Abstract
We identified a dwarf transgenic hybrid poplar (Populus tremula × Populus alba) after screening of 627 independent activation-tagged transgenic lines in tissue culture, greenhouse, and field environments. The cause of the phenotype was a hyperactivated gene encoding GA 2-oxidase (GA2ox), the major gibberellin (GA) catabolic enzyme in plants. The mutation resulted from insertion of a strong transcriptional enhancer near the transcription start site. Overexpression of the poplar GA2ox gene (PtaGA2ox1) caused hyperaccumulation of mRNA transcripts, quantitative shifts in the spectrum of GAs, and similarity in phenotype to transgenic poplars that overexpress a bean (Phaseolus coccineus) GA2ox gene. The poplar PtaGA2ox1 sequence was most closely related to PsGA2ox2 from pea (Pisum sativum) and two poorly known GA2oxs from Arabidopsis (AtGA2ox4 and AtGA2ox5). The dwarf phenotype was reversible through gibberellic acid application to the shoot apex. Transgenic approaches to producing semidwarf trees for use in arboriculture, horticulture, and forestry could have significant economic and environmental benefits, including altered fiber and fruit production, greater ease of management, and reduced risk of spread in wild populations.
Dwarf or semidwarf varieties are widely used in orchard and cereal crops because of the diverse management and yield benefits they provide. Major genes for dwarfism were key enabling technologies for the “green revolution” (David and Otsuka, 1994; Perkins, 1997), where they allowed crops to produce high yields while avoiding lodging (Perkins, 1997). In fruit trees, dwarf and semidwarf varieties are often preferred (Webster, 2002), and breeding efforts have resulted in selection of dwarf scions or dwarfing rootstock varieties in almost all of the main temperate and tropical fruit species (Janick et al., 1996). These cultivars allow dense field cultivation, facilitate mechanized maintenance, increase efficiency of fruit collection, and allow more precise pesticide application, reducing spray drift (Webster, 2002). Most of the dwarfing rootstocks also induce precocious and profuse flowering (Atkinson and Else, 2001).
To our knowledge, semidwarf varieties have not been employed in forestry; however, they may also provide significant advantages (Bradshaw and Strauss, 2001). Stands may have higher biomass productivity due to reduced investment in root mass by shorter trees and lower moisture stress and respiratory surface area of the bole. They may also produce higher quality wood due to reduced bending and leaning (reaction wood) and be less prone to wind-fall. Dwarfing genes might be useful in urban forestry where they could help in producing tree varieties that do less damage to power lines, are less prone to being blown down onto structures, and impose on neighboring homes and yards to lesser degrees.
Because natural selection will act to remove dominant alleles that result in short tree stature in the face of competition for light, healthy dwarf genotypes that are expected to be very rare and, thus, difficult to obtain through classical tree breeding without sacrificing other breeding goals. Therefore, insertion of dominant transgenes may be an important alternative method for obtaining dwarf phenotypes in many genotypes and species. Such transgenes, by strongly reducing tree fitness, will also greatly reduce the propensity of tree progeny to spread in wild and feral populations. This could be very useful for mitigating invasive tendencies of exotic tree species, which can sometimes cause major ecological disruptions (Richardson, 1998), and to greatly reduce the probability of introgression by linked transgenes (Gressel, 1999).
Research in the last several years has demonstrated that dwarfism is commonly associated with deficiencies in GA levels or signaling (Peng et al., 1999; Spielmeyer et al., 2002). GAs are a complex family of tetracyclic diterpenoid plant hormones that play critical roles in plant growth and development (Davies, 1995). The level of bioactive GAs is precisely controlled by several mechanisms, including transcriptional regulation of the genes encoding enzymes from both biosynthetic and catabolic pathways (Olszewski et al., 2002). By modifying regulation of genes controlling GA flux, it is possible to modify processes regulated by GA and, thus, plant form (Hedden and Phillips, 2000b). GA biosynthetic genes have received much more study than catabolic genes, but recently identification and functional analysis of the genes encoding the major catabolic enzyme, GA 2-oxidase (GA2ox), have revealed that the catabolic genes are also important in control of GA levels (Reid et al., 1992; Ross et al., 1995; Thomas et al., 1999; Sakamoto et al., 2001). GA2oxs are encoded by small gene families in Arabidopsis and pea (Pisum sativum; Hedden and Phillips, 2000a; Elliott et al., 2001; Hedden et al., 2001; Schomburg et al., 2002), but only one GA2ox gene has been identified in rice (Oryza sativa; Sakamoto et al., 2001).
