Abstract
The transposition frequency of Tam3 in Antirrhinum majus, unlike that of most other cut-and-paste-type transposons, is tightly controlled by temperature: Tam3 transposes rarely at 25°C, but much more frequently at 15°C. Here, we studied the mechanism of the low-temperature-dependent transposition (LTDT) of Tam3. Our results strongly suggest that LTDT is not likely to be due to either transcriptional regulation or posttranscriptional regulation of the Tam3 TPase gene. We found that temperature shift induced a remarkable change of the methylation state unique to Tam3 sequences in the genome: Higher temperature resulted in hypermethylation, whereas lower temperature resulted in reduced methylation. The methylation state was reversible within a single generation in response to a temperature shift. Although our data demonstrate a close link between LTDT and the methylation of Tam3, they also suggest that secondary factor(s) other than DNA methylation is involved in repression of Tam3 transposition.
Tam3 in Antirrhinum majus is exceptional among cut-and-paste-type transposons (TEs) in that it is the only known TE whose transpositional behavior can be strictly controlled by environmental influence. Tam3 transposition is strongly affected by temperature: It is active at low temperatures (around 15°C) and stable at high temperatures (around 25°C; Harrison and Fincham, 1964). Shifting the temperature controls the activity mitotically and probably meiotically. Based on a study of variegation spots, Carpenter et al. (1987) reported that the Tam3 excision rate was approximately 1,000-fold greater at 15°C than at 25°C. Thus, Tam3 is remarkably sensitive to the growth temperature. This trait has provided a great advantage for the isolation and analysis of a variety of genes involved in pigmentation and development in A. majus (Coen et al., 1989).
Among the regulatory factors associated with TE activity, DNA methylation is widely involved in the inhibition of transpositional events in plant TEs. DNA methylation can regulate TE transposition at the levels of both transposase (TPase) gene expression and the TPase binding process (Schlappi et al., 1994; Ros and Kunze, 2001). Martin et al. (1989) suggested that Tam3 inactivation is also associated with DNA methylation. Kitamura et al. (2001) showed that at high temperatures, the repression of transposition occurs simultaneously for all Tam3 copies in the genome, whereas at low temperatures, the repression is released to various degrees depending on the location of the copies. The degree of the Tam3 transposition activity was found to be influenced by chromosomal position and related to the degree of methylation of the element ends.
Here, we attempted to find differences between the active and inactive states of Tam3 in plants grown at different temperatures. We compared the amounts of the transcript of the TPase gene and compared the enzymatic activity of Tam3 TPase between plants grown at 25°C and 15°C. The results showed that neither the transcription of the TPase gene nor its enzymatic activity were markedly influenced by the temperature shift. Interestingly, the methylation state of the genomic Tam3 elements at different temperatures changed specifically in a manner that paralleled the low-temperature-dependent transposition (LTDT), and the original methylation state could be restored within a single generation after a temperature shift. The mechanism of LTDT of Tam3 appeared to be profoundly linked with the methylation state of the end regions of the element. However, our results do not prove that methylation is a direct cause of Tam3 inactivation, but they suggest that there is secondary factor(s) that suppresses Tam3 transposition in the LTDT process.
RESULTS
Expression of the TPase Gene
The HAM5 line of A. majus, which is homozygous for the nivearecurrens:Tam3 (niv rec:Tam3) allele, showed a number of flower variegation spots, which resulted from the somatic reversion of niv expression, at 15°C, whereas only a few spots were observed at 25°C due to the relative stability of Tam3 at the higher temperature (Fig. 1). This effect of temperature on Tam3 excision was previously described by Harrison and Fincham (1964) and Carpenter et al. (1987). One possible mechanism of LTDT is that the transcriptional level of the Tam3 TPase gene depends on the temperature. Hehl et al. (1991) reported that northern analysis revealed a fairly large (about 5 kb) band that was larger than expected (2.7-3.0 kb) based on the TPase open reading frame. This 5-kb transcript containing Tam3 sequence is unlikely to be translated to a proper Tam3 TPase. We also detected this band in leaf RNA samples isolated from plants grown at either 25°C or 15°C (data not shown). No obvious difference in the 5-kb transcript was seen when the 15°C and 25°C RNA samples were compared.
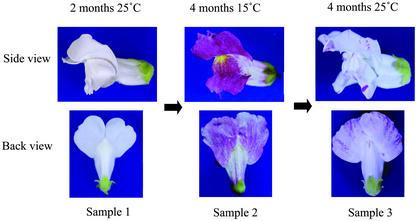
Figure 1.
Changes of flower phenotype in HAM5 after temperature shift. Left, Flower of a plant grown at 25°C has white petals (sample 1). Middle, Flower of the same plant grown at 15°C for 4 months after rearing of seedlings for 2 months at 25°C shows spots of variegation at the lobe (sample 2). Right, Pigmentation of the flowers of the same plant returned to the 25°C condition and grown for 4 months has reverted to that of sample 1, i.e. white petals with a small number of pigmented spots (sample 3). Most of the Tam3 excision events at nivrec:Tam3 result in the reversion (dominance) of the flower pigmentation because the element is present upstream of the transcription start site. RNA or genomic DNA was prepared from plants at each stage, and in further experiments, the genomic DNAs were designated by the sample numbers shown here.
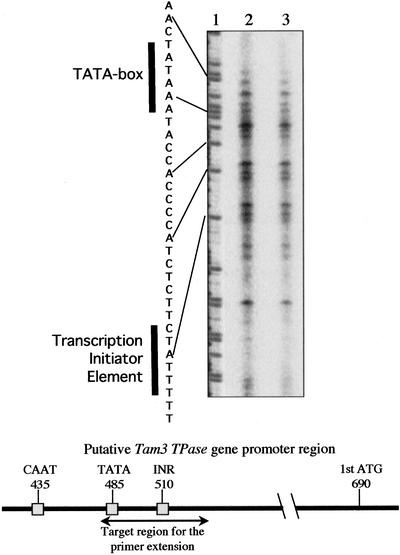
To detect the transcriptional start sites of the TPase gene within the Tam3 sequence, primer extension analysis, which is rather more sensitive than northern analysis, was carried out using the same RNA samples employed in northern analysis. The adenine of the putative first ATG of the Tam3 TPase gene is present at nucleotide 690 of the Tam3 sequence, and the TATA box and transcription initiator element (INR) were predicted to be located between nucleotides 480 and 530 of Tam3 (Fig. 2). Primer Tam3/587 (nucleotides 587-613 of Tam3) generated several extension products in the predicted promoter region of the Tam3 TPase gene (Fig. 2). Corresponding signals were also obtained using a different primer (primer Tam3/825; nucleotides 825-847 of Tam3; data not shown). The multiple signals might have originated from the transcription of different sequences of various Tam3 copies in the genome. The same signals were detected in the 25°C and 15°C RNA samples (Fig. 2). These results indicated that the Tam3 TPase gene was similarly transcribed from the sites proximal to its putative promoter sequence at the two temperatures.
