinositol kinase by phospholipase A2 and activation by a 70 kDa protein. Biochim Biophys Acta 1134: 73-80 [DOI] [PubMed] [Google Scholar]
- Gungabissoon RA, Jiang C-J, Drobak BK, Maciver SK, Hussey PJ (1998) Interaction of maize actin-depolymerising factor with actin and phosphoinositides and its inhibition of plant phospholipase C. Plant J 16: 689-696 [Google Scholar]
- Hama H, Schnieders EA, Thorner J, Takemoto JY, DeWald DB (1999) Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J Biol Chem 274: 34294-34300 [DOI] [PubMed] [Google Scholar]
- Hendricks KB, Wang BQ, Schnieders EA, Thorner J (1999) Yeast homologue of neuronal frequenin is a regulator of phosphatidylinositol-4-OH kinase. Nat Cell Biol 1: 234-241 [DOI] [PubMed] [Google Scholar]
- Hendrix W, Assefa H, Boss WF (1989) The polyphosphoinositides, phosphatidylinositol monophosphate and phosphatidylinositol bisphosphate are present in nuclei isolated from carrot protoplasts. Protoplasma 151: 62-72 [Google Scholar]
- Jones DH, Morris JB, Morgan CP, Kondo H, Irvine RF, Cockcroft S (2000) Type I phosphatidylinositol 4-phosphate 5-kinase directly interacts with ADP-ribosylation factor 1 and is responsible for phosphatidylinositol 4,5-bisphosphate synthesis in the Golgi compartment. J Biol Chem 275: 13962-13966 [DOI] [PubMed] [Google Scholar]
- Jung JY, Kim YW, Kwak JM, Hwang JU, Young J, Schroeder JI, Hwang I, Lee Y (2002) Phosphatidylinositol 3- and 4-phosphate are required for normal stomatal movements. Plant Cell 14: 2399-2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YW, Park DS, Park SC, Kim SH, Cheong GW, Hwang I (2001) Arabidopsis dynamin-like 2 that binds specifically to phosphatidylinositol 4-phosphate assembles into a high-molecular weight complex in vivo and in vitro. Plant Physiol 127: 1243-1255 [PMC free article] [PubMed] [Google Scholar]
- Luckow VA (1995) Baculovirus Expression Systems and Biopesticides. Wiley-Liss, Inc., New York
- Meyers R, Cantley LC (1997) Cloning and characterization of a wortmannin-sensitive human phosphatidylinositol 4-kinase. J Biol Chem 272: 4384-4390 [DOI] [PubMed] [Google Scholar]
- Minogue S, Anderson JS, Waugh MG, dos Santos M, Corless S, Cramer R, Hsuan JJ (2001) Cloning of a human type II phosphatidylinositol 4-kinase reveals a novel lipid kinase family. J Biol Chem 276: 16635-16640 [DOI] [PubMed] [Google Scholar]
- Monks DE, Aghoram K, Courtney PD, DeWald DB, Dewey RE (2001) Hyperosmotic stress induces the rapid phosphorylation of a soybean phosphatidylinositol transfer protein homolog through activation of the protein kinases SPK1 and SPK2. Plant Cell 13: 1205-1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Roeber B, Pical C (2002) Inositol phospholipid metabolism in Arabidopsis: characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol 130: 22-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Irvine RF, Musgrave A (1998) Phospholipid signalling in plants. Biochim Biophys Acta 1389: 222-272 [DOI] [PubMed] [Google Scholar]
- Okpodu CM, Gross W, Burkhart W, Boss WF (1995) Purification and characterization of a soluble phosphatidylinositol 4-kinase from carrot suspension culture cells. Plant Physiol 107: 491-500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson SH, Love J, Tsou P-L, Robertson D, Thompson WF, Boss WF (2002) When a day makes a difference. Interpreting data from endoplasmic reticulum-targeted green fluorescent protein fusions in cells grown in suspension culture. Plant Physiol 128: 341-344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike LJ (1992) Phosphatidylinositol 4-kinases and the role of polyphosphoinositides in cellular regulation. Endocrinol Rev 13: 692-706 [DOI] [PubMed] [Google Scholar]
- Shank KJ, Su P, Brglez I, Boss WF, Dewey RE, Boston RS (2001) Induction of lipid metabolic enzymes during the endoplasmic reticulum stress response in plants. Plant Physiol 126: 267-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommarin M, Sandelius AS (1988) Phosphatidylinositol and phosphatidylinositolphosphate kinases in plant plasma membranes. Biochim Biophys Acta 958: 268-278 [Google Scholar]
- Stevenson JM, Perera IY, Boss WF (1998) A phosphatidylinositol 4-kinase pleckstrin homology domain that binds phosphatidylinositol 4-monophosphate. J Biol Chem 273: 22761-22767 [DOI] [PubMed] [Google Scholar]
- Stevenson JM, Perera IY, Heilmann I, Persson S, Boss WF (2000) Inositol signaling and plant growth. Trends Plant Sci 5: 252-258 [DOI] [PubMed] [Google Scholar]
- Tan Z, Boss WF (1992) Association of phosphatidylinositol kinase, phosphatidylinositol monophosphate kinase, and diacylglycerol kinase with the cytoskeleton and F-actin fractions of carrot (Daucus carota L.) cells grown in suspension culture. Plant Physiol 100: 2116-2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X (2001) Plant phospholipases. Annu Rev Plant Physiol Plant Mol Biol 52: 211-231 [DOI] [PubMed] [Google Scholar]
- Westergren T, Ekblad L, Jergil B, Sommarin M (1999) Phosphatidylinositol 4-kinase associated with spinach plasma membranes: isolation and characterization of two distinct forms. Plant Physiol 121: 507-516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JJ, Boss WF (1987) Polyphosphoinositides are present in plasma membranes isolated from fusogenic carrot cells. Plant Physiol 85: 389-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K, Meyers R, Cantley LC (1997) Subcellular locations of phosphatidylinositol 4-kinase isoforms. J Biol Chem 272: 13236-13241 [DOI] [PubMed] [Google Scholar]
- Xu P, Lloyd CW, Staiger CJ, Drøbak BK (1992) Association of phosphatidylinositol 4-kinase with the plant cytoskeleton. Plant Cell 4: 941-951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue HW, Pical C, Brearley C, Elge S, Muller-Rober B (1999) A plant 126-kDa phosphatidylinositol 4-kinase with a novel repeat structure: cloning and functional expression in baculovirus-infected insect cells. J Biol Chem 274: 5738-5745 [DOI] [PubMed] [Google Scholar]
- Yang W, Boss WF (1994) Regulation of phosphatidylinositol 4-kinase by the protein activator PIK-A49. Activation requires phosphorylation of PIKA49. J Biol Chem 269: 3852-3857 [PubMed] [Google Scholar]
- Yao L, Janmey P, Frigeri LG, Han W, Fujita J, Kawakami Y, Apgar JR, Kawakami T (1999) Pleckstrin homology domains interact with filamentous actin. J Biol Chem 274: 19752-19761 [DOI] [PubMed] [Google Scholar]
- Zhao XH, Bondeva T, Balla T (2000) Characterization of recombinant phosphatidylinositol 4-kinase beta reveals auto- and heterophosphorylation of the enzyme. J Biol Chem 275: 14642-14648 [DOI] [PubMed] [Google Scholar]