Little is known about the developmental role of GA2ox genes in plants. Overexpression of the rice GA2ox gene (OsGA2ox1) causes a dwarf phenotype and delay in reproductive development (Sakamoto et al., 2001). Loss-of-function mutation in a pea PsGA2ox1 results in the hyperelongated slender phenotype (Martin et al., 1999). Similarly, in Arabidopsis, overexpression of two GA2ox genes (AtGA2ox7 and AtGA2ox8) causes dwarfing and delayed flowering under noninductive short days (Schomburg et al., 2002), and loss-of-function mutations of the same genes led to increased hypocotyl elongation and higher levels of active GAs. RNA in situ localizations of OsGA2ox1 in rice revealed a ring-like expression pattern in the basal region of leaf primordia, near the vegetative shoot apical meristem (Sakamoto et al., 2001). Very low mRNA levels were detected in the inflorescence meristem. This expression pattern suggests that OsGA2ox1 may prevent GA flux into the shoot apical meristem, but OsGA2ox1 is not expressed during flower development when high GA levels are needed. No GA2ox genes have yet been cloned from a tree, and their developmental function and regulation during woody plant development is completely unknown.
Here, we demonstrate that hyperactivation of a poplar (Populus tremula × Populus alba) gene encoding a GA catabolic enzyme GA2ox has dramatic effects on tree form, suggesting that this gene and related family members could provide major new tools for research and genetic engineering of tree stature.
RESULTS
Mutant Isolation and Characterization
From the 627 independent activation-tagged poplar lines, nine (>1%) exhibited an obvious morphological phenotype that had never been seen among the thousands of transgenic poplars produced in our laboratory. One of these nine lines displayed extremely short internodes and dark-green leaves with a stiff, leathery texture (Fig. 1). The mutant, which we call stumpy for its short, stout form, was approximately 4-fold shorter than WT but had a similar number of internodes (Table I). Stem diameter at the top of the plant was proportionally larger in the mutant, indicating that these plants had less stem taper than WT. Branch number and length were also substantially reduced in the mutant.
Figure 1.
Wild-type (WT) and mutant (stumpy) plants at the same age and grown under the same conditions. Stumpy is on the left-hand side of each frame; WT is on the right. A, Tree size after 14 months of growth in a greenhouse. B, Short internodes of stumpy. C, Dark foliage of stumpy.
Table I.
Morphological characterization of WT and stumpya
| Trait | WT | Stumpy |
|---|---|---|
| Leaf length (mm) | 9.80 (0.26) | 6.61 (0.11)* |
| Diameter (base; mm) | 17.82 (1.41) | 13.20 (0.69), NS |
| Diameter (one-half; mm) | 9.15 (1.15) | 8.27 (0.44), NS |
| Diameter (top; mm) | 1.49 (0.01) | 3.39 (0.26)* |
| Internode no. | 133.50 (0.87) | 128.33 (2.60), NS |
| Height (cm) | 326.39 (2.21) | 89.54 (2.26)**** |
| Branch length (cm) | 46.36 (1.50) | 3.18 (0.31)*** |
| Branch no. | 12.00 (0.47) | 5.00 (0.47)*** |
Measurements were taken from three WT and stumpy plants grown for 20 months in the same location in a greenhouse. Stem diameters were measured at three positions: stem base (base), one-half of total stem height (one-half), and at the second internode from the top (top). The length of the longest branch was measured. SES are in parentheses. Statistically significant differences are indicated at the 5% (*), 0.001(***), and 0.0001(****) probability levels. NS, Nonsignificant (>5%; unpaired Student's t test, unequal variances).
