Abstract
Acyl-activating enzymes are a diverse group of proteins that catalyze the activation of many different carboxylic acids, primarily through the formation of a thioester bond. This group of enzymes is found in all living organisms and includes the acyl-coenzyme A synthetases, 4-coumarate:coenzyme A ligases, luciferases, and non-ribosomal peptide synthetases. The members of this superfamily share little overall sequence identity, but do contain a 12-amino acid motif common to all enzymes that activate their acid substrates using ATP via an enzyme-bound adenylate intermediate. Arabidopsis possesses an acyl-activating enzyme superfamily containing 63 different genes. In addition to the genes that had been characterized previously, 14 new cDNA clones were isolated as part of this work. The protein sequences were compared phylogenetically and grouped into seven distinct categories. At least four of these categories are plant specific. The tissue-specific expression profiles of some of the genes of unknown function were analyzed and shown to be complex, with a high degree of overlap. Most of the plant-specific genes represent uncharacterized aspects of carboxylic acid metabolism. One such group contains members whose enzymes activate short- and medium-chain fatty acids. Altogether, the results presented here describe the largest acyl-activating enzyme family present in any organism thus far studied at the genomic level and clearly indicate that carboxylic acid activation metabolism in plants is much more complex than previously thought.
Carboxylic acid activation plays a vital role in numerous metabolic pathways in all living organisms. Activation of carboxylic acids provides the precursors for pathways that lead to the biosynthesis and/or breakdown of many types of important metabolites, including lipids, amino acids, sugars, and a variety of secondary metabolites. Given the chemical diversity of substrates requiring activation, several types of carboxylic acid activating enzymes have evolved to fulfill this role. Although some of these enzymes couple carboxyl groups to amines or alcohols, most acyl-activating enzymes are acid-thiol ligases (EC 6.2.1). Even within this smaller group, the similarity in the types of products formed is not strictly reflected in the routes by which these enzymes carry out their respective reactions: at least three different catalytic mechanisms are used to create the various thioester products (Groot et al., 1976; Stein et al., 1996; Sánchez et al., 2000). We are particularly interested in the acyl-activating enzymes (AAEs). This group of enzymes has previously been called the acyl adenylate-forming (Conti et al., 1996; Chang et al., 1997) or AMP-binding protein (Fulda et al., 1997) superfamily. In this report, we define AAEs as those enzymes that first activate their respective carboxylic acid substrates through the pyrophosphorylysis of ATP, forming an enzyme-bound acyl-AMP intermediate called an adenylate. The carbonyl carbon of the adenylate most commonly then undergoes nucleophilic attack by the free electrons of a thiol group from the acyl acceptor, forming the relevant thioester and releasing AMP (Bar-Tana et al., 1973). The identity of the ultimate acyl acceptor may vary considerably.
AAEs have been described in every class of organism on earth. Dozens, if not hundreds, of genes have been cloned, and the respective enzymes have been shown to participate in an immense variety of anabolic and catabolic pathways. Most organisms contain one or more of the different types of acyl-CoA synthetases, which includes enzymes that activate acetate, medium-chain, long-chain, and very long-chain fatty acids (for review, see Watkins, 1997). Other acid:CoA ligases synthesize CoA thioesters of such diverse acids as hydroxy- and chlorobenzoate (Chang et al., 1997), cinnamic acid (and several derivatives of cinnamate; Lee et al., 1995), citrate, malate, and malonate (An et al., 1999). Microbial multidomain enzyme complexes that contain AAE domains carry out synthesis of many peptide antibiotics and polyketides. These domains transfer the activated acyl substrate to the thiol group of an enzyme-bound 4-phosphopantetheine group (Conti et al., 1997). The firefly Photinus pyralis produces the chemiluminescent compound luciferin. This molecule is produced by the unusual AAE enzyme luciferase, which catalyzes the oxidative decarboxylation of an acyl-AMP using molecular oxygen, thus forming the chemiluminescent compound luciferin (Conti et al., 1996). Luciferase also binds coenzyme A, although its role in the reaction is not clear (Conti et al., 1996).
Despite the extraordinary differences in their substrates, products, and sizes, most AAEs share some conserved structural elements. One motif in particular appears to be absolutely necessary for binding of ATP and adenylate formation (Conti et al., 1996). This motif (PROSITE PS00455) is very highly conserved in many of the enzymes that catalyze the general two-step, adenylate intermediate-containing reaction (Conti et al., 1997). The presence of this motif unites this group of AAEs into a large superfamily.
The investigation of acid activation in plants has lagged behind that of animals, yeast, and bacteria. Although extensive reviews regarding mammalian and bacterial acid activation have been written (Watkins, 1997), only a few reports that characterize fatty acid-activating enzymes from plants currently exist. These studies have primarily focused on the long-chain acyl-CoA synthetases (LACSs), a class of enzymes that provide the acyl-CoA pools used to synthesize the phospholipids and triacylglycerols found in plant vegetative and seed tissues (Fulda et al., 1997; Pongdontri and Hills, 2001; Shockey et al., 2002). Recently, we characterized the gene family for LACS in Arabidopsis. This group consists of nine genes whose enzymes activate 14 to 20 carbon fatty acids (Shockey et al., 2002). Extended analyses of individual members of the LACS gene family has begun to reveal important information about the roles of peroxisomal (Fulda et al., 2002) and plastidial (Schnurr et al., 2002) isoforms of this enzyme. However, we felt that the completion of sequencing of the Arabidopsis genome created a unique opportunity to extend our knowledge of carboxylic acid activation beyond LACS enzymes alone. Using the conserved AAE sequence elements as sequence analysis tools, we identified, compared, and analyzed a surprisingly large number of other putative AAE genes.
Careful reviews of the plant literature revealed very few other biochemical descriptions of carboxylic acid activation reactions (Orchard and Anderson, 1996; Behal et al., 2002). Also rare were descriptions of cloned cDNAs or purified enzymes that catalyze these reactions (Lee et al., 1995; Ehlting et al., 1999). The glaring discrepancy between the large number of new AAE genes identified in our analyses and the low number of defined biochemical reactions or pathways that use AAE-derived reaction products compelled us to investigate the AAE superfamily much more closely. We conclude that the family of genes described here constitutes the entire set of ATP-dependent AAEs in Arabidopsis. Many of these genes had not been previously cloned, and the biochemical functions of most of them were unknown. Through a combination of sequence analyses and biochemical characterizations, we have determined the functions of some of the newly characterized sequences, including that of an entirely new subclass of plant-specific AAE genes. These data are summarized in this report. This information will be used as a framework from which rational investigations into the subcellular locations and physiological roles of different subsets of these genes can be undertaken.
