Abstract
We previously demonstrated that overexpression of the horseradish (Armoracia rusticana) peroxidase prxC1a gene stimulated the growth rate of tobacco (Nicotiana tabacum) plants. Here, the cauliflower mosaic virus 35S::prxC1a construct was introduced into hybrid aspen (Populus sieboldii × Populus grandidentata). The growth rate of these transformed hybrid aspen plants was substantially increased under greenhouse conditions. The average stem length of transformed plants was 25% greater than that of control plants. There was no other obvious phenotypic difference between the transformed and control plants. Fast-growing transformed hybrid aspen showed high levels of expression of prxC1a and had elevated peroxidase activities toward guaiacol and ascorbate. However, there was no increase of the endogenous class I ascorbate peroxidase activities in the transformed plants by separate assay and activity staining of native polyacrylamide gel electrophoresis. Furthermore, calli derived from the transformed hybrid aspen grew faster than those from control plants and were resistant to the oxidative stress imposed by hydrogen peroxide. Therefore, enhanced peroxidase activity affects plant growth rate and oxidative stress resistance.
Peroxidases (EC 1.11.1.7, donor: hydrogen peroxide oxidoreductase) are widely found in animals, plants, and microbes and oxidize a vast array of compounds (electron donors) in the presence of hydrogen peroxide (H2O2). Forty-two expected sequence tags encoding different peroxidase isoenzymes are found in rice (Oryza sativa; Hiraga et al., 2001). Hoyle (1977) found 42 isoenzymes and/or isoforms in commercial preparations of horseradish (Armoracia rusticana) peroxidase (HRP). The plant peroxidase superfamily is divided into three classes based on differences in primary structure (Welinder, 1992). Class I plant peroxidases include the intracellular enzymes in plants, bacteria, and yeast (Saccharomyces cerevisiae), such as microbial cytochrome c peroxidase (EC 1.11.1.5), bacterial catalase-peroxidase (EC 1.11.1.6), and ascorbate peroxidase (EC 1.11.1.11). Class II plant peroxidases are extracellular peroxidases from fungi, including lignin peroxidase (EC1.11.1.14) and Mn2+-independent peroxidase (EC 1.11.1.13). Class III plant peroxidases (EC 1.11.1.7) were originally described as peroxidases and are secreted outside of the cells or transported into vacuoles. HRP prxC1a of this study is a member of the class III peroxidases. To date, four genomic DNAs that encode HRP (Fujiyama et al., 1990) and four cDNAs have been isolated (Fujiyama et al., 1988; Bartnek-Roxa et al., 1991). All of the genes consist of four exons and three introns, and the number of amino acid residues deduced from nucleotide sequences varies from 327 to 353. Nucleotide sequence homologies in the coding regions were found to be 90% between prxC1a and prxC1b, 71% between prxC1a and prxC2, and 66% between prxC1a and prxC3. The prxC2 gene is induced by wounding and functional analysis of the prxC2 promoter has been reported previously (Kawaoka et al., 1994b; Kaothien et al., 2000, 2002). Furthermore, its transcription factors, TFHP1 and Ntlim1, have also been identified (Kawaoka et al., 1994c, 2000). Because the amino acid sequence deduced from the prxC1a gene contains the same sequence as that determined for the purified C isoenzyme of HRP by Welinder (1974), 30 and 15 amino acid residues of the N terminus and C terminus coded on prxC1a are putative signal sequences, respectively (Fujiyama et al., 1988).
Several physiological functions for class III plant peroxidases have been suggested: for example, removal of H2O2, oxidation of toxic reductants, biosynthesis and degradation of lignin (Grisebach, 1981), defensive responses to wounding (Espelie et al., 1986), and catabolism of auxin (Hinnman and Lang, 1965). They have extremely broad substrate specificity and exist in a multitude of isoenzyme forms, making it difficult to ascertain their precise functions. The tools of recombinant DNA technology have been applied to the analysis of the physiological functions of plant genes.
Previously, we reported that transformed tobacco (Nicotiana tabacum), overexpressing prxC1a under the control of the cauliflower mosaic virus (CaMV) 35S promoter and HRP prxC2 promoter, grew significantly faster than control plants (Kawaoka et al., 1994a). Compared with the control plants, the time to flowering was reduced by approximately 20%. However, the underexpression of the homologous anionic peroxidase gene using antisense RNA suppresses endogenous enzymatic activity and, thus, affects the growth rate (Lagrimini et al., 1997b). This implies that alteration of peroxidase activity influences plant growth rate. Here, the HRP prxC1a gene, driven by the CaMV 35S promoter, was introduced into hybrid aspen (Populus sieboldii × Populus grandidentata) trees, and its effects were characterized. We demonstrate that overproduction of the HRP prxC1a stimulates the vegetative growth rate of hybrid aspen and confers resistance to oxidative stresses.
RESULTS
Preparation of Transformed Hybrid Aspen and Growth Rate of in Vitro-Cultured Transformed Plants
The 1.1-kb DNA fragment of a horseradish neutral peroxidase prxC1a cDNA was ligated to the CaMV 35S promoter. The resulting chimeric gene was introduced into hybrid aspen plants, and transformed plants were selected on 100 mg L–1 kanamycin. Kanamycin-resistant shoots were placed in a rooting medium. Over 40 transformed plants were generated, and the presence of the transgene in the kanamycin-resistant plants was confirmed by PCR using specific primers for the peroxidase gene (data not shown).