Plasmid Rescue of Genomic Sequence Flanking Left Borders and Right Borders
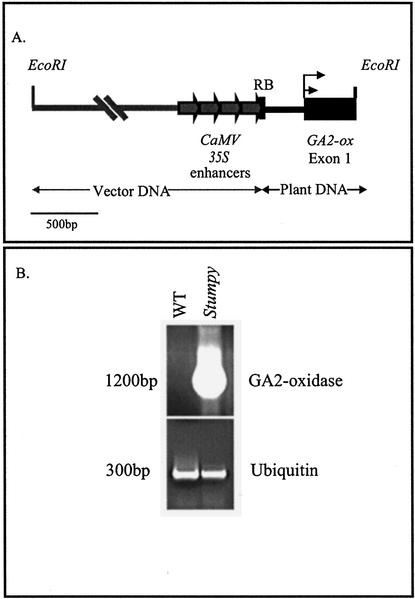
Restriction mapping of the plasmids rescued from the left- and right-hand borders in approximately 30 clones suggested that only a single transgene insert was present. Homology searches with the sequences recovered from the right border showed a high level of similarity to GA2oxs from pea and other species (Lester et al., 1999; Martin et al., 1999; Thomas et al., 1999). Therefore, we fully sequenced this clone (Fig. 2A). The translation initiation codon of the putative poplar GA2ox gene (PtaGA2ox1) appears to be 333 bp downstream of the four 35S enhancers; the recovered sequence spans 371 bp and is not interrupted by introns.
Figure 2.
Map of the EcoRI-rescued activation tagging plasmid from stumpy (A) and PtaGA2ox1 accumulation in stumpy and WT poplars (B).
Expression of GA 2-oxidase Is Markedly Elevated in the Mutant
We used reverse transcription (RT)-PCR and primers designed to start at the translation initiation codon to assess the expression of the gene in rapidly elongating shoots and leaves from WT and mutant plants (Fig. 2B). We could not visually detect PtaGA2ox1 transcript in the WT plant but observed very strong expression in the mutant. In contrast, a poplar ubiquitin gene showed approximately equal expression levels in the mutant and WT plants.
Isolation of a Putative GA 2-oxidase cDNA
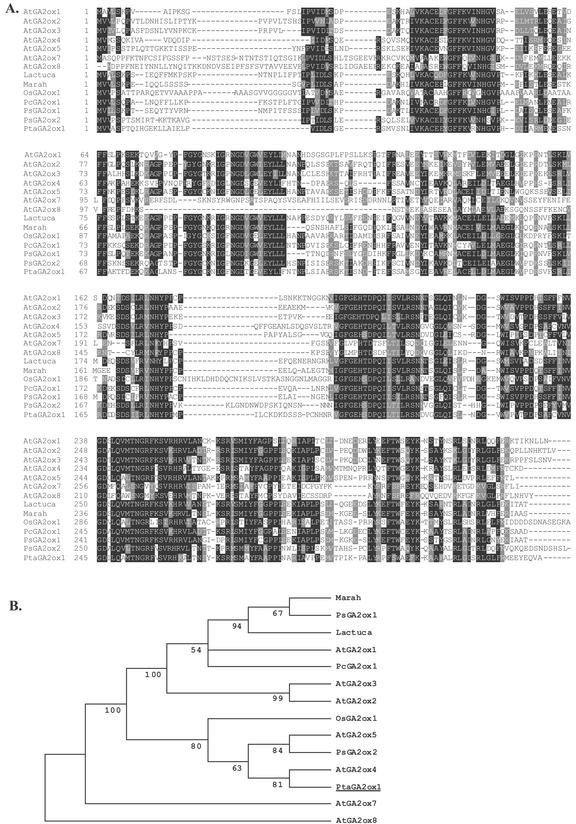
We amplified and fully sequenced a cDNA fragment from the mutant. Alignment of the cDNA with the 371-bp genomic sequence showed perfect correspondence (data not shown). The cloned cDNA fragment is 1,246 bp [excluding the poly(A+) tail] and encodes an open reading frame of 335 amino acids. Homology searches with the derived amino acid sequence identified strong homology to GA2oxs from pea (Lester et al., 1999), Arabidopsis (Thomas et al., 1999), rice (Sakamoto et al., 2001), bean (Phaseolus coccineus; Thomas et al., 1999), lettuce (Lactuca sativa), and wild cucumber (Marah macrocarpus). Amino acid alignment revealed high conservation across these species (Fig. 3A), and phylogenetic analysis based on this alignment showed that the poplar sequence is most closely related to PsGA2ox2 from pea and two poorly known GA2oxs from Arabidopsis (AtGA2ox4 and AtGA2ox5; Fig. 3B).