Figure 2.
Primer extension analysis to detect transcriptional start sites of the TPase genes. The fluorescent-labeled antisense primer, primer-Tam3/587, corresponding to nucleotides 587 to 609 from the Tam3 5′ end, was used for detection of transcriptional start sites of the TPase genes (lanes 2 and 3) and “A” ladder sequencing (lane 1). Lane 2, Leaf RNA sample extracted from a plant grown at 15°C. Lane 3, Leaf RNA sample extracted from the same plant grown at 25°C. The sequence of nucleotides 483 to 515 from the Tam3 5′ end is shown on the left of the “A” ladder. The TATA box and INR are indicated by bold vertical lines, and the positions of the CAAT-box, TATA-box, INR, and first ATG on the Tam3 sequence are indicated on the horizontal bar.
In Vivo Activity of the Tam3 TPase
To analyze the Tam3 TPase activity, we prepared plasmid constructs containing a Tam3 element in which there was a deletion of the internal sequence. If Tam3 TPase activity is present in a plant cell, the Tam3 element is expected to be excised from the plasmid DNA after integration into the cell. A transient assay for Ac activity used to demonstrate Ds excision after bombardment of transgenic barley (Hordeum vulgare) callus lines containing Ac has been reported (McElroy et al., 1997). Using this assay, we delivered the plasmid pBS18-10E, carrying a 1.6-kb partial Tam3 DNA sequence, into leaf cells through particle bombardment. After rescue of the plasmids, the size was checked by agarose gel electrophoresis. The frequencies of Tam3-excised plasmids from randomly selected colonies are shown in Table I. We could not statistically distinguish difference in the frequencies of Tam3-excised plasmids between the leaves of plant grown at 25°C and the leaves of plant grown at 15°C. On the other hand, no Tam3-excised plasmid was obtained from tobacco, which possesses no Tam3 TPase activity. The HAM3 line, which has the Stabilizer gene (which represses Tam3 activity), had a very low frequency of excision of Tam3 from the plasmid (Table I). These results showed that the TPase activity is not influenced by temperature. As a consequence, we concluded that LTDT of Tam3 is not associated with the control of TPase gene expression. In addition, our data interestingly suggested that Stabilizer might function to suppress Tam3 TPase activity (perhaps via posttranscriptional gene silencing).
Table I.
Frequency of Tam3 excision from plasmid p18-10E after the bombardment
| Bombarded
Plantsa
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trial | HAM5 25 °C
|
HAM5 15 °C
|
HAM3 25 °C
|
Tobacco (Nicotiana tabacum)
|
||||||||
| Reb | Exc | Frd | Re | Ex | Fr | Re | Ex | Fr | Re | Ex | Fr | |
| 1 | 60 | 3 | 5.0 | 27 | 1 | 3.7 | 96 | 0 | 0 | 48 | 0 | 0 |
| 2 | 48 | 4 | 8.3 | 48 | 3 | 6.3 | 96 | 1 | 1.0 | 48 | 0 | 0 |
| 3 | 42 | 4 | 9.5 | 48 | 4 | 8.3 | 96 | 0 | 0 | - | - | - |
| 4 | 48 | 7 | 14.6 | 48 | 1 | 2.1 | - | - | - | - | - | - |
| 5 | 56 | 3 | 5.4 | 48 | 3 | 6.3 | - | - | - | - | - | - |
| 6 | - | - | - | 48 | 4 | 8.3 | - | - | - | - | - | - |
| Total | 254 | 21 | 8.3e | 267 | 16 | 6.0e | 267 | 1 | 0.03 | 96 | 0 | 0 |
All target materials for the bombardment were leaves of the respective plants. b No. of rescued clones. c No. of Tam3-excised clones. d Frequency (%). e Both the averages of the frequencies were calculated as t = 2.26ns, which indicates no statistically difference.
Methylation of Tam3 Sequences in Genomic DNA
Next, to examine the methylation state of the Tam3 sequences in the HAM5 genome, we performed hybridization analysis using a methylation-sensitive enzyme, HpaII, and a partially methylcytosine-sensitive isoschizomer, MspI. We examined plants exposed to different temperature conditions: The seeds were reared at 25°C for 2 months (sample 1), then the plants were shifted to 15°C and grown for 4 months (sample 2), and then they were shifted back to 25°C for 4 months (sample 3; Fig. 1). DNA sampling of the HAM5 leaves was performed before the end of the growth period at each temperature. The three probes employed were designed from the region containing the first ATG (probe A), the middle region (Tam3 nucleotides 1,419-1,799), and the region containing the stop codon of the putative Tam3 TPase gene (Fig. 3). The blotting results represent the whole Tam3 family in the A. majus genome, which contains about 50 copies of Tam3. A chloroplast DNA fragment (Kishima et al., 1995) was used as a control probe to confirm the completeness of digestions (data not shown).
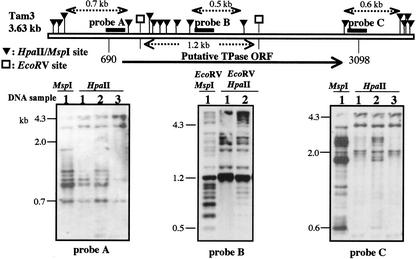
Figure 3.
Methylation state of the Tam3 end regions in the genomic DNAs extracted from the plants grown at different temperatures. The upper map corresponds to a representative Tam3 sequence with the indicated HpaII/MspI (black triangle) sites. The regions of the three amplified PCR probes are shown in the physical map by thick lines (probes A-C). The putative TPase gene is marked, and the start and stop codons are indicated as Tam3 positions 690 and 3,098, respectively. The DNA samples were extracted from the leaves of the plants designated by numbers that correspond with those of the flower samples in Figure 1 (sample 1, first 2 months/25°C; sample 2, next 4 months/15°C; and sample 3, last 4 months/25°C). Due to accidental loss of the genomic DNA of sample 3, the fourth lane for probe B was not present. The DNA was electrophoresed on a 1.5% (w/v) agarose gel. For the blot with probe B, DNAs were first digested with EcoRV (a methylation-insensitive enzyme) to restrict HpaII/MspI sites within the TPase coding region, and then a secondary digestion was performed with HpaII or MspI. The left-most lane shows the MspI digestion pattern, and the other lanes show the HpaII digestion patterns. The minimum lengths of the hybridized bands generated are indicated above the map.