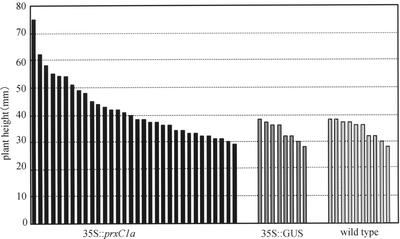
Meristematic tissue samples were cut from the top 20 mm of the stems of the transformed hybrid aspens. Samples were transferred to culture tubes (130 × 40 mm) containing freshly prepared rooting medium without kanamycin. Rooting was observed in most of the plants within 10 d of planting. The growth rates of 32 independently transformed plants with 35S::prxC1a were compared with those of eight transformed plants with pBI121 containing the 35S promoter and β-glucuronidase (GUS) structural gene and 10 non-transformed plants as controls by measuring stem lengths for 60 d after transferring them to the culture tubes. After this period, the average stem lengths of transformed with 35S::prxC1a, 35S::GUS, and wild-type plants were 42.0, 33.9, and 34.4 mm, respectively (Fig. 1). Several transformed plants with 35S::prxC1a were significantly taller than the control plants (P ≤ 0.05). There were no obvious differences between the growth rates of the transformed and control plants (data not shown).
Figure 1.
Growth rate of in vitro cultured hybrid aspen. The top parts of the stem, containing the shoot apex, in 32 transformed plants with 35S::prxC1a, eight transformed plants with 35S::GUS, and 10 wild-type plants were transferred into rooting medium, and stem lengths were measured after 60 d.
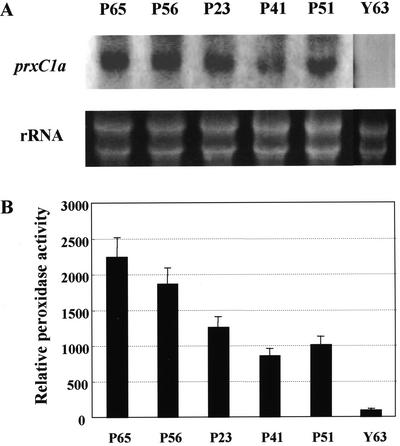
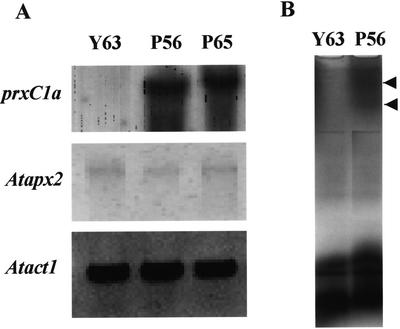
Total RNA was extracted from leaves taken from the five tallest transformed plants with 35S::prxC1a (labeled as P65, P56, P23, P41, and P51) that had exhibited the highest growth rate during in vitro culturing (Fig. 2). Northern-blot analysis was performed using the full-length prxC1a cDNA as a probe. Strong expression was observed in the leaves and stems of both transformed plants P56 and P65, whereas no detectable transcript was found in the wild-type plant Y63 (Fig. 2A). The guaiacol peroxidase activities of the soluble fractions extracted from the leaves of these plants were also measured. These fast-growing plants exhibited peroxidase activities more than 10 times higher than those in control plants (Fig. 2B). Because of their high levels of peroxidase activity, the transformed plants P56 and P65 were used subsequently in further experiments.
Figure 2.
Expression levels of the transgene and peroxidase activities in the transformed plants. A, RNA gel blot of the prxC1a gene product. Total RNA extracted from leaves in the in vitro cultured transformed (P56, P65, P23, P41, and P51) and control (Y63) plants were fractionated by agarose gel. Hybridization was performed using a 1.1-kb prxC1a cDNA fragment as a probe. Ribosomal RNA (rRNA) was stained with ethidium bromide. B, Relative peroxidase activities in the in vitro-cultured transformed plants. The guaiacol peroxidase activities of the soluble fractions extracted from leaves in the plants were measured. The enzyme activities were relative to the specific activity in the wild-type plant Y63 (=100%, 140.8 nmol min–1 mg protein–1). Each data point represents the average of three replicates.
Growth Rate of Peroxidase-Overproducing Plants under Greenhouse Conditions
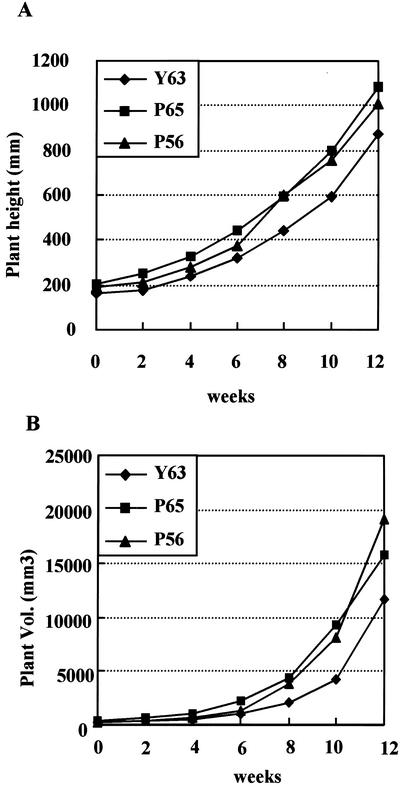
Plants P56 and P65 had two and three copies, respectively, of the transgene inserted by Southern hybridization (data not shown). At least five 100-mm-tall micropropagated plants of each line were transferred to pots (280 × 280 mm) containing Metromix-350 (Scotts, Marysville, OH), and their growth rates were evaluated under greenhouse conditions (see “Materials and Methods”). After a 4-week period of acclimation, their stem lengths and basal diameters were measured every 2 weeks (Fig. 3). Twelve weeks after acclimation, the average stem lengths were 1,010 ± 87 mm, 1,086 ± 80 mm, and 870 ± 93 mm for P56, P65, and Y63 lines, respectively (Fig. 3A). Therefore, the average stem length in the P65 line was 25% greater than that of the Y63 control line (P ≤ 0.05), and there was a corresponding 1.3-fold increase in stem volume of transformed plants compared with that of the control plants (Fig. 3B).