RESULTS
Identification and Phylogenetic Analysis of AAE Superfamily in Arabidopsis
Our initial efforts to investigate carboxylic acid activation in plants centered on the search for LACS genes in Arabidopsis. The LACS gene family ultimately was found to contain nine genes, as determined by the cloning and functional expression of the corresponding cDNAs in yeast and Escherichia coli (Shockey et al., 2002). In addition to these genes, however, our searches revealed a large number of other genes that bore significant sequence similarity to known LACSs from other organisms. The predicted amino acid sequences of each of these genes contain numerous conserved motifs; however, one motif in particular was very strictly maintained in all the sequences. As such, this motif acts as the unifying feature of the AAE gene superfamily. The AAE consensus motif is represented in the PROSITE PS00455 consensus sequence [LIVMFY]-X-X-[STG]-[STAG]-G-[ST]-[STEI]-[SG]-X-[PASLIVM]-[KR]. Searches of the annotated Arabidopsis genome identified 44 genes that contained the AAE consensus motif and possessed high sequence similarity to known AAEs from other organisms. This set of genes included the family of nine LACSs, three known 4-coumarate:CoA ligases (4CLs), and one acetyl-CoA synthetase. In addition, we found 31 genes of unknown function.
However, during these studies, Staswick et al. (2002) described the cloning and partial characterization of a family of 19 cDNAs that are functionally related to the AAEs described here. Biochemical analysis indicated a related enzymatic function for these enzymes, so their sequences were included in our analysis.
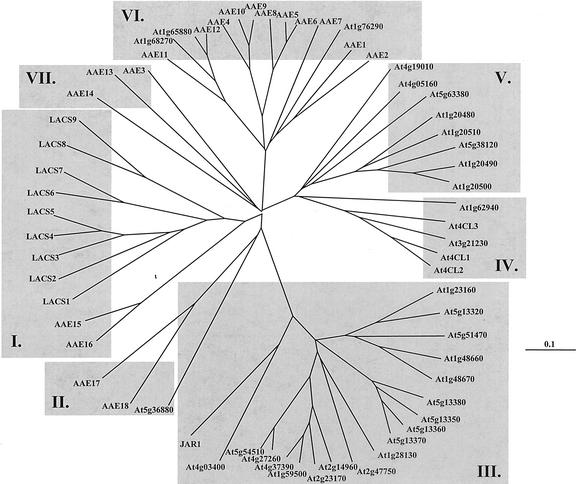
The degree of sequence similarity between the sequences of all 63 AAEs was assessed by phylogenetic comparisons. Using the neighbor-joining algorithm (Saitou and Nei, 1987), the sequences were aligned and compared using the ClustalX program (Thompson et al., 1997). The resulting multiple sequence alignment was displayed graphically as an unrooted phylogenetic tree, as shown in Figure 1. The names, GenBank accessions, Munich Information Center for Protein Sequences (MIPS) codes, and other information pertinent to each of the AAE superfamily genes are summarized in Table I.
Figure 1.
Phylogenetic analysis of the Arabidopsis AAEs. The complete protein sequences of the AAEs were aligned using ClustalX and were displayed graphically using TREEVIEW as described in the text. The branch lengths are proportional to the degree of divergence, with the scale of “0.1” representing 10% change.
Table I.
The members of the Arabidopsis AAE superfamily
| Clade | Gene Name/Description | MIPS Code | Genbank Accession No. | Cloned/Reference |
|---|---|---|---|---|
| I | LACS1 | At2g47240 | AF503751 | Shockey et al. (2002) |
| LACS2 | At1g49430 | AF503752 | Shockey et al. (2002) | |
| LACS3 | At1g64400 | AF503753 | Shockey et al. (2002) | |
| LACS4 | At4g23850 | AF503754 | Shockey et al. (2002) | |
| LACS5 | At4g11030 | AF503755 | Shockey et al. (2002) | |
| LACS6 | At3g05970 | AF503756 | Shockey et al. (2002) | |
| LACS7 | At5g27600 | AF503757 | Shockey et al. (2002) | |
| LACS8 | At2g04350 | AF503758 | Shockey et al. (2002) | |
| LACS9 | At1g77590 | AF503759 | Shockey et al. (2002) | |
| AAE15 | At4g14070 | AF503770 | Shockey et al. (2002) | |
| AAE16 | At3g23790 | AF503771 | Shockey et al. (2002) | |
| II | Acetyl-CoA synthetase | At5g36880 | AF036618 | Ke et al. (2000) |
| AAE17 | At5g23050 | AY250844 | This study | |
| AAE18 | At1g55320 | AY250845 | This study | |
| III | JAR1, JA adenylase | At2g46370 | - | Staswick et al. (2002) |
| Putative hormone adenylase | At4g03400 | - | Staswick et al. (2002) | |
| IAA adenylase | At5g54510 | - | Staswick et al. (2002) | |
| IAA/SA adenylase | At4g27260 | - | Staswick et al. (2002) | |
| IAA adenylase | At4g37390 | - | Staswick et al. (2002) | |
| IAA adenylase | At1g59500 | - | Staswick et al. (2002) | |
| IAA adenylase | At2g23170 | - | Staswick et al. (2002) | |
| Putative hormone adenylase | At2g14960 | - | Staswick et al. (2002) | |
| IAA adenylase | At2g47750 | - | Staswick et al. (2002) | |
| IAA adenylase | At1g28130 | - | Staswick et al. (2002) | |
| Putative hormone adenylase | At5g13370 | - | Staswick et al. (2002) | |
| Putative hormone adenylase | At5g13360 | - | Staswick et al. (2002) | |
| Putative hormone adenylase | At5g13350 | - | Staswick et al. (2002) | |
| Putative hormone adenylase | At5g13380 | - | Staswick et al. (2002) | |
| Putative hormone adenylase | At1g48670 | - | Staswick et al. (2002) | |
| Putative hormone adenylase | At1g48660 | - | Staswick et al. (2002) | |
| Putative hormone adenylase | At5g51470 | - | Staswick et al. (2002) | |
| Putative hormone adenylase | At5g13320 | - | Staswick et al. (2002) | |
| Putative hormone adenylase | At1g23160 | - | Staswick et al. (2002) | |
| IV | At4CL1 | At1g51680 | U18675 | Lee et al. (1995) |
| At4CL2 | At3g21240 | AF106085 | Ehlting et al. (1999) | |
| At4CL3 | At1g65060 | AF106087 | Ehlting et al. (1999) | |
| 4CL homolog | At3g21230 | AY250837 | This study | |
| 4CL homolog | At1g62940 | AY250836 | This study | |
| V | 4CL-like protein | At1g20500 | - | - |
| 4CL-like protein | At1g20490 | - | - | |
| 4CL-like protein | At5g38120 | AY250832 | This study | |
| 4CL-like protein | At1g20510 | AY250838 | This study | |
| 4CL-like protein | At1g20480 | AY250833 | This study | |
| 4CL-like protein | At5g63380 | AY250835 | This study | |
| 4CL-like protein | At5g05160 | AY250839 | This study | |
| 4CL-like protein | At4g19010 | AY250834 | This study | |
| VI | AAE1 | At1g20560 | AF503760 | Shockey et al. (2002); this study |
| AAE2 | At2g17650 | AF503761 | Shockey et al. (2002); this study | |
| AAE4 | At1g77240 | AF503763 | Shockey et al. (2002); this study | |
| AAE5 | At5g16370 | AF503764 | Shockey et al. (2002); this study | |
| AAE6 | At5g16340 | AF503765 | Shockey et al. (2002); this study | |
| AAE7 | At3g16910 | AF503766 | Shockey et al. (2002); this study | |
| AAE8 | At1g75960 | AF503767 | Shockey et al. (2002); this study | |
| AAE9 | At1g21540 | AF503768 | Shockey et al. (2002); this study | |
| AAE10 | At1g21530 | AF503769 | Shockey et al. (2002); this study | |
| AAE11 | At1g66120 | AY250841 | This study | |
| AAE12 | At1g65890 | AY250840 | This study | |
| Putative AAE | At1g65880 | - | - | |
| Putative AAE | At1g68270 | - | - | |
| Putative AAE | At1g76290 | - | - | |
| VII | AAE3 | At3g48990 | AF503762 | Shockey et al. (2002); this study |
| AAE13 | At3g16170 | AY250842 | This study | |
| AAE14 | At1g30520 | AY250843 | This study |
Genes are grouped by clade
As shown in Figure 1, the AAE superfamily segregated into seven distinct subfamilies. Clade I contains the LACS subfamily, consisting of the nine genes active against long-chain fatty acids, plus two genes (AAE15 and AAE16) that are highly similar to the LACSs but do not produce LACS enzyme activity in vitro. These genes have been characterized in greater detail previously (Shockey et al., 2002). As yet, the enzyme activities encoded by AAE15 and AAE16 remain a mystery.