Figure 3.
Alignment (A) and phylogenetic tree (B) of putative poplar PtaGA2ox1 with GA2oxs from Arabidopsis, rice, pea, bean, wild cucumber, and lettuce. Sequences were aligned using ClustalW (http://www.ebi.ac.uk/clustalw/), and the output was produced using BoxShade (http://www.ch.embnet.org/software/BOX_form.html). Black shading indicates identity, and gray shading indicates similarity, of amino acid residues. Phylogenetic analysis was performed using MEGA2 software (http://www.megasoftware.net/). Sites containing alignment gaps were excluded from further analysis, and the distance between sequences represent the proportion of amino acid sites at which the two sequences compared are different. The unrooted tree was constructed using the neighbor-joining method. The bootstrap percentage indicated at each joint point was created from 1,000 data samples. Nodes with less than 50% bootstrap confidence were collapsed.
Dwarfism Is Reversed by GA3 Application
GA3 is a bioactive form of GA but cannot be metabolized by GA2ox (Sakamoto et al., 2001). Thus, if the mutant has low bioactive GA levels due to a high rate of inactivation by GA2ox, application of GA3 should be able to rescue the mutant phenotype. After only one application of 3 mm GA3, we observed reversion of the mutant phenotype (Fig. 4). Not only was normal stem growth restored, but the leaves returned to normal color, size, and texture. After GA3 application was discontinued, the plants reverted to the mutant phenotype (data not shown).
Figure 4.
Reversion of mutant phenotype in greenhouse-grown plants by application of GA3. The photograph represents a typical response observed in five experimental plants in each of the GA and control treatments.
Altered GA Content in the Mutant
We quantified some GAs in leaves that might be affected by ectopic expression of the putative PtaGA2ox1 (Table II). Consistent with our expectations, the main bioactive GAs (GA1 and GA4) were substantially reduced in mutant compared with WT plants (Table II). GA8 and GA34, which are the inactive, C-2 hydroxylated catabolites of GA1 and GA4, respectively, were 6.1- and 4.5-fold higher in the mutant plants than in WT. An increase in GA29 levels, which is the C-2 hydroxylated catabolite of GA20 (the immediate precursor of GA1), was also detected. The levels of GA20, however, were nearly unaffected.
Table II.
GA content in leaves (blades and petioles) of stumpy and WT plantsa
| GA | WT | Stumpy | Ratio |
|---|---|---|---|
| ng g dry wt-1 | |||
| GA1 | 3.0 (0.4) | 0.8 (0.1) | 0.3 |
| GA4 | 8.2 (1.4) | 1.4 (0.2) | 0.2 |
| GA20 | 6.4 (1.4) | 6.7 (1.2) | 1.0 |
| GA8 | 10.5 (1.4) | 64.4 (6.8) | 6.1 |
| GA34 | 6.0 (1.3) | 26.9 (5.2) | 4.5 |
| GA29 | 7.6 (1.2) | 13.5(1.5) | 1.8 |
GAs above the dashed line were diminished, and those below the line elevated, in the mutant. Results are expressed as means of three independent measurements, with SES in parentheses. All differences are significant at the 5% probability level (unpaired Student's t test, unequal variances), except for GA20.