Figure 3.
Growth rate of the hybrid aspen in the greenhouse. A, Growth curve of stem height (sample sizes: Y63, n = 5; P56, n = 10; and P65, n = 10). B, Growth curves for plant volume. Growth rate was determined by measuring stem length from the top of the shoot apex to the base of the stem.
Characterization of Fast-Growing Transformed Plants
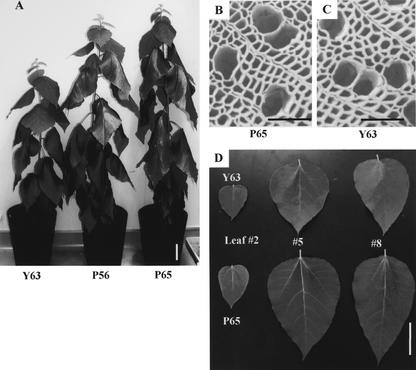
After growing under greenhouse conditions for 12 weeks, the only remarkable phenotypic difference between the transformed and the control plants was in their growth rates (Fig. 4A). In addition, the leaves of transformed plants were larger than those of the controls (Fig. 4D). Scanning electron microscopy revealed the shape and size of stem xylem fibers and vessel elements of transformed plants to be similar to those of control plants (Fig. 4, B and C). The lengths of the internodes were almost the same; however, the transformed aspens possessed on average 1.2 times more internodes than the control plants, confirming the fast-growing characteristics of the peroxidase-overexpressing hybrid aspen trees (Table I).
Figure 4.
Phenotype of the transformed hybrid aspen. A, Photograph of 12-week-old transformed plants: P56 (middle), P65 (right), and control Y63 (left). Bar = 100 mm. B, Scanning electron micrograph of stem cross section at the 10th internode in the P65 plant. Bar = 100 μm. C, Scanning electron micrograph of stem cross section at the 10th internode in the Y63 plant. Bar = 100 μm. D, Leaves of Y63 (upper) and P65 (lower), second (left), eighth (middle), and 15th (right) leaves from the apex. Bar = 100 mm.
Table I.
Phenotypic characterization of transformed hybrid aspen
Greenhouse-grown 12-week-old transformed plants P56 and P65 and control plant Y63 were investigated. Results are the means of five measurements.
| Phenotype | Y63 | P56 | P65 |
|---|---|---|---|
| Internode no. | 24.5 ± 1.6 | 31.0 ± 1.6 | 29.2 ± 2.8 |
| Internode length (mm) | 35.8 ± 5.1 | 36.5 ± 5.7 | 36.8 ± 8.6 |
| Plant height (mm) | 870.0 ± 92.7 | 1,010.0 ± 87.0 | 1,085.7 ± 80.3 |
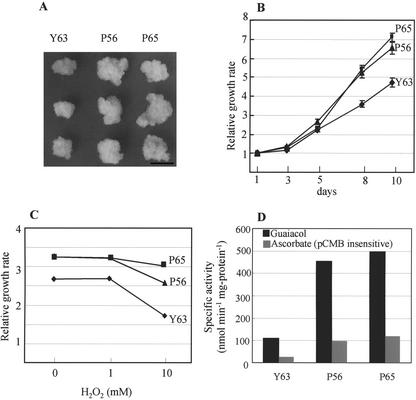
The soluble protein fraction was extracted from the leaves of the 12-week-old hybrid aspen plants, and peroxidase activities toward guaiacol and ascorbate as electron donors were investigated by a separate assay method using a p-chloromercuribenzoate (pCMB) as a specific inhibitor of ascorbate peroxidase (Amako et al., 1994). The transformed plants exhibited 7 to 10 times higher guaiacol peroxidase activity than the control plants (Table II). Using the separate assay, we found that the endogenous ascorbate peroxidase activities in each line were similar. There were no differences in the transcript levels of class I ascorbate peroxidase between the transformed and control plants, whereas the transcripts of prxC1a were produced in much higher levels in transformed plants (Fig. 5A). Visualization of guaiacol peroxidase activity by native PAGE showed two bands at low migrated positions in the transformed plant P56. These two bands might have arisen as the results of a difference in glycosilation. No induction of endogenous peroxidase in the transformed plant was observed (Fig. 5B). However, pCMB-insensitive peroxidase activities toward ascorbate as an electron donor in the transformed plants P56 and P65 were 6- to 7-fold higher than in the control plant Y63 (Table II). These results suggest that the transformed aspen plant containing the introduced prxC1a gene exhibits a high level of peroxidase activity toward ascorbate.
Table II.