Previous reports described a single gene for acetyl-CoA synthetase (At5g36680; Ke et al., 2000). Analysis of Figure 1 reveals two new genes (AAE17 and AAE18) with limited sequence similarity to acetyl-CoA synthetase. These three sequences form clade II. The amino acid sequence of the known acetyl-CoA synthetase shares only 28% identity with that of either of the two related genes, compared with 50% to 55% identity between the Arabidopsis acetyl-CoA synthetase and the homologous enzymes from Brewer's yeast (Saccharomyces cerevisiae), fruitfly (Drosophila melanogaster), or Caenorhabditis elegans (data not shown). Definitive assignment of function to AAE17 and AAE18 will require biochemical assays and other types of analysis.
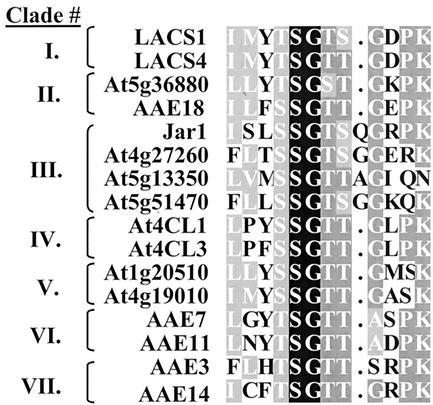
The largest clade (clade III) within the AAE superfamily is made up of the adenylases described by Staswick and coworkers (2002). These enzymes are thought to participate in plant hormone signaling pathways, through ATP-dependent adenylation of jasmonate, salicylate, and/or indole-3-acetic acid (IAA). The identity of the acyl acceptor(s), if any, is not yet known. Activation of these substrates had never been observed previously. The unusual nature of the acyl group substrate suggested that the sequences of these enzymes were unique compared with the other types of known AAEs. This hypothesis was confirmed by sequence homology comparisons as shown in Figure 1. Clade III contains an exceptionally long base branch compared with the other clades. Comparisons to other types of AAEs revealed very limited similarity between the amino acid sequences. JAR1 and the other clade III genes share only 5% to 20% amino acid identity with the members of the other clades. Even the AMP-binding/adenylate-formation motifs in these enzymes were unusual. Instead of the typical 12 amino acid residue motif found in all 44 other Arabidopsis sequences, the hormone adenylases contained an extra residue inserted between the ninth and tenth residues of the consensus sequence. The sequences of this motif from representatives of each of the other subfamilies are compared with those of the hormone adenylases in Figure 2. The discovery of these genes is significant in that the search for AAEs must now be expanded to include those enzymes that contain a non-canonical AMP-binding/adenylate-forming motif.
Figure 2.
Alignment of the PROSITE PS00455 AMP-binding motif of representative enzymes from each of the seven phylogenetic clades. The enzymes from clade III contain an extra residue relative to the other enzymes, which match the traditional 12-residue consensus sequence.
The phylogenetic comparison of this superfamily also revealed several new sequences related to the 4CLs. 4CL produces CoA thioesters of a variety of hydroxy- and methoxy-substituted cinnamic acids, which are used to synthesize several phenylpropanoid-derived compounds, including anthocyanins, flavonoids, lignin, and coumarins. Genes for 4CL have been cloned from numerous plant species, including three from Arabidopsis. These three genes had been assumed to comprise the complete 4CL gene family in Arabidopsis (Cukovic et al., 2001). Clade IV contains these three 4CLs, but also two new genes (At1g62940 and At3g21230) that are closely related to the coumarate-CoA ligases and therefore probably encode 4CL or related activities as well. Eight other new sequences that possessed considerable sequence similarity to the cloned 4CLs were also discovered. These genes (At4g19010, At4g05160, At5g63380, At1g20510, At5g38120, At1g20480, At1g20490, and At1g20500) formed clade V.
Perhaps the most intriguing observation made concerning the phylogenetic comparisons summarized in Figure 1 was the discovery of the new undescribed 14-member subfamily of genes that make up clade VI. The deduced proteins in this group contained the consensus AMP-binding motif and possessed approximately equal degrees of sequence identity to the members of the LACS and 4CL clades (approximately 20%–25% amino acid identity), including much higher levels of identity in several other conserved amino acid motifs (data not shown). These data strongly suggested that these genes encode some type of acid:CoA ligase. As described in more detail later in this report, database searches revealed that homologs for these genes could only be found in other plant species. Therefore, this class of genes seemed to represent a plant-specific branch of CoA-dependent acid activation biochemistry not previously investigated.
Finally, clade VII of Figure 1 contains three genes of unknown function. These genes are quite widely diverged; the sequences bear only slightly more similarity to each other than to any of the other genes in the family. These genes are more similar to the 4CLs than to the LACSs, the acetyl-CoA synthetase, or any of the genes in clade VI, but only slightly so (approximately 25% amino acid identity to 4CL1, compared with 20%–22% for the other sequences). On the basis of this analysis, the biological role of each of these three genes may be unique, unlike the high degree of overlapping function expected from many of the other groups in this family that contain several related orthologous genes.
Comparison of AAE Superfamily Complexity between Arabidopsis and Other Organisms
To assess the relative complexity of the AAE superfamily in Arabidopsis, representative members of each of the different clades of genes shown in Figure 1 were used to search the completed genome databases of various other eukaryotic and prokaryotic organisms. The total number of AAE genes in each genome was determined by conducting BLAST searches (Altschul et al., 1997) with representative protein sequences. Table II summarizes this information. Where possible, the genes were separated into different functional categories. Some of the eukaryotic genomes, in particular, have not yet been completely annotated. Therefore, the numbers and types of genes presented in Table II represent the most accurate data that we could determine through database searches, but some of the specific information may change as the AAE genes in these organisms are analyzed in more detail. The AAE genes that had not been characterized specifically or did not contain an irrefutably high level of sequence identity to a particular type of AAE were designated unknown function AAEs.
Table II.