The blotting patterns obtained using the three Tam3 probes showed a common trend (Fig. 3). The MspI (first lane) and HpaII (second lane) digests of DNA from sample 1 showed different digestion patterns, indicating that the Tam3 sequences were considerably methylated in the HAM5 genomic DNA at 25°C. Preferential digestion of DNA from sample 2 (third lane) compared with DNA from sample 1 was observed with HpaII, indicating that the Tam3 sequences reduced the methylated level at 15°C relative to 25°C. The digestion patterns obtained for sample 3 had reverted to those of sample 1 (fourth lane). These results showed that the methylation state of Tam3 changes reversibly depending on the temperature.
The methylation state varied depending on the region within Tam3. As shown in each panel of Figure 3, the results obtained with probes B and C revealed relatively heavier methylation of the Tam3 TPase coding sequence at both the temperatures, compared with the 5′ region of the gene as revealed by the hybridization of probe A with the smallest possible HpaII fragment (0.7 kb). This implies that the TPase coding sequence is highly methylated at 25°C despite production of the transcript.
Methylation of the End Regions in Representative Copies
We then investigated the methylation state of the end regions of the Tam3 copies inserted in the S-6, S-78, and S-99 loci (Kishima et al., 1999). These copies have identical structures but show very different excision frequencies in the active condition: Tam3: S-6 is relatively inactive, Tam3: S-99 has modest excision activity, and Tam3: S-78 is very active (relative excision frequencies: S-6:S-99:S-78 = 1:28:125; Kitamura et al., 2001). These Tam3 transposition frequencies were clearly correlated with the methylation state of the end regions in plants grown at 15°C. DNA samples from plant samples 1 to 3 described in the above section were examined (see Fig. 1). PCR fragments from both flanking regions of each copy were used as probes (see maps in Fig. 4). First, DNA was digested with MboI, and then a secondary digestion was performed with HpaII. All the probes gave rise to a single band or a few bands when the 15°C sample was digested with MboI (insensitive to cytosine methylation; Fig. 4, left-most lane in each panel). Some of the HpaII sites are clustered in the end regions, with the outermost sites located 33 and 31 bp from the 5′ and 3′ ends, respectively. There are no HpaII sites in the sequences immediately flanking these three Tam3 copies (see maps in Fig. 4).
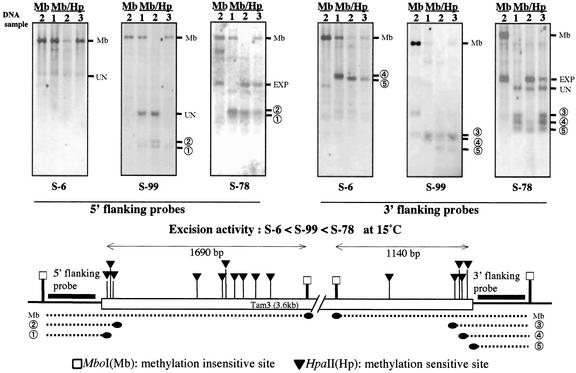
Figure 4.
Methylation state of Tam3 at three representative loci in the genomic DNAs extracted from plants grown at different temperatures. The end regions of Tam3 at three loci, S-6, S-99, and S-78, were analyzed. These copies have identical sequences. The transposition activity of the three copies varies: the order of the activity is S-6 < S-99 < S-78 in the active condition, as noted in the middle of the figure. The leaf DNA samples are designated by numbers that correspond to those of the flower samples in Figure 1 (sample 1, first 2 months/25°C; sample 2, next 4 months/15°C; and sample 3, last 4 months/25°C). We used the restriction enzymes MboI (left-most lane) and MboI+HpaII (the other three lanes). Clusters of HpaII sites are present in both end regions of the Tam3 sequence, and MboI sites flank these clusters. The amplified probes hybridized with the two flanking regions and are marked by thick lines. The DNA was electrophoresed on a 1% (w/v) agarose gel. The hybridized fragments are indicated by dotted lines with the respective numbers under the map. The probes also hybridized with several bands that were unrelated to the target sites of the probes: These are marked “UN” (unknown fragment). In the blots of S-78, intense bands were detected at sizes that were not related to those of the methylated bands: These were due to de novo excision products of Tam3 and are marked “EXP” (excision products). Extensive methylation at HpaII sites present within the MboI fragment gave rise to a fragment of the same size as the MboI fragment (Mb).
The hybridization patterns in Figure 4 revealed the methylation states of the end regions in the three copies. In each panel, the MboI/HpaII digestion patterns of samples 1 and 3 (the two 25°C samples) were similar, whereas the digestion pattern of the 15°C sample (sample 2) was different except in the case of the inactive Tam3 at S-6. With MboI/HpaII double digestion, most of the 25°C samples produced the same largest sized bands as produced by MboI single digestion, but in the 15°C samples, these bands disappeared in the active copies at the S-78 and S-99 loci. The largest bands in the 25°C samples resulted from extensive and heavy methylation of HpaII sites in the MboI fragment. The methylation occurred in the initial 25°C growth condition, and it recurred when the temperature was shifted back to 25°C after growth at 15°C. The reversion to the high degree of methylation within single generation is a unique phenomenon for the plant genome.
Smaller sized bands, which indicated a less methylated state at the Tam3 end, arose in the active Tam3 copies at the S-78 and S-99 loci in the 15°C condition. In the 3′ region of Tam3: S-78, the most active copy (extra bands due to Tam3 excision reflect the activity at S-78; see Fig. 4), the temperature-dependent alteration of the methylation pattern was limited in the outermost sites of the HpaII cluster. The results showed that methylation of the Tam3 end regions also parallels the temperature change, like methylation of the Tam3 internal regions, and is a somatically reversible reaction. Taken together, the results showed that the degree of methylation state of the Tam3 sequences of the high-temperature samples was much heavier than that of the low-temperature sample and that the change of the Tam3 methylation state was well correlated with the LTDT response.