Guaiacol and ascorbate peroxidase activities in transformed hybrid aspen
Soluble fractions were extracted from the leaves of the 12-week-old transformed and control plants, and separate assays were carried out (see “Materials and Methods”). Values given are the means of at least three samples.
| Electron donor | Y63 | P56 | P65 |
|---|---|---|---|
| nmol min-1 mg protein-1 | |||
| Guaiacol | 167.8 ± 19.3 | 1,297.0 ± 73.7 | 1,630.3 ± 96.2 |
| Ascorbate (pCMB sensitive) | 40.7 ± 5.4 | 37.5 ± 4.5 | 45.0 ± 6.8 |
| Ascorbate (pCMB insensitive) | 25.9 ± 5.2 | 166.3 ± 11.2 | 149.4 ± 10.2 |
Figure 5.
Expression levels of prxC1a and apx products and active staining by native PAGE in the transformed hybrid aspen and control plants. A, Total RNAs extracted from leaves were blotted onto a nylon membrane and hybridized with each probe. Atprx2 full-length cDNA (X98275) was used to detect ascorbate peroxidase transcripts. B, Activity staining was carried out using the soluble fraction from each plant. Arrows show the putative introduced gene products.
Class III peroxidase plays a role in lignin biosynthesis (Whetten et al., 1998). We determined the lignin content of the cell wall residue (CWR) of stem xylem tissues in transformed and control plants using the gravimetric Klason method. Lignin content was measured from samples taken from the lower part of the stem of plants after 12 weeks of growth in the greenhouse. There was no obvious difference between transformed and control plants (Table III).
Table III.
Lignin content in transformed hybrid aspen
Lignin content is expressed as a percentage (w/w) of CWRs.
| Y63 | P56 | P65 | |
|---|---|---|---|
| Klason lignin | 18.3 ± 0.2 | 18.4 ± 0.1 | 18.3 ± 0.3 |
Analysis of Oxidative Stress Resistance in Transformed Plants
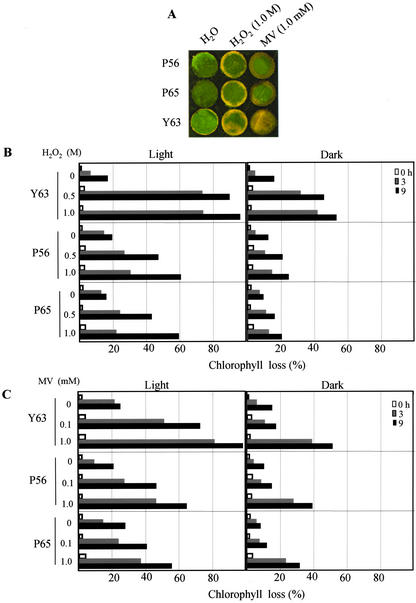
Elevated peroxidase activity level in plants is highly correlated with increased resistance to abiotic stresses, including oxidative stress (Hiraga et al., 2001). We examined whether peroxidase-overproducing plants show increased resistance to oxidative stress. Oxidative stress was generated by exposing plant leaf tissues to H2O2 and methyl viologen (MV), which produces O2–. Oxidative damage was evaluated as the percentage loss of chlorophyll after these treatments under strong light and dark growth conditions. The control plants were visibly damaged within 3 h of receiving either 1.0 m H2O2 or 1.0 mm MV treatment (Fig. 6A). Leaf discs from plants grown in strong light showed over twice the level of damage compared with those from plants grown in darkness, a fact that was true for both the control and transformed plants. However, the transformed plants showed a marked increase in their ability to withstand these treatments and had a substantially higher resistance (Fig. 6, B and C). These results strongly suggest that peroxidase-overproducing plants have an increased resistance to oxidative stress.
Figure 6.
Enhanced oxidative stress tolerance of the transformed plants overproducing peroxidase, grown under greenhouse conditions. A, Photograph of leaf samples floated on distilled water, 1.0 mm MV, and 1.0 m H2O2 for 3 h under light conditions. B, Resistance as a percentage of chlorophyll pigment lost after the 0.5 and 1.0 m H2O2 treatments. C, Resistance as a percentage of chlorophyll pigment lost after the 0.1 and 1.0 mm MV treatment under light (left) and dark (right) conditions. Results represent the mean of five measurements, with sds less than 5%.
Callus Growth of Transformed Cells
To investigate the growth rate of the peroxidase-overproducing plants, we induced callus growth from the stem segments of in vitro-cultured transformed plants. White calli formed on both the transformed and control plants within 4 weeks of being transferred to the callus-inducing medium (CIM). Growth rates were assessed by measuring the fresh weight of the calli (Fig. 7A). After 10 d, the weights of calli from plants P56 and P65 had increased by 6.5- and 7.0-fold respectively. However, calli derived from the control plant Y63 showed less than a 5-fold gain in fresh weight during the same growth period. Thus, the growth rates of peroxidase-overproducing calli are approximately 30% faster than normal (Fig. 7B).
Figure 7.
Growth rate of calli derived from the transformed hybrid aspens P56 and P65 and the control plant Y63. A, Photograph of the calli in each line, grown on the CIM for 8 d under dark conditions. Bar = 10 mm. B, Relative growth rates of calli derived from transformed and control plants. Fresh weights of at least five calli in each line were weighed. C, Relative growth rates of hybrid aspen calli after incubation with H2O2 for 6 d. D, Specific peroxidase activities toward guaiacol and ascorbate (pCMB insensitive) in control (Y63) and transformed calli (P56 and P65). Results represent the mean of five measurements, with sds less than 5%.