Comparison of genomic complexity to AAE superfamily size in various completed eukaryotic and prokaryotic genomes
| Organism | Genome Complexity (Approximate Gene No.) | AAE Gene No. | AcCS | MACS | LACS | NRPS | SBCL | Other | Unknown Function AAE |
|---|---|---|---|---|---|---|---|---|---|
| Prokaryotes | |||||||||
| H. influenzae | 1,740 | 3 | 1 | - | 1 | - | 1 | - | - |
| Synechocystis sp. | 3,170 | 3 | 1 | - | 1 | - | - | - | 1 |
| PCC6803 | |||||||||
| P. aeruginosa | 5,570 | 28 | 2 | - | 2 | 4 | - | - | 20 |
| E. coli K12 | 4,300 | 9 | 1 | - | 1 | 2 | 1 | 1 | 3 |
| Bacillus subtilis 168 | 4,100 | 23 | 1 | - | 1 | 14 | 1 | - | 6 |
| Eukaryotes | |||||||||
| Homo sapiens (Human) | 35,300 | ≥14 | 2 | 2 | 5 | - | - | 5 | - |
| Fruitfly | 13,500 | ≥32 | 1 | - | - | - | - | 5 | 26 |
| C. elegans | 19,000 | ≥30 | 1 | - | - | - | - | 2 | 27 |
| Brewer's yeast | 7,200 | 9 | 2 | - | 4 | - | 1 | 2 | - |
| Arabidopsis | 25,000 | 63 | 2 | 1 | 9 | - | - | 12 | 39 |
Values not given (-) indicate not present or exact number not known. AcCS, acetyl-CoA synthetase; MACS, medium-chain acyl-CoA synthetase; NRPS, non-ribosomal peptide synthetase; SBCL, O-succinylbenzoate-CoA ligase
All organisms investigated contained members of the AAE superfamily, as expected. All, however, contained far fewer AAE genes than Arabidopsis. Hemophilus influenzae and Synechocystis sp. PCC6803 possessed the two smallest genomes analyzed (approximately 1,740 and 3,168 genes, respectively; Fleischmann et al., 1995; Kaneko et al., 1996) and the fewest AAE genes (three each). Pseudomonas aeruginosa, on the other hand, contained both the largest bacterial genome (Stover et al., 2000) and the largest number of AAE genes in a prokaryote. Analysis of the eukaryotic genomes revealed that genome complexity and AAE gene number are not always directly correlated. Fruitfly and C. elegans, which contain about 14,000 and 19,000 genes, respectively (Stein and Thierry-Mieg, 1998; Adams et al., 2000), contained at least 30 different AAE genes each. The human genome (approximately 35,000 genes) contained only about 14 AAEs. However, annotation of the human genome is not yet complete, and the true number of AAE genes in the human genome may actually be much higher.
The data presented in Table II provide an interesting perspective on the dynamic process of genome evolution and how the numbers and types of AAEs have evolved to meet the physiological needs of each species. However, the most salient point to be derived from Table II is that Arabidopsis, with a genome of 120 Mbp and approximately 25,000 genes, has evolved an AAE superfamily of far greater size and complexity than any other organism whose genome has been sequenced thus far. These data, combined with the phylogenetic groupings shown in Figure 1, suggest that Arabidopsis may contain classes of carboxylic acid-activating enzymes that have not yet been discovered or characterized in any way.
Analysis of AAE Gene Expression Profiles
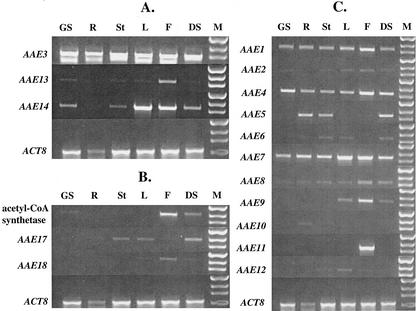
Deciphering the function of an unknown gene typically requires a specific determination of the timing and location of gene expression. Gene expression for the newly cloned genes from clades II, VI, and VII was measured by semiquantitative reverse transcriptase (RT)-PCR (Shockey et al., 2002). The results are shown in Figure 3. The actin ACT8 gene (An et al., 1996) was used as a control.
Figure 3.
Determination of the tissue-specific RNA expression patterns of Arabidopsis AAE genes from clades II (B), VI (C), and VII (A). Gene-specific primer pairs for each gene were used to assess the relative expression levels of each gene by RT-PCR starting from total RNA as described in the “Materials and Methods.” GS, Germinating seedling; R, root; St, stem; L, leaf; F, flower; DS, developing seed; M, Mr marker.
Figure 3B shows the expression patterns for the genes in clade II. The acetyl-CoA synthetase gene was expressed in germinating seedlings and developing seeds, albeit rather weakly. Acetyl-CoA synthetase has been investigated as a source of the acetyl-CoA starting material needed for fatty acid synthesis (Ke et al., 2000). Although it is not known to what extent transcript level correlates to enzyme activity level for this gene, the low levels of expression of this gene in seedlings and developing seeds is consistent with other recent findings that argue against acetyl-CoA synthetase playing a significant role in fatty acid synthesis (Behal et al., 2002). The two other genes in clade II, AAE17 and AAE18, were expressed in similar, but partially nonoverlapping patterns, both of which are distinct from acetyl-CoA synthetase. Both genes were expressed at low levels in developing seeds. AAE18 was expressed in flowers and leaves. AAE17 was expressed in stems and leaves but was one of the few genes in the entire superfamily that did not show detectable expression in flowers. The function of these two genes is unknown. Sequence homology alone does not clearly indicate a likely purpose; although belonging to the same clade of genes as the acetyl-CoA synthetase, the level of similarity between the acetyl-CoA synthetase and either AAE17 or AAE18 is low. Although sharing 54% amino acid identity between them, AAE17 and AAE18 are only 28% identical to acetyl-CoA synthetase.
The expression patterns of the genes in clade VII are shown in Figure 3A. AAE3 was expressed ubiquitously throughout the plant and at the highest levels observed for any of the genes tested in Figure 3. These results are consistent with the analysis of the Arabidopsis expressed sequence tag collections, which showed that AAE3 is represented by more than 50 different expressed sequence tags, far more than any of the other genes in the AAE superfamily. The expression patterns for AAE13 and AAE14 were very similar. Each was expressed throughout the plant, with lowest levels in roots and highest levels in leaves and flowers. The similar transcriptional profiles for these two genes may suggest some similarity or overlap in function, despite the low level of sequence identity between them.
Figure 3C summarizes the results of the analysis for the genes in clade VI. Like AAE17, AAE18, and all three genes from clade VII, all 14 genes in clade VI are plant specific. Database searches revealed undeniable homologs in several different plant species but none were found in the genomes of bacteria, humans, mice, or yeast (data not shown). This large subfamily showed a highly varied and complex pattern of gene expression. Like the LACS subfamily, several of the genes were expressed throughout the plant (Shockey et al., 2002). AAE7 in particular was expressed at high levels in all tissues tested. AAE11 was unique within all three sets of genes analyzed here in that its expression appeared to be flower specific. Although nearly all other genes in the superfamily, including the LACS genes, are expressed in flowers, only AAE11 and LACS5 are flower specific (Shockey et al., 2002). Collectively, the data summarized in Figure 3 do not clearly indicate a particular function for any of the genes analyzed. They do indicate though that several of the genes, such as AAE1, AAE3, AAE4, and AAE7, provide a ubiquitous housekeeping function throughout the plant, whereas the specificity in timing and location of expression of other genes, such as AAE9, AAE10, and AAE11, suggests a more specialized role for these enzymes.