However, except in the case of the 5′ end of S-6, the smaller bands, indicative of hypomethylation, did not disappear in the 25°C samples, implying that methylation at the terminal sites was not complete in the inactive condition of Tam3. This leads us to suppose that methylation in the end regions might not be the sole determinant of the suppression of Tam3 transposition at the higher temperature, although the varying transposition activities among the copies appeared to be related to the methylation level at the terminal sites of each locus.
Methylation Detected by Sodium Bisulfite Sequencing
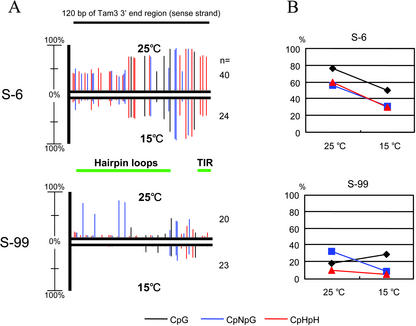
We performed 5-methylcytosine analysis using sodium bisulfite sequencing to confirm the above results. The genomic DNAs from plant samples 1 and 2 (see Fig. 1) were examined, and primers were designed to amplify the 3′ regions of Tam3 at the S-6 and S-99 loci (unfortunately, we were unable to amplify the 5′ regions of the two Tam3 loci). The sense strands of 120 nucleotides from the 3′ end of Tam3 that contains six sites of CpG, 15 sites of CpNpG (where N is either A, C, G, or T), and 22 asymmetric (CpHpH, where H is either A, C, or T) cytosine sites were analyzed. As expected in the above results, the 3′ regions of both the Tam3 loci in the 25°C sample exhibited the hypermethylation state compared with that of each locus in the 15°C samples (Fig. 5). The difference in the methylation levels at the two loci also reflected the results obtained by Southern hybridization analysis (Fig. 4) where the methylation state of Tam3 of S-6 was heavier than that of S-99. In the S-6 locus, about 65% of the total cytosines in the 3′ end region were methylated in the plant grown at 25°C, and the methylcytosines in the plant grown at 15°C were reduced to one-half of those in the plant grown at 25°C. In the Tam3 3′ end region of S-99, 20% of the overall cytosines in the 25°C sample were methylated, whereas in the 15°C sample, only 10% of cytosines were detected as methylcytosine. These results also show that the methylation level remarkably varies among Tam3 copies, which inserted into different genomic locations. As shown in Figure 5A, the regions where the methylation levels were affected by a temperature change corresponded between S-6 and S-99. However, no specific relationships seem to be present between the change of methylation level and CpG, CpNpG, and CpHpH sites (Fig. 5B). The methylcytosines that responded to the temperature shift were localized on the terminal inverted repeat and the hairpin loop cluster in the 3′ subterminal region of Tam3 (Yamashita et al., 1999). These regions correspond with sequence forming secondary structure that may also contain possible motifs for the TPase-binding sites.
Figure 5.
Distribution and quantity of methylcytosine in the Tam3 3′ end regions of S-6 and S-99 loci at the two temperatures. A, Sodium bisulfite sequencing was performed for the sense strand of the Tam3 3′ end regions (120 nucleotides) at the S-6 and S-99 loci. Vertical lines indicate the positions of methylcytosines, with the height of each line representing the percentage of the methylcytosine at the position. The upper pattern exhibits the methylation state of the sample 1 (see Fig. 1) genomic DNA isolated from the plant grown at 25°C, and the lower pattern exhibits the methylation state of the sample 2 (see Fig. 1) genomic DNA isolated from the plant grown at 15°C. n, Clone number analyzed for each plot. The green horizontal bars correspond with the terminal inverted repeat (TIR) and the hairpin loop cluster region in the Tam3 3′ subterminal sequence. The black, blue, and red lines indicate cytosines in the context of CpG, CpNpG, and CpHpH, respectively. B, Quantitative change of methylcytosines in the context of CpG, CpNpG, and CpHpH resulted from the temperature shift. Based on A, the percentages of the methylcytosines in CpG, CpNpG, and CpHpH were estimated. The two different temperatures affected quantity of methylcytosine of these sequences.
Methylation of Other Repetitive Sequences in the Genome
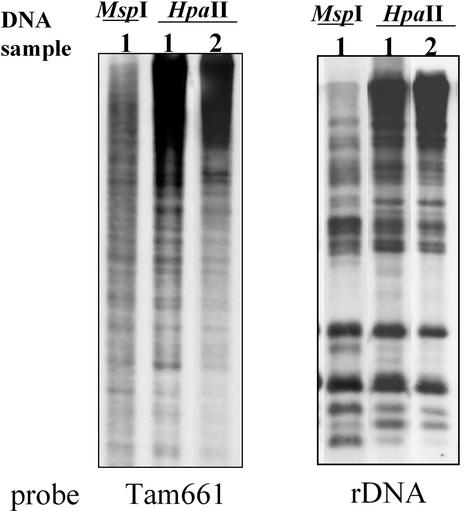
To examine whether the methylation state of other repetitive sequences in the A. majus genome changes depending on the temperature, we used the Southern-blot method using two different repetitive sequences as probes (Fig. 6). A TE-like short sequence in the A. majus genome, Tam661 (Yamashita et al., 1998), and the 25S-17S rDNA region, which has often been used as a standard to evaluate the overall methylation state of the genome (Ellis et al., 1990), were employed as probes. Both of these repetitive sequences contain HpaII sites. As indicated in Figure 6, each repetitive sequence was methylated in the genome, as indicated by the different hybridization patterns produced by digestion of the samples with MspI versus HpaII, but the repetitive probes did not produce different digestion patterns for the genomic DNAs isolated from plants grown at 25°C versus 15°C. This suggests that there was no significant difference in the methylation state of the repetitive sequences at the two temperatures. Although analysis of additional repetitive sequences would be necessary to assess the overall methylation state in various regions of the genome, the present results show that the methylation state of Tam3 in the genome changes specifically in response to a temperature shift.
Figure 6.
Methylation state of two repetitive sequences in the genomic DNAs extracted from plants grown at different temperatures. DNAs from samples 1 and 2 (see Fig. 1) were probed with two repetitive sequences, Tam661 and rDNA. Tam661 is a transposon-like sequence (Yamashita et al., 1998). The DNA samples were digested with HpaII, for which restriction sites are present in both of the sequences. The DNA was electrophoresed on a 1% (w/v) agarose gel. The blotting patterns of the two repetitive sequences showed little difference between the samples.