We also examined the influence of H2O2 concentration on growth rate by adding various concentrations of H2O2 to the CIM and assessing relative growth rates of calli after a period of 6 d of incubation (Fig. 7C). In the absence of H2O2, peroxidase-overproducing calli grew at a relative rate of approximately 3.3, whereas growth in the control group was approximately 2.7. Peroxidase activities toward both guaiacol and (pCMB insensitive) ascorbate as electron donors were approximately 4 times higher in the peroxidase-overproduced calli than in the control group (Fig. 7D). Although the presence of 1 mm H2O2 did not affect growth rates in either the transformed or control calli, a concentration of 10 mm H2O2 resulted in noticeably lower growth rates.
DISCUSSION
Some class III peroxidase isoenzymes originate from divergent genes, which can differ by more than 50% in their peptide sequences (Welinder, 1992). Other isoenzymes might originate from the same gene product and differ only in their carbohydrate moiety (Lagrimini, 1992). These constraints make it difficult to develop a model for peroxidase function in plant growth and development. However, several studies have analyzed the extent of ectopically expressed peroxidase function using sense and antisense methods. In transformed tobacco overexpressing the tobacco anionic peroxidase TOBPXDLF, lateral root formation is suppressed, probably because of enhanced indole-3-acetic acid (IAA) degradation (Lagrimini et al., 1997a). The underproduction of TOBPXDLF results in a higher growth rate (Lagrimini et al., 1997b) and a reduction in root growth, which gives rise to a leaf-wilting phenotype at the flowering stage due to the reduced ability of plants to sequester water (Lagrimini, 1992). Some studies have also indicated the possible involvement of TOBPXDLF in lignification and polyphenol metabolism (Lagrimini et al., 1997a), suggesting that this anionic peroxidase has multiple roles during growth and development in the tobacco plant. Here, we showed that the overexpression of HRP prxC1a resulted in higher growth rates in hybrid aspen (Fig. 4), confirming the possibility of multiple roles for peroxidases. The cells of hybrid aspen with overexpressed prxC1a grew faster than those of control plants (Fig. 7) and showed higher levels of tolerance to oxidative stresses (Fig. 6). However, there was no abnormal increase of the endogenous class I ascorbate peroxidase and class III guaiacol peroxidase activities in the transformed plants by separate assay and activity staining of native PAGE (Table II; Fig. 5). MV primarily induces photooxidative damage in leaves and rapidly inactivates chloroplastic ascorbate peroxidase (Mano et al., 2001). As shown in Figure 6, MV damaged the leaves of wild-type hybrid aspen grown under strong light conditions. MV-mediated increases in the transcript level of cytosolic ascorbate peroxidase have been observed in pea (Pisum sativum) leaves (Donahue et al., 1997). The transformed plants P56 and P65 clearly possessed a high level of resistance to two oxidative stresses, H2O2 and MV. Our data therefore suggest that the overexpressed peroxidase probably functions as a strong scavenger against a broad range of oxidative stresses in the cell.
What is the mechanism associated with the rapid growth of hybrid aspen plants containing the HRP prxC1a gene? There are three possible explanations concerning the physiological roles of peroxidase. The first is auxin catabolism: peroxidase oxidizes IAA, resulting in the degradation of auxin (Hinnman and Lang, 1965). The ratio of auxin to cytokinin is thought to be important for plant growth and development (Romano et al., 1991). However, this seems an implausible mechanism because northern-blot analysis and enzyme assays in transformed plants have revealed much higher levels of auxin catabolism compared with the control plants (Fig. 5; Table II). Moreover, there were no obvious phenotypic changes between the transformed and control plants (Fig. 4). If the IAA were degraded in large quantities in transformed plants, abnormal growth of lateral roots or axial shoots would be observed. Morphological differences in root growth were not observed between the transformed and control plants.
Second, peroxidase is associated with the process of lignification during the last step of monolignol deposition (Whetten et al., 1998). To date, many studies have investigated the modification of lignin content or composition in transformed plants. Modified expression of the genes involved in lignin biosynthesis, such as Phe ammonia-lyase, coumaroyl CoA reductase, and caffeoyl-CoA O-methyl transferase, has been reported that severely reduces lignin content, resulting in abnormal phenotypes (Elkind et al., 1990; Piquemal et al., 1998; Zhong et al., 1998). In quaking aspen (Populus tremuloides), which has a lower lignin content, the suppression of 4-coumarate: CoA ligase (4CL) promoted increased growth rates under greenhouse conditions (Hu et al., 1999). Hu et al. (1999) suggested that the growth enhancement resulted from a shift between primary and secondary metabolism by changing the 4CL enzymatic activity in transgenic 4CL down-regulated aspen. In the present study, the transformed hybrid aspen trees had the same levels of Klason lignin as the controls, even those with elevated peroxidase activity (Tables II and III). Overexpression of the peroxidase gene prxC1a did not affect lignin content in plant stems.