Figure 3 clearly demonstrates the complexity of the Arabidopsis AAE superfamily and how many different transcriptional patterns exist for individual genes or subsets of genes. From these results, it is also clear that the large size of the AAE superfamily is not due to the presence of several unexpressed pseudogenes. These data reiterated to us how little we know about the numbers and types of organic acid activation reactions in plants, and about the enzymes that catalyze these reactions. Therefore, we attempted to learn more about the functions of some of the newly cloned AAE genes. Given their seclusion to the plant kingdom, and their lack of substantial homology to any other characterized group of AAE genes, we concentrated on the genes of clade VI.
Biochemical Analysis of Clade VI AAEs in Vivo and in Vitro
Biology contains well-known examples of genes of unrelated structure being recruited to serve the same function. With this in mind, we considered the possibility that the clade VI genes might represent a second family of LACSs. To address this possibility, nine representative cDNAs from clade VI (AAE1, AAE2, and AAE4–AAE10) were cloned into the Galinducible yeast expression vector pYES2 and transformed into competent cells of the YB525 mutant of Brewer's yeast. As described previously (Knoll et al., 1995), when cultured in the presence of fatty acids as the sole carbon source, and in the presence of cerulenin, which inhibits endogenous fatty acid synthesis, YB525 will not grow unless complemented by an active, non-peroxisomally targeted LACS. Although all seven non-peroxisomal LACS constructs restored growth of the mutant to wild-type levels (Shockey et al., 2002), none of the clade VI AAE constructs complemented the mutant phenotype (data not shown). These results strongly argued against the possibility that the clade VI AAE genes were a second class of LACSs.
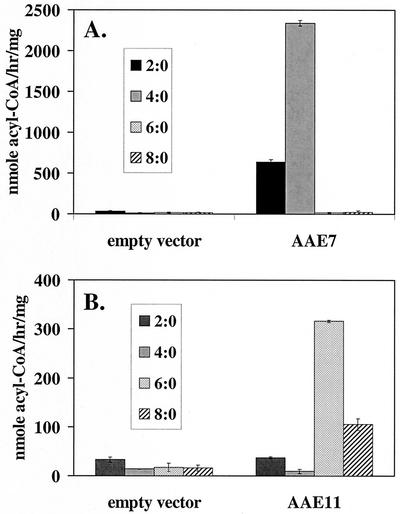
To extend the analysis of the possible substrate specificity of the clade VI AAE genes further, representative members of this subfamily were cloned into prokaryotic expression vectors and overexpressed in E. coli. Membrane fractions from isolates expressing AAE1, AAE2, AAE7, AAE9, AAE11, or AAE12 were tested for the ability to attach coenzyme A to all straight-chain acid substrates ranging in length from C2 (acetate) to C14 (myristate). The results of these assays are shown in Figure 4. Most of the enzymes were not active against any of the substrates tested. However, AAE7 and AAE11 were selectively active against some of the short- and medium-chain substrates. AAE7 activated butyrate at nearly 2,400 nmol h-1 mg-1 of total membrane protein and acetate at approximately one-fourth that rate. This enzyme showed no activity against any substrates longer than C4. AAE11, on the other hand, displayed activity against C6 and C8 acids, with no measurable activity against acetate or butyrate.
Figure 4.
Biochemical analysis of the short- and medium-chain acyl-CoA synthetase activities expressed from AAE7 (A) and AAE11 (B). Prokaryotic expression constructs were introduced into E. coli and protein overproduction induced with isopropylthio-β-galactoside. Membrane fractions from the induced bacterial cultures were used as enzyme sources in acyl-CoA synthetase assays using radioactive fatty acid substrates. Membranes from E. coli containing the empty vector were used as the negative controls.
DISCUSSION
The study of yeast and mammalian AAEs has identified important and specific roles for carboxylic acid-activating enzymes in disease (Watkins et al., 2000), metabolite transport (Faergeman et al., 2001), and signal transduction (Johnson et al., 1994), whereas only preliminary cloning and characterization of the first plant enzymes of this type, the LACSs, has been achieved (Fulda et al., 1997; Pongdontri and Hills, 2001; Shockey et al., 2002). During our efforts to clone the LACS gene family, it became obvious that Arabidopsis contained a very large number of genes with substantial levels of sequence identity to the known LACS genes of other organisms. The main purpose of this study was to try to categorize this entire family of genes.
The Arabidopsis genome contains 63 genes that possess a close match to the AMP-binding/adenylate-forming consensus defined by PROSITE motif PS00455. This set included nine LACS genes (Shockey et al., 2002) and 19 plant hormone adenylases, including JAR1 (Staswick et al., 2002). Although JAR1 and the other hormone adenylase genes are evolutionarily quite distant to the other members of this superfamily (see Figs. 1 and 2), the predicted folding of these proteins was very similar to that of firefly luciferase, another member of the general AAE superfamily (Staswick et al., 2002). Also, the ability to catalyze adenylation reactions demanded the inclusion of the hormone adenylases sequences in the AAE superfamily. As a whole, the 63 Arabidopsis genes represent by far the largest AAE superfamily in any organism, eukaryotic or prokaryotic, whose genome has been sequenced to date.
The superfamily formed into seven phylogenetically distinct clades (Fig. 1). Database homology searches with these sequences suggested a surprising level of novelty in plant AAEs. True homologs from non-plant species could only be found with the genes from clades I (LACS) and II (acetyl-CoA synthetase), while potential matches to some members of clade V also did exist. The other four clades were plant specific, because very closely related sequences for each could be found in a variety of plant species (data not shown). A large majority of the genes in the AAE superfamily had not previously been cloned; therefore very little information was available to suggest potential functions. Aside from the LACS and hormone adenylase enzymes, only a single acetyl-CoA synthetase (Ke et al., 2000) and three 4CLs (Lee et al., 1995; Ehlting et al., 1999) had been characterized biochemically.
Acetyl-CoA synthetase activity has been measured in purified chloroplasts (Roughan and Ohlrogge, 1994), and as such was initially thought to be the main supplier of acetyl-CoA for fatty acid synthesis. Recent studies however, suggest that although plant cells can use acetate for fatty acid synthesis, little carbon is channeled through this pathway under normal conditions (Behal et al., 2002; Rawsthorne, 2002). Therefore, the true role(s) of acetyl-CoA synthetase remains unclear. AAE17 and AAE18 share about 28% amino acid identity with the acetyl-CoA synthetase protein sequence. The enzymes encoded by these two genes also have sequence similarity to the SA gene, a mammalian mitochondrial medium-chain acyl-CoA synthetase. Altered SA expression has been associated with hypertension and elevated plasma cholesterol and triglyceride levels (Iwai et al., 2002). However, the precise role of SA and its homologs in other organisms (Karan et al., 2001) has not been deduced. Additional work will likewise be necessary to determine the biological function of AAE17 and AAE18.