DISCUSSION
We showed here that LTDT of Tam3 was coordinated with a change of the methylation level of the Tam3 sequence. One striking finding of our study was that shifting the temperature affected the DNA methylation level of a specific sequence during a single generation of plant growth. Although genomic methylation in mammals undergoes resetting early in development, the methylation state of plant DNA is generally well maintained throughout the lifespan of the organism, and methylation at some loci has been reported to be heritably stable over a number of generations (Cubas et al., 1999). Particular interesting phenomenon observed here is a rapid reversion from the reduced methylation state after the rising of the temperature. Although the number of studies concerning methylation in the plant genomes have analyzed the aspect of the reducing level, the studies on increasing level of methylation were few. In Arabidopsis, hypomethylation induced by ddm1 mutation did not readily revert even in the dominant DDM1 background, and the methylation level slowly restored through several generations (Kakutani et al., 1999). The rapid reversion of the methylation level of Tam3 implicates that Tam3 is a preferential target for methylation and demethylation.
DNA methylation seems to be a key factor for repressing the transposition of plant TEs. A number of studies have addressed the relationship between DNA methylation and TPase gene expression. An increased level of DNA methylation of the promoter regions of autonomous elements such as Ac, Spm, and MuDR tends to reduce the production of TPase transcripts and the transposition frequency (Chandler and Walbot, 1986; Schwartz and Dennis, 1986; Bennetzen, 1987; Chomet et al., 1987; Kunze et al., 1987, 1988; Brown and Sundaresan, 1992; Brutnell and Dellaporta, 1994; Schlappi et al., 1994). On the other hand, the methylation at TPase-binding sites has been addressed by in vitro binding assays (Kunze and Starlinger, 1989), which showed that the Ac TPase specifically binds to AAACGG, of which multiple copies are present in both ends of Ac, and that cytosine methylation of this sequence almost completely inhibits binding of the TPase. Recently, in vivo assays demonstrated that transposition of hemimethylated Ac/Ds occurred more frequently than transposition of the fully methylated element (Ros and Kunze, 2001). The Tam3 transposition activity at the low temperature depends on the chromosomal position, and this effect is also related to the degree of methylation of the copy's ends (Kitamura et al., 2001). The position effect seems also to be determined, in part, by the binding affinity of TPase to the ends of the element, including the binding affinity to the terminal inverted repeat motifs.
Several possible mechanisms of regulating the level of methylation in the Tam3 ends can be suggested, including the following: (a) Tam3 DNA contains a total of 16 firm hairpin structures with high GC content in sequences that lie within 500 bp from each end of the element (Yamashita et al., 1999). These hairpins might be targets for DNA methylation. The human (Homo sapiens) fragile X DNA triplet repeats with hairpin structures cause hypermethylation resulting in the suppression of the FMR-1 gene (Chen et al., 1995; Godde et al., 1996). It has been suggested that the hairpin formation could account for the specific methylation of the CpG Island in the fragile X DNA repeats. Similarly, it is reasonable to predict that the hairpins in Tam3 may facilitate recognition of a temperature-sensitive DNA methyltransferase because there are locus-specific methyltransferase genes (Cao and Jacobsen, 2002). (b) Alternatively, considering that no recognition sequences (neither for symmetrical nor for asymmetrical sites such as CpG, CpNpG, and so on) of DNA methyltransferases have been identified so far, protein(s) that are expressed at low temperature might prevent interaction of a DNA methyltransferase with the Tam3 end regions or might trap a DNA methyltransferase. Gorbunova et al. (2000) reported that some plant nuclear proteins bind to motifs that are included in the terminal regions of Ac/Ds and Mu elements but that these host proteins did not appear to be essential at least for excision of Ds; thus, the function of the binding proteins remains unknown. Similar proteins might compete with DNA methyltransferase for binding to the Tam3 terminal regions at low temperature. (c) At present, we cannot rule out the presence of an enzyme with DNA demethylation activity (Wolffe et al., 1999) in A. majus. (d) Finally, factor(s) could modulate the local chromatin structures containing Tam3 in a temperature-dependent manner, and these factor(s) might simultaneously influence both the transposition and methylation state. Mammalian histone deacetylases HDAC1 and 2, whose activities alter the chromatin state, resulting in transcriptional repression, are recruited by the methyl-CpG-binding protein MeCP2 (Nan et al., 1998). In addition, HDAC1 has been found to associate with a DNA methyltransferase, Dnmt1 (Fuks et al., 2000; Robertson et al., 2000). The possible involvement of these factors associated with DNA methyltransferases in LTDT of Tam3 should be considered.
The arrest of Tam3 transposition at the high temperature might not be determined solely by methylation. This possibility is suggested by two features of the stable condition of Tam3: The transcriptional and posttranscriptional activities of the Tam3 TPase gene were detectable (Figs. 2 and 3), and methylation was not complete at the end regions of the active Tam3 copies (Figs. 4 and 5). Although the correlation between the Tam3 activity and the methylation state accords well with the fact that DNA methylation is often found in inactive transposons, our results raise the question of whether methylation is a direct cause of LTDT or an event that occurs simultaneously with LTDT. Evidence that methylation is not essential for repression of the TE activity was reported for the retrotransposon MAGGY of Magnaporthe grisea (Nakayashiki et al., 2001). We must take into account the other possibility that DNA methylation alone is not responsible for Tam3 inactivation in the LTDT process.
There is evidence that temperature affects the transposition of plant TEs in several descriptions of unstable pigmentation phenotypes: petal flaking frequency in Portulaca grandyflora (Beale and Faberge, 1941), speckled red-on-white sectors at the v locus in Nicotiana spp. (Sand, 1957), dot frequency in the Dotted-a1 system of maize (Zea mays; Rhoades, 1941), and the frequency of purple sectors determined by the mutable allele e in Primula sinensis (Harrison and Fincham, 1964). Although all these examples show a much weaker effect of temperature on the variegation frequency than examples involving Tam3, in all of these cases, an increase in temperature tends to decrease the frequency of pigmented cells. A common mechanism might underlie these LTDTs if they are due to TEs.