Finally, peroxidase has broad substrate specificity, and class III peroxidase has an activity toward ascorbate at a low rate (Kvaratskhelia et al., 1997). Plant cell growth imposes a unique set of constraints on cell division and elongation (Jacobs, 1997): Cell elongation occurs after cell division stops, several genes control cell division (i.e. cyclin; Jacobs, 1997) and cell elongation (i.e. expansin; Cosgrove, 1996), and ascorbate peroxidase produces monodehydroascorbate from ascorbate after the peroxidation of H2O2. It has been proposed that the expression of ascorbate oxidase and the production of dehydroascorbate are under the control of the cell cycle and that ascorbate oxidase might function in apoplast as an ascorbate oxidizer in the process of cell elongation (Kato and Esaka, 1999). Monodehydroascorbate is the primary peroxidation product from ascorbate in either class I or III peroxidase reaction (Yamazaki and Piette, 1961; Chen and Asada, 1990). In the case of ascorbate oxidase, the primary oxidation product is also monodehydroascorbate (Yamazaki and Piette, 1961). Dehydroascorbate is one of the spontaneous disproportionate products of monodehydroascorbate. The ratio of ascorbate to dehydroascorbate is purported to be functionally important for plant cell division and cell wall elongation (Kato and Esaka, 1999). The pCMB-insensitive peroxidase activity toward ascorbate was increased in leaves by the overexpression of the prxC1a protein although the endogenous class I ascorbate peroxidase activity was not changed (Table II). Furthermore, calli from transformed plants with higher peroxidase activity stimulated growth rates and were highly tolerant to oxidative stress, such as that imposed by H2O2 (Fig. 6). The results imply that the introduced gene product prxC1a has an activity toward ascorbate and that its overexpression confers oxidative stress tolerance.
The cytosolic ascorbate peroxidase activity of spinach (Spinacia oleracea) increases dramatically in response to oxidative stresses (Yoshimura et al., 2000). Class I ascorbate peroxidase is ubiquitous in higher plants, and its major physiological function is the scavenging of H2O2 in chloroplasts (Welinder, 1992). In a recent review, Asada (2000) pointed out that the water-water cycle scavenges active oxygen ions in chloroplasts. It seems likely that the induction of cytosolic ascorbate peroxidase expression against oxidative stresses plays an important role in removing H2O2 and minimizing photooxidative damage (Donahue et al., 1997). The overexpression of prxC1a increases peroxidase activity toward ascorbate, and this might result in a change in the ratio of ascorbate to dehydroascorbate in the apoplast. Calli derived from transformed plants grew faster than those derived from control plants (Fig. 6B). To know the precise physiological function of prxC1a, analysis of its subcellular localization is important. To determine the subcellular localization of the prxC1a protein, we constructed a chimeric gene in which the green fluorescent protein gene was fused to the nucleotide sequences corresponding to both the N- and C-terminal signal peptides of prxC1a under the control of the CaMV 35S promoter. This chimeric construct was introduced into Bright-Yellow 2 tobacco cultured cells with a kanamycin-resistant gene. Under fluorescence microscopy, the transformed Bright-Yellow 2 cells were observed to exhibit the green fluorescent signal primarily in the vacuole, with a weak signal also diffused into the cytoplasm (T. Matsui, K. Yoshida, and A. Shinmyo, unpublished data). We are currently conducting experiments to examine the subcellular localization of overproduced prxC1a in transformed plant (cells).
We have shown that the overexpression of the prxC1a gene in hybrid aspen resulted in higher peroxidase activity levels toward guaiacol and ascorbate in the cytosol. Growth rates and resistance to oxidative stress of transformed plants under greenhouse conditions increased. Woody plants have longer growth periods than herbaceous plants, and field studies of the growth rate of hybrid aspen with prxC1a overexpression are required. Our results suggest that the overexpression of HRP in woody plants can be an efficient strategy for producing biomass in the forestry, textile, pulp, and paper industries.
MATERIALS AND METHODS
Plant Materials
Hybrid aspen (Populus sieboldii × Populus grandidentata Y63) were grown in a greenhouse under 16-h daily light periods (300 μmol m–2 s–1) at 25°C. Plants were maintained in Metromix-350 medium.
Construction of the Chimeric Gene
The chimeric gene between the CaMV 35S promoter and a 1.1-kb DNA fragment containing cDNA of horseradish (Armoracia rusticana) neutral peroxidase prxC1a was constructed as described previously (Kawaoka et al., 1994a).
Transformation of Hybrid Aspen
Hybrid aspen plants were transformed with Agrobacterium tumefaciens LBA4404 using the stem transformation method. The stem of a flask-grown plant was cut into 5-mm sections and then inoculated with the A. tumefaciens LBA4404 with 35S::prxC1a and 35S::GUS. Transformants were selected on a modified Murashige and Skoog medium that changed the nitrogen source composition to 10 mm ammonium nitrate and 30 mm potassium nitrate, containing 100 mg L–1 kanamycin and 0.5 mg L–1 t-zeatin. Kanamycin-resistant shoots were placed on a medium (two-thirds-strength Murashige and Skoog medium, 2% [w/v] Suc, 0.25% [w/v] Gelrite, and 0.05 mg L–1 indole-butyric acid) for root regeneration.
Callus Formation and Cultivation of Hybrid Aspen
For callus induction in transformed and wild-type hybrid aspen plants, the stem segments of in vitro-cultured plants were placed on a basal medium containing 1.0 mg L–1 naphthylacetic acid and 0.5 mg L–1 benzyladenine at 25°C under dark conditions. After 4 weeks, calli were generated and cultivated at 25°C under dark conditions in the same medium used for callus induction.
RNA Isolation and Northern-Blot Hybridization
Total RNA was extracted from aspen leaves using the method described by Chomczynski and Sacchi (1987). Twenty micrograms of total RNA was subjected to electrophoresis on an agarose gel that contained 0.66 m formaldehyde, and bands of RNA were transferred to a Hybond N+ membrane (Amersham, Buckinghamshire, UK). The digoxigenin-labeled hybridization probe was prepared by random priming following the manufacturer's protocol (Roche Diagnostics, Mannheim, Germany). The cDNA fragments Atapx2 (X98275) and Atact1 (AY064043) were isolated by PCR using the specific primers.