Clade III contains the hormone adenylases. After overexpression in E. coli, some of these enzymes could catalyze isotope exchange between 32PPi and ATP when incubated with jasmonic acid, salicylic acid, and/or IAA. Isotope exchange is indicative of the adenylation half-reaction carried out by all other types of AAE enzymes (Bar-Tana et al., 1973). Interestingly, addition of CoA to the enzyme assays did not cause release of AMP (Staswick et al., 2002), so it is unlikely that these enzymes are acyl-CoA synthetases. Jasmonate, salicylate, and IAA activity levels are regulated by conjugation to various amino acids and sugars. The clade III enzymes may play a role in formation of these conjugates. Definitive assignment of functions to these enzymes also awaits additional analysis.
The three known 4CLs are contained in clade IV. These enzymes provide thioesters of the various hydroxy- and methoxy-substituted cinnamic acids needed to synthesize numerous phenylpropanoid-derived compounds including flavonoids, lignin, coumarins, and cell wall-bound phenolics (Ehlting et al., 1999). The regulation of gene expression and the biochemical properties of the enzymes encoded by the three 4CLs are well established (Lee et al., 1995; Ehlting et al., 1999; Cukovic et al., 2001; Stuible and Kombrink, 2001). These genes have also been characterized in several other plant species, most of which seem to contain 4CL gene families containing between two (Lee and Douglas, 1996) and four (Lindermayr et al., 2002) members. Arabidopsis contains 13 genes with significant sequence identity to other plant 4CLs. Clade IV contains the three previously characterized 4CL genes plus one additional gene (At3g21230) that is very closely related to them (65%–68% amino acid identity to At4CL1 and At4CL2). At3g21230 represents another gene duplication event, given its close chromosomal alignment with At4CL2 (At3g21240), and therefore probably encodes 4CL activity as well. At1g62940 nearly bridges the divide between the clade IV 4CL genes and the 4CL-like genes of clade V; its predicted amino acid sequence shares 38% to 40% identity with members of both clades. As such, its enzymatic activity is less certain, and will require biochemical verification. The 4CL-like genes of clade V represent intriguing variations on the structural properties common to the definitive 4CLs. All eight deduced amino acid sequences of the clade V genes contain type I peroxisome targeting sequences at their respective C termini. None of the five clade IV enzymes contain these sequences. Peroxisomal isoforms of LACS provide long chain acyl-CoA substrates for β-oxidation, but coumarate is activated in the cytosol, and essentially nothing is known about peroxisomal metabolism of this acid or the other related acids activated by 4CL, namely caffeate, ferulate, and sinapate. The peroxisomal 4CL-like isoforms may in fact catalyze the CoA activation of smaller subsets of structurally related compounds, such as cinnamic acid and benzoic acid. Several recent reports have identified plant enzyme activities that catalyze the CoA activation of these two acids, or use the resulting thioesters as substrates in other reactions (Abd El-Mawla et al., 2001; Beuerle and Pichersky, 2002). The biosynthesis of several important hormones and other secondary metabolites depend on benzoyl- and/or cinnamoyl-CoA as substrates (Yang et al., 1997; Ribnicky et al., 1998; Graser et al., 2001), so characterization of the 4CL/4CL-like genes from clades IV and V may help to expand our understanding of the biosynthesis of these compounds. Alternatively, some or all of the clade V enzymes might participate in the metabolism of very long-chain (>C22) fatty acids. Very long-chain fatty acids, and the esters, alcohols, ketones, and aldehydes derived from them, are major components of the surface wax layer that covers the aerial parts of all terrestrial plants. The mouse very long-chain acyl-CoA synthetase mmVLCS2 (Berger et al., 1998) displays relatively higher levels of sequence identity to some of the members of clade V than to other Arabidopsis AAEs. BLAST analysis showed E value scores of 7e-018 and 3e-017 for At5g63380 and At1g20510, respectively, compared with values greater than 1e-05 for any of the nine LACS enzymes. Therefore, it may be interesting to investigate the clade V enzymes with regard to activation of very long chain fatty acids.
The genes in clades VI and VII were the biggest surprises uncovered in the present study. Clade VI contains at least two members with short-chain or medium-chain acyl-CoA synthetase activity, plant enzyme activities that has received very little previous notice in plants, and with no clear metabolic purpose as yet. Orchard and Anderson (1996) studied short-chain acyl-CoA synthetase enzyme activity in Monterey pine (Pinus radiata) and postulated that it may play a role in metabolizing propionate and acrylate to acetate. The biosynthesis of the branched-chain amino acids Val, Leu, and Ile involves an acid: CoA ligation step but via a completely different mechanism than that used by the enzymes described here. This step in the pathway, catalyzed by branched-chain α-keto acid dehydrogenase does not use ATP, requires oxidized NAD, and ultimately decarboxylates the acid substrate (Zolman et al., 2001). Therefore, it is highly unlikely that the short-chain/medium-chain acyl-CoA synthetase activities associated with AAE7 and AAE11 are reflective of a role in amino acid metabolism. Additional studies of this subfamily should yield some entirely new and probably unexpected insights regarding activation and metabolism of short- and medium-chain fatty acids.
The genes from clade VII almost defy categorization in a single clade altogether. All three sequences are quite divergent relative to all other members of the superfamily. No obvious enzymatic activity can yet be ascribed to any of the clade VII enzymes; none of the three was active in acyl-CoA synthetase assays using any of the straight chain acids from acetate to myristate (data not shown). The Capsicum annuum homolog of AAE3 was recently cloned, and shown to be rapidly up-regulated by treatment of pepper leaves with either salicylic acid or the pathogenic bacterium Xanthomonas campestris (Lee et al., 2001). These intriguing results suggest a role for the enzyme in CoA-dependent acid activation as a part of a pathogen defense response pathway. It is not known whether Arabidopsis AAE3, AAE13, or AAE14 is regulated in a similar manner. The level of divergence between these three genes may indicate specific roles for each gene, with little or no functional overlap, as might be expected from the genes in the other more tightly clustered groups of the superfamily. If this is true, analysis of T-DNA mutants or antisense-suppressed lines for AAE3, AAE13, and AAE14 may provide direct clues as to their functions.
The diversity of potential carboxylic acid-activating genes uncovered in this study provides an exciting opportunity. Future studies that combine the initial characterizations and comparisons described here with the various other useful genomic, proteomic, and metabolomic tools available in Arabidopsis hold great potential. These studies will soon provide powerful new insight and understanding of the various primary and secondary metabolic pathways that depend on carboxylic acid activation by AAEs.
MATERIALS AND METHODS
Sequencing and Sequence Homology Analysis
All DNA sequencing was conducted in the Macromolecular Analysis Laboratory at Washington State University using automated sequencing equipment (Applied Biosystems, Foster City, CA). Sequences were assembled and modified using the GCG suite of programs (Wisconsin Package v10.0, Genetics Computer Group, Madison, WI). Database homology searches were conducted using the database maintained at The Arabidopsis Information Resource (http://www.Arabidopsis.org/). MIPS designations refer to the nomenclature used at the MIPS Arabidopsis database (http://mips.gsf.de/proj/thal/db/search/search_frame.html). Protein sequence alignments were conducted using the ClustalX program (Thompson et al., 1997) using the default gap creation and gap extension penalty scores. Phylogenetic trees were drawn from the alignments using the TREEVIEW program (Page, 1996). All other sequence analysis was carried out using GCG (Wisconsin Package v10.0, Genetics Computer Group).