MATERIALS AND METHODS
Plant Materials and Extraction of DNA and RNA
We used the HAM5 and HAM3 lines of Antirrhinum majus, which were kindly provided by Dr. Cathie Martin (John Innes Center, Norwich, UK). HAM5, derived from the nivearecurrens:Tam3/stabilizer– line, was initially grown at 25°C for 2 months, and subsequently shifted to a 15°C growth chamber and grown for 4 months, then shifted back to 25°C and grown for 4 months. The HAM5 isogenic line HAM3, which carries the stabilizer+ allele, was grown at 25°C. DNA was extracted from young leaves (3-4 cm in length) of A. majus plants. The procedure of the DNA extraction was modified from the one described by Murray and Thompson (1980), i.e. to exclude RNA debris, aliquot of RNase A was added and incubated at 65°C for 20 min before chloroform extraction. RNA was extracted from the young leaves according to Martin et al. (1989).
Northern-Blot Hybridization
Twenty micrograms of each RNA sample was electrophoresed on a 1% (w/v) agarose gel containing formaldehyde and transferred onto a nylon membrane (Positively Charged, Boehringer Mannheim/Roche, Basel). A 1,300-bp (nucleotide positions 1,700-3,000 in the Tam3 sequence) region of the Tam3 TPase coding region was employed as the template for the probe. The Tam3 probe was prepared using a PCR-based labeling system with PCR DIG labeling mix (Roche) and the following primers: Tam3-1700F, 5′GTTGCATTACCGCACATTGG3′; and Tam3-C, 5′TCTCTATATTGTTGGTCGAGCATGTCT3′. Hybridization was carried out overnight at 65°C, and detection was performed using a DIG Nucleic Acid Detection Kit (Roche).
Primer Extension
For determination of the transcription initiation sites of the Tam3 TPase gene, primer extension experiments were performed using polyadenylated poly(A+) RNA isolated from the leaves. Poly(A+) RNA was isolated using the PolyATtract mRNA isolation systems (Promega, Madison, WI). Oligonucleotide primers 5′-CACCGTGGAGGTATGAC-3′ (primer-Tam3/587), and 5′-GTGAATCTTCATATGGTGTATC-3′ (primer-Tam3/825), corresponding to sequences starting at nucleotide positions 587 and 825 of Tam3, respectively, were end labeled with IRD800 and IRD700 (LI-COR, Lincoln, NE), respectively. Poly(A+) RNA (2 μg) was hybridized with the labeled primers in a solution containing 250 mm KCl, 10 mm Tris/HCl (pH 8.3), and 1 mm dithiothreitol at 85°C for 10 min and then at room temperature for 90 min. The extension reaction was carried out in 50 mm Tris/HCl (pH 8.3), 75 mm MgCl2, 1 mm dithiothreitol, each dNTP at 0.25 mm, 1 unit of RNAsin (Takara, Kyoto), and 5 μL of reverse transcriptase (100 units μL–1 MMLV Reverse Transcriptase RNaseH–, TOYOBO, Tokyo, Japan) at 42°C for 1h. After RNase treatment (16 μg mL–1, 37°C for 30 min), the cDNA products were extracted with phenol and chloroform and precipitated with ethanol. The DNA was analyzed by electrophoresis on a 4.5% (w/v) polyacrylamide sequencing gel. Dideoxy sequencing products primed with the corresponding primers were electrophoresed in parallel for size comparison. The fluorescent signals were detected using a DNA Sequencer model LIC400 (LI-COR).
In Vivo Assay of Tam3 Excision by the Plasmid Rescue Method
A plasmid, pBS18-10E, carrying a 1,370-bp partial Tam3 sequence with an internal deletion of the 2,260-bp region from the BalI to the TthHB81 site was used for monitoring Tam3 excision. This plasmid was constructed by combining the vector pBluescriptSK with the Tam3 5′ region from AG1400 (SK; from the XbaI to the BalI site) and the Tam3 3′ region from Tam3:S-18 (SK; from the TthHB81 to the EaeI site). The Tam3 5′ and 3′ regions were ligated to each other and then treated by filling in with T4 DNA polymerase. This plasmid does not have 8-bp duplications at the ends of Tam3 and, therefore, does not undergo loop-out recombination between the target site duplications. Thus, the plasmid produced pBS18-10E, which was delivered via the bombardment method into leaves isolated from plants grown at different temperatures. For the bombardment delivery, surface-sterilized A. majus leaves were placed directly on agar plates containing 0.8% (w/v) agar in water. Gold particles (2 mg, 1.0-μm diameter, Bio-Rad Laboratories, Hercules, CA) were coated with a 6:1 (w/v) ratio of the reporter plasmid carrying the internally deleted Tam3 to the reference plasmid carrying the renilla luciferase reporter gene. The plant material was bombarded with 1 μg of DNA-coated gold particles using a helium-driven Biolistic PDS 1000 System (Bio-Rad Laboratories) with a 28-mmHg vacuum. The distance between the rupture disc and the macrocarrier was 1 cm. The bombarded leaves were incubated for 16 h at 15 or 25°C. Leaves from HAM5, HAM3, and tobacco (Nicotiana tabacum cv SR1) were obtained from plants grown at 15°C or 25°C. The leaf DNA was extracted after incubation at 15°C or 25°C for 16 h. To recover the plasmid, an aliquot of the DNA solution was used for transformation of Escherichia coli (DH5α strain, TOYOBO), and the transformed E. coli were grown in Luria-Bertani medium containing 50 μg mL–1 ampicillin. Two hundred colonies were randomly selected, and the plasmids were collected from them. To examine whether the plasmid had lost Tam3, its size was checked by 1.5% (w/v) agarose gel electrophoresis after SacI digestion. The relative activity of Tam3 TPase was calculated as the ratio of the number of colonies with the altered-size plasmid to the total number of plasmid-carrying colonies.