Activity Staining
A native PAGE was carried out using a MiniProtean system (Bio-Rad, Hercules, CA). The gels were stained for peroxidase activity in a 4:1 (v/v) solution of buffer A (150 mm NaCl in 50 mm Tris-HCl [pH 7.5]) and buffer B (0.3% [v/v] guaiacol, 0.1% [v/v] H2O2, and 85% [v/v] methanol).
Growth Rate Measurement
Growth rate was determined by measuring stem length from the top of the shoot apex to the base of the stem. Plant volume was approximated as a cone shape by measuring stem length and the diameter of the basal part of the stem.
Oxidative Stress Treatment and Chlorophyll Determination
Samples (approximately 1.0 cm2) cut from fully expanded leaves of plants grown in the greenhouse were floated on either H2O2, MV, or distilled water at 25°C in strong light conditions (1,600 μmol m–2 s–1). The chlorophyll a and b contents of the leaf tissues were determined by spectrophotometry (Lichtenthaler and Wellburn, 1983).
Enzyme Assays
Separate assays on peroxidase activities toward guaiacol and ascorbate were carried out following the method of Amako et al. (1994). Leaf tissue from greenhouse-grown control and transformed aspen plants with the peroxidase gene prxC1a was homogenized and extracted in a buffer containing 1 mm ascorbate, 1 mm EDTA, and 50 mm potassium phosphate (pH 7.0). The homogenate was centrifuged, and the supernatant was passed through a gel filtration column of Sephadex G-25 (1.0 × 5.0 cm). The filtrate was concentrated using Ultrafree C3-GV (Millipore, Bedford, MA). Peroxidase activity with guaiacol or ascorbate as the reducing substrate was determined in a reaction mixture containing 50 mm potassium phosphate (pH 7.0), 1 mm guaiacol or ascorbate, and 0.5 mm H2O2. Oxidation of guaiacol or ascorbate was followed by a change in A470 (ε = 26.6 mm–1 cm–1) or A290 (ε = 2.8 mm–1 cm–1), respectively (Amako et al., 1994).
Lignin Determination
Lignin content was determined from the dried insoluble CWRs of samples extracted with toluene/ethanol, ethanol, and water. Klason lignin was measured using the method described by Effland (1977).
Acknowledgments
We are grateful to Dr. Yasushi Sato and Mr. Takeshi Matsui for technical advice regarding the native PAGE experiment.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.102.019794.
References
- Amako K, Chen G-X, Asada K (1994) Separate assay specific for ascorbate peroxidase and guaiacol peroxidase and for chloroplastic and cytosolic isoenzymes of ascorbate peroxidase in plants. Plant Cell Physiol 35: 497–504 [Google Scholar]
- Asada K (2000) The water-water cycle as alternative photon and electron sinks. Philos Trans R Soc Lond B 355: 1419–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartnek-Roxa E, Eriksson H, Mattiasson B (1991) The cDNA sequence of a neutral horseradish peroxidase. Biochim Biophys Acta 1088: 245–250 [DOI] [PubMed] [Google Scholar]
- Chen G-X, Asada K (1990) Hydroxyurea and p-aminophenol are the suicide inhibitors of ascorbate peroxidase. J Biol Chem 265: 2775–2781 [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (1996) Cellular mechanisms underlying growth asymmetry during stem gravitropism. Planta 203: 130–135 [DOI] [PubMed] [Google Scholar]
- Donahue JL, Okpodu CM, Cramer CL, Grabau EA, Alscher RG (1997) Responses of antioxidants to paraquat in pea leaves. Plant Physiol 113: 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effland MJ (1977) Modified procedure to determine acid insoluble lignin in wood and pulp. Tech Assoc Pulp Paper Ind J 60: 143–144 [Google Scholar]
- Elkind Y, Edwards R, Mavandad M, Hendrick SA, Dixon RA, Lamb CJ (1990) Abnormal plant development and down-regulation of phenylpropanoid biosynthesis in transgenic tobacco containing a heterogous phenylalanine ammonia-lyase gene. Proc Natl Acad Sci USA 87: 9057–9061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelie KE, Franceschi VR, Kolattukudy PE (1986) Immunocytochemical localization and time course of appearance of an anionic peroxidase associated with suberization in wound-healing potato tuber tissue. Plant Physiol 81: 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama K, Takemura H, Shibayama S, Kobayashi K, Choi J-K, Shinmyo A, Takano M, Okada H (1988) Structure of the horseradish peroxidase isozyme C genes. Eur J Biochem 173: 681–687 [DOI] [PubMed] [Google Scholar]
- Fujiyama K, Takemura H, Shinmyo A, Okada H, Takano M (1990) Genomic DNA structure of two new horseradish-peroxidase-encoding genes. Gene 89: 163–169 [DOI] [PubMed] [Google Scholar]
- Grisebach H (1981) Lignins. In EE Conn, ed, The Biochemistry of Plants, Vol 7. Academic Press, New York, pp 457–478 [Google Scholar]
- Hinnman RL, Lang J (1965) Peroxidases catalyzed oxidation of indole-3-acetic acid. Biochemistry 4: 144–158 [DOI] [PubMed] [Google Scholar]