Identification and Cloning of AAE cDNAs
The cDNA clones for the LACS subfamily and several other AAE genes from clade VI were cloned as described previously (Shockey et al., 2002). The remaining AAE sequences were identified either by sequence homology to known LACS and AAE sequences or through PatMatch searches of the annotated protein sequences. Oligonucleotide primers corresponding to the start ATG and stop codons for each gene were used to generate full-length RT-PCR products for cloning into the prokaryotic expression vectors pET24c or pET24d (Novagen, Madison, WI). First-strand cDNA was produced from mRNA isolated from Arabidopsis cv Wassilewskija (for those cDNAs from clades II, IV, V, and VII) or Arabidopsis cv Columbia (clade VI).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes. No unreasonable restrictions or conditions will be placed on the use of any materials described in this paper that would limit their use in noncommercial research purposes.
Expression of Arabidopsis AAEs in Brewer's Yeast (Saccharomyces cerevisiae) and Escherichia coli
AAE yeast expression constructs were introduced into the YB525 mutant of Brewer's yeast (Knoll et al., 1995) and analyzed for LACS activity on minimal media containing fatty acids and cerulenin as described previously (Shockey et al., 2002).
AAE pET24 plasmids were transformed into competent E. coli cells and selected on kanamycin-containing media. For those predicted proteins that contained paired AGA/AGG Arg codons (Schenk et al., 1995), E. coli BL21-RIL (DE3) cells (Invitrogen, Carlsbad, CA) were used to increase the efficiency of full-length protein translation. All other constructs were introduced into the LACS-deficient K27 mutant of E. coli that expressed T7 RNA polymerase, via the chromosomal integration of the DE3 prophage (Shockey et al., 2002). Liquid cultures were induced with 0.7 mm isopropylthio-β-galactoside and grown with vigorous shaking overnight at room temperature, and the cells collected by centrifugation. The washed cell pellet was suspended in lysis buffer (50 mm Tris-HCl, pH 7.5 + protease inhibitors) and disrupted by sonication. After low-speed centrifugation (5,000g for 20 min) to remove cellular debris, the membrane and soluble fractions were separated by centrifugation at 100,000g for 1 h. These fractions were suspended in lysis buffer containing 25% (v/v) glycerol, aliquoted, frozen at -20°C, and used as enzyme sources for the various assays.
Acyl-CoA Synthetase Enzyme Assays
LACS assays were conducted as described previously (Shockey et al., 2002). For all acid substrates between C2 and C12, the assay conditions and DE81 paper-binding detection method of Roughan and Ohlrogge (1994) was used instead.
Gene Expression Analysis
Gene expression levels were analyzed by relative quantitative RT-PCR as described previously (Shockey et al., 2002), using total RNA preparations from developing seeds, 1-d-old germinating seedlings, roots, young leaves, stems, and flowers as templates for reverse transcription.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.020552.
This work was supported in part by a National Science Foundation postdoctoral fellowship to J.S. (grant no. BIR–9627559), by Dow Chemical Company/Dow AgroSciences (grant to J.B.), by the U.S. Department of Agriculture (grant no. USDA–NRI 2001–35318–10186 to J.B.), and by the Agricultural Research Center at Washington State University.
References
- Abd El-Mawla AM, Schmidt W, Beerhues L (2001) Cinnamic acid is a precursor of benzoic acids in cell cultures of Hypericum androsaemum L. but not in cell cultures of Centaurium erythraea RAFN. Planta 212: 288-293 [DOI] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF (2000) The genome sequence of Drosophila melanogaster. Science 287: 2185-2195 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JH, Lee GY, Jung JW, Lee W, Kim YS (1999) Identification of residues essential for a two-step reaction by malonyl-CoA synthetase from Rhizobium trifolii. Biochem J 344: 159-166 [PMC free article] [PubMed] [Google Scholar]
- An YQ, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10: 107-121 [DOI] [PubMed] [Google Scholar]
- Bar-Tana J, Rose G, Brandes R, Shapiro B (1973) Palmitoyl-coenzyme A synthetase: mechanism of reaction. Biochem J 131: 199-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behal RH, Lin M, Back S, Oliver DJ (2002) Role of acetyl-coenzyme A synthetase in leaves of Arabidopsis thaliana. Arch Biochem Biophys 402: 259-267 [DOI] [PubMed] [Google Scholar]
- Berger J, Truppe C, Neumann H, Forss-Petter S (1998) A novel relative of the very-long-chain acyl-CoA synthetase and fatty acid transporter protein genes with a distinct expression pattern. Biochem Biophys Res Commun 247: 255-260 [DOI] [PubMed] [Google Scholar]
- Beuerle T, Pichersky E (2002) Purification and characterization of benzoate: coenzyme A ligase from Clarkia breweri. Arch Biochem Biophys 400: 258-264 [DOI] [PubMed] [Google Scholar]
- Chang KH, Xiang H, Dunaway-Mariano D (1997) Acyl-adenylate motif of the acyl-adenylate/thioester-forming enzyme superfamily: a sitedirected mutagenesis study with the Pseudomonas sp. strain CBS3 4-chlorobenzoate:coenzyme A ligase. Biochemistry 36: 15650-15659 [DOI] [PubMed] [Google Scholar]
- Conti E, Franks NP, Brick P (1996) Crystal structure of firefly luciferase throws light on a superfamily of acyl-activating enzymes. Structure 4: 287-298 [DOI] [PubMed] [Google Scholar]
- Conti E, Stachelhaus T, Marahiel MA, Brick P (1997) Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J 16: 4174-4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukovic D, Ehlting J, VanZiffle JA, Douglas CJ (2001) Structure and evolution of 4-coumarate:coenzyme A ligase (4CL) gene families. Biol Chem 382: 645-654 [DOI] [PubMed] [Google Scholar]
- Ehlting J, Buttner D, Wang Q, Douglas CJ, Somssich IE, Kombrink E (1999) Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J 19: 9-20 [DOI] [PubMed] [Google Scholar]
- Faergeman NJ, Black PN, Zhao XD, Knudsen J, DiRusso CC (2001) The Acyl-CoA synthetases encoded within FAA1 and FAA4 in Saccharomyces cerevisiae function as components of the fatty acid transport system linking import, activation, and intracellular utilization. J Biol Chem 276: 37051-37059 [DOI] [PubMed] [Google Scholar]
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM et al. (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269: 496-512 [DOI] [PubMed] [Google Scholar]
- Fulda M, Heinz E, Wolter FP (1997) Brassica napus cDNAs encoding fatty acyl-CoA synthetase. Plant Mol Biol 33: 911-922 [DOI] [PubMed] [Google Scholar]
- Fulda M, Shockey J, Werber M, Wolter FP, Heinz E (2002) Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid beta-oxidation. Plant J 32: 93-103 [DOI] [PubMed] [Google Scholar]