Southern Blotting to Detect Methylation
The methylation state of Tam3 and the other repetitive elements was investigated by Southern-blot analysis using the C-methylation-sensitive enzyme HpaII, partially sensitive enzyme MspI (isoschizomer of HpaII), and insensitive enzymes EcoRV and MboI. HAM5 genomic DNA was isolated from the plants as described above. The following probes were prepared using a PCR-based labeling system with PCR DIG labeling mix (Roche): Tam3 probe A, CCTCACATTTTTATTTCTTAGTG + GGGTCGGTACTTGGAACTCC; Tam3 probe B, CACCGTGGAGGTATGAC + GAGGACATTGTGGCATCGCG; Tam3 probe C, CTAACCCCTGTCTTGGC + ACGCGTCGACGCAACTACAACAAAGGTGC; 5′-flanking sequence of (5′ fla.) S-6, CAAACAGGTTCAGCTCTCC + GTATCTACACCAATAACTGCG; 3′ flanking sequence of (3′ fla.) S-6, AAAATATGTCATCTTGGTCACTGGTTGC + GTGTACTTACTGCATAGCGTTCCTT; 5′ fla. S-78, CTTGTTCGTGGATTGGTTGGTGGTCGCCTG + GTTGTAGCATAGTGTAGTTAG; 3′ fla. S-78, GCAATAGATACAACAATAGCAGG + GATCATGAACCAATTTCAAAACTCTCC; 5′ fla. S-99, AACTTCCTCCTACGATATTGCTC + CCCTTTAATTGAGTGGTCATCTCTC; and 3′ fla. S-99, ACAGTGGACTATGTCTCCTAGTATAC + AATTCGCGGCCGCT. The DNA templates (the plasmid clones; Kitamura et al., 2001) and primers were denatured at 94°C for 30 s, annealed at a few degrees above the melting temperature calculated for each primer and extended at 72°C for 30 s for a total of 35 cycles. Fifteen micrograms of each digested DNA sample was electrophoresed on 1% or 1.5% (w/v) agarose gel and transferred onto a nylon membrane (Positively Charged, Roche). The probes for Tam661 and rDNA (a rice [Oryza sativa] 17S-25S rDNA fragment) were prepared from plasmid clones (Sano and Sano, 1990; Yamashita et al., 1998). Hybridization was performed using the ECL gene detection system (Amersham, Buckinghamshire, UK). To verify that complete digestion was achieved by the enzymes, a chloroplast DNA fragment, 5.2-kb Sma-8 of buckwheat (Fagopyrum esculentum; Kishima et al., 1995), was used as a control probe.
Sodium Bisulfite Sequencing to Detect Methylcytosine
Before the sodium bisulfite reaction, the genomic DNAs of samples 1 and 2 (see Fig. 1) were digested with MboI to facilitate the reaction. The sodium bisulfite modification procedures were adapted as described by Frommer et al. (1992). In brief, approximately 10 μg of the digested genomic DNA was denatured in 0.2 m NaOH for 10 min. As a control, 100 ng of EcoRV-digested Bluescript plasmid was added in the denature treatment. Thirty microliters of 10 mm hydroquinone (Sigma, St. Louis) and 270 μL of 3 m sodium bisulfite (Sigma) at pH 5, both freshly prepared, were added and mixed, and samples were incubated under mineral oil at 55°C for 16 h. Modified DNA was purified using the Geneclean spin kit (Bio 101, Vista, CA) according to the manufacturer's recommendations and eluted into 80 μL. Bisulfite-modified DNA was amplified with specific primers for five nested PCR reactions. Primers used here were as follows: Tam3 5′ of S-6 antisense strand first primer, TCTTCTTCARCTCAAAGCCCCTATTA+AATTGAAAAATGTTATGAAGGTATTGTGTG; Tam3 5′ of S-6 antisense strand second primer, CCTAATAAAACAACAATTARCACACCTA+GGCTAGAAGTAGATTAGAATTGG; GGGGTTGGAATAGGAGAGTGTATGAGGGAG+CCATAAACTCCACCCCCCTAACAAACATC; Tam3 5′ of S-99 antisense strand first primer, AACTTCCTCCTACRATATTRCTCTA+AATTGAAAAATGTTATGAAGGTATTGTGTG; Tam3 5′ of S-99 antisense strand second primer, AAARTCAAACRCACACRAR+GGCTAGAAGTAGATTAGAATTGG; Tam3 3′ of S-6 sense strand first primer, TTTTTTTTTTTATGCAGGGGTTTTTTTTTT+ATACACTTCACCTCARACCCAAAARCAAAAC; Tam3 3′ of S-6 sense strand second primer, GGGTTGAATTGGTTGGAGAAATGG+TTCTAATTCACAAACGTAATACAAAAAATT; Tam3 3′ of S-99 sense strand first primer, TTTTTTTTTTTATGCAGGGGTTTTTTTTTT+AATATTTATTAAACACATATTTTA; Tam3 3′ of S-99 sense strand second primer, GGGTTGAATTGGTTGGAGAAATGG+AACACATATACTAACAAACACATATTTTAT; and pBluescript primer, GGGTTGAATTGGTTGGAGAAATGG+AACACATATACTAACAAACACATATTTTAT. PCR reactions were performed as follows: two cycles of 95°C for 30 s, 60°C for 1 min, 72°C for 1 min; two cycles of 95°C for 30 s, 58°C for 1 min, 72°C for 1 min, 40 cycles of 95°C for 30 s, 56°C for 1 min, 72°C for 1 min; and termination at 72°C for 7 min. Amplified PCR products were cloned by pBluescript SK (Stratagene, La Jolla, CA) vector. Individual clones were sequenced by an ABI377 Automated DNA Sequencer (PE-Applied Biosystems, Foster City, CA).
Acknowledgments
We are especially grateful to Dr. Cathie Martin (John Innes Center, Norwich, UK) for valuable comments on the manuscript. We thank Prof. Yoshio Sano and Dr. Tomohiko Kubo (Hokkaido University, Faculty of Agriculture, Japan) for providing a sequencing machine and technical advice, and Dr. Cathie Martin for gifts of the A. majus seeds. Part of this work was done at the Research Center for Molecular Genetics, Hokkaido University.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.017533.