- Hiraga S, Sasaki K, Ito H, Ohashi Y, Matsui H (2001) A large family of Class III plant peroxidases. Plant Cell Physiol 42: 462–468 [DOI] [PubMed] [Google Scholar]
- Hoyle MC (1977) High resolution of peroxidase-indoleacetic acid oxidase isozymes from horseradish by isoelectrofocusing. Plant Physiol 60: 787–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W-J, Harding SA, Popko JL, Ralph J, Stokke DD, Tsai C-J, Chiang VL (1999) Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol 17: 808–812 [DOI] [PubMed] [Google Scholar]
- Jacobs T (1997) Why do plant cells divide? Plant Cell 9: 1021–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaothien P, Kawaoka A, Ebinuma H, Yoshida K, Shinmyo A (2002) Ntlim1, a PAL-box binding factor, controls promoter activity of the horseradish wound-inducible peroxidase gene. Plant Mol Biol 49: 591–599 [DOI] [PubMed] [Google Scholar]
- Kaothien P, Shimokawatoko Y, Kawaoka A, Yoshida K, Shinmyo A (2000) A cis-element containing PAL-box functions in the expression of the wound-inducible peroxidase gene of horseradish. Plant Cell Rep 19: 558–562 [DOI] [PubMed] [Google Scholar]
- Kato N, Esaka M (1999) Changes in ascorbate oxidase gene expression and ascorbate levels in cell division and cell elongation in tobacco cells. Physiol Plant 105: 321–329 [Google Scholar]
- Kawaoka A, Kaothien P, Yoshida K, Endo S, Yamada K, Ebinuma H (2000) Functional analysis of tobacco LIM protein Ntlim1 involved in lignin biosynthesis. Plant J 22: 289–301 [DOI] [PubMed] [Google Scholar]
- Kawaoka A, Kawamoto T, Moriki H, Ohta H, Sekine M, Takano M, Shinmyo A (1994a) Growth stimulation of tobacco plant introduced the horseradish peroxidase gene prxC1a. J Ferment Bioeng 78: 49–53 [Google Scholar]
- Kawaoka A, Kawamoto T, Ohta H, Sekine M, Takano M, Shinmyo A (1994b) Wound-induced expression of horseradish peroxidase. Plant Cell Rep 13: 149–154 [DOI] [PubMed] [Google Scholar]
- Kawaoka A, Kawamoto T, Sekine M, Yoshida K, Takano M, Shinmyo A (1994c) A cis-acting element and trans-acting factor involved in the wound-induced expression of a horseradish peroxidase gene. Plant J 6: 87–97 [DOI] [PubMed] [Google Scholar]
- Kvaratskhelia M, Winkel C, Thorneley RNF (1997) Purification and characterization of a novel class III peroxidase isoenzyme from tea leaves. Plant Physiol 114: 1237–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrimini ML, Bradford S, Rothestein S (1992) Peroxidase-induced wilting in transgenic tobacco plants. Plant Cell 2: 7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrimini ML, Gings V, Finger F, Rothestein S, Liu T-TY (1997a) Characterization of antisense transformed plants deficient in the tobacco anionic peroxidase. Plant Physiol 114: 1187–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrimini ML, Joly RJ, Dunlap JR, Liu T-TY (1997b) The consequence of peroxidase overexpression in transgenic plants on root growth and development. Plant Mol Biol 33: 887–895 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR (1983) Determination of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem Soc Trans 11: 591–592 [Google Scholar]
- Mano J, Ohno C, Domae Y, Asada K (2001) Chloroplastic ascorbate peroxidase is the primary target of methylviologen-induced photooxidative stress in spinach leaves: its relevance to monodehydroascorbate radical detected with in vivo ESR. Biochim Biophys Acta 1504: 275–287 [DOI] [PubMed] [Google Scholar]
- Piquemal J, Lapierre C, Myton K, O'Connell A, Schuch W, Grima-Pettenati J, Boudet A-M (1998) Down-regulation of cinnamoyl-CoA reductase induces significant changes of lignin profiles in transgenic tobacco plants. Plant J 13: 71–83 [Google Scholar]
- Romano CP, Hein MB, Klee HJ (1991) Inactivation of auxin in tobacco transformed with the indoleacetic acid-lysine synthetase gene of Pseudomonas savastoni. Genes Dev 5: 438–446 [DOI] [PubMed] [Google Scholar]
- Yamazaki I, Piette L (1961) Mechanism of free radical formation and disappearance during the ascorbic acid oxidase and peroxidase reactions. Biochim Biophys Acta 50: 62–69 [DOI] [PubMed] [Google Scholar]
- Yoshimura K, Yabuta Y, Ishikawa T, Shigeoka S (2000) Expression of spinach ascorbate peroxidase isoenzymes in response to oxidative stress. Plant Physiol 123: 223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welinder KG (1974) Covalent structure of the glycoprotein horseradish peroxidase (EC1.11.1.7). FEBS Lett 72: 19–23 [DOI] [PubMed] [Google Scholar]
- Welinder KG (1992) Superfamily of plant, fungal and bacterial peroxidases. Curr Opin Struct Biol 2: 388–393 [Google Scholar]
- Whetten RW, MacKay JJ, Sederoff RR (1998) Recent advances in understanding lignin biosynthesis. Annu Rev Plant Physiol Plant Mol Biol 49: 585–609 [DOI] [PubMed] [Google Scholar]
- Zhong R, Morrison WH, Negrel J, Ye Z-H (1998) Dual methylation pathway in lignin biosynthesis. Plant Cell 10: 2033–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]