- Graser G, Oldham NJ, Brown PD, Temp U, Gershenzon J (2001) The biosynthesis of benzoic acid glucosinolate esters in Arabidopsis thaliana. Phytochemistry 57: 23-32 [DOI] [PubMed] [Google Scholar]
- Groot PH, Scholte HR, Hulsmann WC (1976) Fatty acid activation: specificity, localization, and function. Adv Lipid Res 14: 75-126 [DOI] [PubMed] [Google Scholar]
- Iwai N, Katsuya T, Mannami T, Higaki J, Ogihara T, Kokame K, Ogata J, Baba S (2002) Association between SAH, an acyl-CoA synthetase gene, and hypertriglyceridemia, obesity, and hypertension. Circulation 105: 41-47 [DOI] [PubMed] [Google Scholar]
- Johnson DR, Knoll LJ, Levin DE, Gordon JI (1994) Saccharomyces cerevisiae contains four fatty acid activation (FAA) genes: an assessment of their role in regulating protein N-myristoylation and cellular lipid metabolism. J Cell Biol 127: 751-762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y Miyajima N, Hirosawa M, Sugiura M, Sasamoto S (1996) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803: II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res 3: 109-136 [DOI] [PubMed] [Google Scholar]
- Karan D, David JR, Capy P (2001) Molecular evolution of the AMP-forming acetyl-CoA synthetase. Gene 265: 95-101 [DOI] [PubMed] [Google Scholar]
- Ke J, Behal RH, Back SL, Nikolau BJ, Wurtele ES, Oliver DJ (2000) The role of pyruvate dehydrogenase and acetyl-coenzyme A synthetase in fatty acid synthesis in developing Arabidopsis seeds. Plant Physiol 123: 497-508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll LJ, Johnson DR, Gordon JI (1995) Complementation of Saccharomyces cerevisiae strains containing fatty acid activation gene (FAA) deletions with a mammalian acyl-CoA synthetase. J Biol Chem 270: 10861-10867 [DOI] [PubMed] [Google Scholar]
- Lee D, Douglas CJ (1996) Two divergent members of a tobacco 4-coumarate: coenzyme A ligase (4CL) gene family. cDNA structure, gene inheritance and expression, and properties of recombinant proteins. Plant Physiol 112: 193-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Ellard M, Wanner LA, Davis KR, Douglas CJ (1995) The Arabidopsis thaliana 4-coumarate:CoA ligase (4CL) gene: stress and developmentally regulated expression and nucleotide sequence of its cDNA. Plant Mol Biol 28: 871-884 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Suh MC, Kim S, Kwon JK, Kim M, Paek KH, Choi D, Kim BD (2001) Molecular cloning of a novel pathogen-inducible cDNA encoding a putative acyl-CoA synthetase from Capsicum annuum L. Plant Mol Biol 46: 661-671 [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Mollers B, Fliegmann J, Uhlmann A, Lottspeich F, Meimberg H, Ebel J (2002) Divergent members of a soybean (Glycine max L.) 4-coumarate:coenzyme A ligase gene family. Eur J Biochem 269: 1304-1315 [DOI] [PubMed] [Google Scholar]
- Orchard SG, Anderson JW (1996) Substrate specificity of the short chain fatty acyl-coenzyme A synthetase of Pinus radiata. Phytochemistry 6: 1465-1472 [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357-358 [DOI] [PubMed] [Google Scholar]
- Pongdontri P, Hills M (2001) Characterization of a novel plant acyl-CoA synthetase that is expressed in lipogenic tissues of Brassica napus L. Plant Mol Biol 47: 717-726 [DOI] [PubMed] [Google Scholar]
- Rawsthorne S (2002) Carbon flux and fatty acid synthesis in plants. Prog Lipid Res 41: 182-196 [DOI] [PubMed] [Google Scholar]
- Ribnicky DM, Shulaev VV, Raskin II (1998) Intermediates of salicylic acid biosynthesis in tobacco. Plant Physiol 118: 565-572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughan PG, Ohlrogge JB (1994) On the assay of acetyl-CoA synthetase activity in chloroplasts and leaf extracts. Anal Biochem 216: 77-82 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406-425 [DOI] [PubMed] [Google Scholar]
- Sánchez LB, Galperin MY, Müller M (2000) Acetyl-CoA synthetase from the amitochondriate eukaryote Giardia lamblia belongs to the newly recognized superfamily of acyl-CoA synthetases (nucleoside diphosphate-forming). J Biol Chem 275: 5794-5803 [DOI] [PubMed] [Google Scholar]
- Schenk PM, Baumann S, Mattes R, Steinbiss HH (1995) Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare ArgtRNAs. Biotechniques 19: 196-198 [PubMed] [Google Scholar]
- Schnurr JA, Shockey JM, De Boer GJ, Browse JA (2002) Fatty acid export from the chloroplast: molecular characterization of a major plastidial acyl-coenzyme a synthetase from Arabidopsis. Plant Physiol 129: 1700-1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey JM, Fulda MS, Browse JA (2002) Arabidopsis contains nine long-chain acyl-coenzyme A synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol 129: 1710-1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I, Rowe ML (2002) Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14: 1405-1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Thierry-Mieg J (1998) Scriptable access to the Caenorhabditis elegans genome sequence and other ACEDB databases. Genome Res 8: 1308-1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein T, Vater J, Kruft V, Otto A, Wittmann-Liebold B, Franke P, Panico M, McDowell R, Morris HR (1996) The multiple carrier model of nonribosomal peptide biosynthesis at modular multienzymatic templates. J Biol Chem 271: 15428-15435 [DOI] [PubMed] [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M et al (2000) Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406: 959-964 [DOI] [PubMed] [Google Scholar]
- Stuible HP, Kombrink E (2001) Identification of the substrate specificityconferring amino acid residues of 4-coumarate:coenzyme A ligase allows the rational design of mutant enzymes with new catalytic properties. J Biol Chem 276: 26893-26897 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876-4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins PA (1997) Fatty acid activation. Prog Lipid Res 36: 55-83 [DOI] [PubMed] [Google Scholar]
- Watkins PA, Lu JF, Braiterman LT, Steinberg SJ, Smith KD (2000) Disruption of a yeast very-long-chain acyl-CoA synthetase gene simulates the cellular phenotype of X-linked adrenoleukodystrophy. Cell Biochem Biophys 32: 333-337 [DOI] [PubMed] [Google Scholar]
- Yang Q, Reinhard K, Schiltz E, Matern U (1997) Characterization and heterologous expression of hydroxycinnamoyl/benzoyl-CoA:anthranilate N-hydroxycinnamoyl/benzoyltransferase from elicited cell cultures of carnation, Dianthus caryophyllus L. Plant Mol Biol 35: 777-789 [DOI] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Thompson B, Hawes JW, Krukenberg KA, Matsuda SP, Bartel B (2001) chy1, an Arabidopsis mutant with impaired beta-oxidation, is defective in a peroxisomal betahydroxyisobutyryl-CoA hydrolase. J Biol Chem 276: 31037-31046 [DOI] [PubMed] [Google Scholar]