References
- Beale GH, Faberge AC (1941) Effect of temperature on the mutation rate of an unstable gene in Portulaca grandiflora. Nature 147: 356-357 [Google Scholar]
- Bennetzen JL (1987) Covalent DNA modification and the regulation of Mutator element transposition in maize. Mol Gen Genet 208: 45-51 [Google Scholar]
- Brown J, Sundaresan V (1992) Genetic study of the loss and restoration of Mutator transposon activity in maize: evidence against dominant-negative regulator associated with loss of activity. Genetics 130: 889-898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell TP, Dellaporta SL (1994) Somatic inactivation and reactivation of Ac associated with changes in cytosine methylation and transposase expression. Genetics 138: 213-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE (2002) Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA 99; 16491-16498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter R, Martin C, Coen E (1987) Comparison of genetic behaviour of the transposable element Tam3 at two unlinked pigment loci in Antirrhinum majus. Mol Gen Genet 207: 82-89 [Google Scholar]
- Chandler VL, Walbot V (1986) DNA modification of a maize transposable element correlates with loss of activity. Proc Natl Acad Sci USA 83: 1767-1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Mariappan SV, Catasti P, Ratliff R, Moyzis RK, Laayoun A, Smith SS, Bradbury EM, Gupta G (1995) Hairpins are formed by the single DNA strands of the fragile X triplet repeats: structure and biological implications. Proc Natl Acad Sci USA 92: 5199-5203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomet PS, Wessler S, Dellaporta SL (1987) Inactivation of the maize transposable element Activator (Ac) is associated with its DNA modification. EMBO J 6: 295-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Robbins TP, Almeida J, Hudson A, Carpenter R (1989) Consequences and mechanisms of transposition in Antirrhinum majus. In DE Berg, MH Howe, eds, Mobile DNA. American Society for Microbiology, Washington, DC, pp 413-436
- Cubas P, Vincent C, Coen E (1999) An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401: 157-161 [DOI] [PubMed] [Google Scholar]
- Ellis TH, Delseny M, Lee D, Burcham KW (1990) Methylated and under-methylated rDNA repeats are interspersed at random in two higher plant species. Plant Mol Biol 14: 73-80 [DOI] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA 89: 1827-1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T (2000) DNA methyltransferase Dnm1 associates with histone deacetylase activity. Nat Genet 24: 88-91 [DOI] [PubMed] [Google Scholar]
- Godde JS, Kass SU, Hirst MC, Wolffe AP (1996) Nucleosome assembly on methylated CGG triplet repeats in the Fragile X Mental Retardation gene 1 promoter. J Biol Chem 271: 24325-24328 [DOI] [PubMed] [Google Scholar]
- Gorbunova V, Ramos C, Hohn B, Levy AA (2000) A nuclear protein that binds specifically to several maize transposons is not essential for Ds1 excision. Mol Gen Genet 263: 492-497 [DOI] [PubMed] [Google Scholar]
- Harrison BJ, Fincham JRS (1964) Instability at the pal locus in Antirrhinum majus: I. Effects of environment on frequencies of somatic and germinal mutation. Heredity 19: 237-258 [Google Scholar]
- Hehl R, Nacken WK, Krause A, Saedler H, Sommer H (1991) Structural analysis of Tam3, a transposable element from Antirrhinum majus, reveals homologies to the Ac element from maize. Plant Mol Biol 16: 369-371 [DOI] [PubMed] [Google Scholar]
- Kakutani T, Munakata K, Richards EJ, Hirochika H (1999) Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics 15: 831-838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishima Y, Ogura K, Mizukami K, Mikami T, Adachi T (1995) Chloroplast DNA analysis in buckwheat species: phylogenetic relationships, origin of the reproductive systems and extended inverted repeats. Plant Sci 108: 173-179 [Google Scholar]
- Kishima Y, Yamashita S, Martin C, Mikami T (1999) Structural conservation of the transposon Tam3 family in Antirrhinum majus and estimation of the number of copies able to transpose. Plant Mol Biol 39: 299-308 [DOI] [PubMed] [Google Scholar]
- Kitamura K, Hashida S, Mikami T, Kishima Y (2001) Position effect of the excision frequency of the Antirrhinum transposon Tam3: implication for the degree of position-dependent methylation in the ends of the element. Plant Mol Biol 47: 475-490 [DOI] [PubMed] [Google Scholar]
- Kunze R, Starlinger P (1989) The putative transposase of transposable element Ac from Zea mays L. interacts with subterminal sequences of Ac. EMBO J 8: 3177-3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze R, Starlinger P, Schwartz D (1988) DNA methylation of the maize transposable element Ac interferes with its transcription. Mol Gen Genet 214: 325-327 [DOI] [PubMed] [Google Scholar]
- Kunze R, Stochaj U, Laufs J, Starlinger P (1987) Transcription of transposable element Activator (Ac) of Zea mays L. EMBO J 6: 1555-1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Prescott A, Lister C, MacKay S (1989) Activity of the transposon Tam3 in Antirrhinum and tobacco: possible role of DNA methylation. EMBO J 8: 997-1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy D, Louwerse DJ, McElory SM, Lemaux PG (1997) Development of a simple transient assay for Ac/Ds activity in cells of intact barley tissue. Plant J 11: 157-165 [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321-4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki H, Ikeda K, Hashimoto Y, Tosa Y, Mayama S (2001) Methylation is not the main force repressing the retrotransposon MAGGY in Magnaporthe grisea. Nucleic Acids Res 29: 1278-1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393: 386-389 [DOI] [PubMed] [Google Scholar]
- Rhoades M (1941) The genetic control of mutability in maize. Cold Spring Harbor Symp Quant Biol 9: 138-144 [Google Scholar]
- Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP (2000) Dnmt1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet 25: 338-342 [DOI] [PubMed] [Google Scholar]
- Ros F, Kunze R (2001) Regulation of activator/dissociation transposition by replication and DNA methylation. Genetics 157: 1723-1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sand SA (1957) Phenotypic variability and the influence of temperature on somatic instability in cultures derived between Nicotiana langsdorffii and N. sanderae. Genetics 42: 685-703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano Y, Sano R (1990) Variation of the intergenic spacer region of ribosomal DNA in cultivated and wild rice species. Genome 33: 209-218 [Google Scholar]
- Schlappi M, Raina R, Fedoroff N (1994) Epigenetic regulation of the maize Spm transposable element: novel activation of a methylated promoter by TnpA. Cell 77: 427-437 [DOI] [PubMed] [Google Scholar]
- Schwartz D, Dennis E (1986) Transposase activity of the Ac controlling element in maize is regulated by its degree of methylation. Mol Gen Genet 205: 476-482 [Google Scholar]
- Wolffe AP, Jones PL, Wade PA (1999) DNA demethylation. Proc Natl Acad Sci USA 96: 5894-5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S, Mikami T, Kishima Y (1998) Tam3 in Antirrhinum majus is exceptional transposon in resistant to alteration by abortive gap repair: identification of nested transposons. Mol Gen Genet 259: 468-474 [DOI] [PubMed] [Google Scholar]
- Yamashita S, Shimizu-Takano T, Kitamura K, Mikami T, Kishima Y (1999) Resistance to gap repair of the transposon Tam3 in Antirrhinum majus: a role of the end regions. Genetics 153: 1899-1908 [DOI] [PMC free article] [PubMed] [Google Scholar]