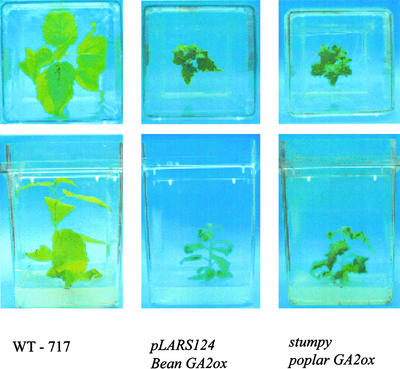
Overexpression of a Bean GA2ox Gene Causes a Similar Phenotype
We transformed a 35S promoter fused to bean PcGA2ox1 cDNA into the same poplar genotype as had been used for activation tagging. Bean PcGA2ox1 is highly similar to poplar PtaGA2ox1, although it is as yet unclear if it is the true poplar ortholog (Table III; Fig. 3B). Approximately 10% of the recovered transgenic lines displayed a mutant phenotype— dark-green leaves and much reduced stem elongation—similar to that in mutant plants (Fig. 5).
Table III.
Comparison of poplar GA2ox (PtaGA2ox1) with GA2oxs from six other plant species. Values indicate percentage identity (above the diagonal) and similarity (below the diagonal) and were obtained by using GENEDOC from the sequence alignment shown in Figure 3A. See “Materials and Methods” for accession nos.
| Enzyme | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. AtGA2ox1 | - | 52 | 48 | 40 | 40 | 19 | 19 | 54 | 58 | 33 | 53 | 53 | 41 | 40 |
| 2. AtGA2ox2 | 71 | - | 68 | 37 | 40 | 20 | 19 | 54 | 53 | 33 | 59 | 51 | 42 | 39 |
| 3. AtGA2ox3 | 67 | 84 | - | 40 | 40 | 19 | 21 | 49 | 50 | 33 | 53 | 49 | 42 | 40 |
| 4. AtGA2ox4 | 56 | 53 | 54 | - | 48 | 20 | 20 | 40 | 44 | 40 | 40 | 41 | 47 | 55 |
| 5. AtGA2ox5 | 55 | 59 | 57 | 59 | - | 20 | 18 | 40 | 45 | 41 | 40 | 41 | 54 | 51 |
| 6. AtGA2ox7 | 36 | 38 | 38 | 38 | 39 | - | 35 | 21 | 20 | 18 | 19 | 19 | 21 | 21 |
| 7. AtGA2ox8 | 34 | 37 | 34 | 34 | 31 | 54 | - | 17 | 19 | 16 | 17 | 19 | 18 | 23 |
| 8. Lettuce | 70 | 73 | 70 | 56 | 58 | 42 | 36 | - | 66 | 34 | 61 | 64 | 41 | 42 |
| 9. Wild cucumber | 73 | 71 | 66 | 57 | 59 | 40 | 34 | 81 | - | 34 | 62 | 68 | 45 | 45 |
| 10. OsGA2ox1 | 49 | 49 | 48 | 53 | 57 | 35 | 29 | 53 | 53 | - | 34 | 34 | 44 | 40 |
| 11. PcGA2ox1 | 71 | 73 | 68 | 54 | 57 | 40 | 36 | 77 | 76 | 49 | - | 57 | 44 | 44 |
| 12. PsGA2ox1 | 69 | 72 | 66 | 53 | 58 | 40 | 34 | 79 | 82 | 51 | 76 | - | 41 | 42 |
| 13. PsGA2ox2 | 58 | 59 | 59 | 61 | 68 | 36 | 33 | 59 | 60 | 60 | 60 | 57 | - | 55 |
| 14. PtaGA2ox1 | 57 | 58 | 58 | 69 | 65 | 41 | 35 | 60 | 64 | 60 | 61 | 60 | 73 | - |
Figure 5.
Overexpression of bean GA 2-oxidase (PcGA2ox1, accession no. AJ132438) causes stumpy-like phenotype. Photographs represent plants grown for 1 month in tissue culture under the same conditions. Lower and upper tiers represent the same plants photographed from above and sideways.
DISCUSSION
The vegetative characteristics of the stumpy mutant are similar to the phenotype of GA-deficient Arabidopsis mutants that contain defective GA biosynthetic genes. They share severely reduced stem elongation, decreased leaf size, and dark-green foliage color (Sun and Kamiya, 1994; Helliwell et al., 1998; Yamaguchi et al., 1998). Similar phenotypes were also observed in bioactive GA-deficient rice overexpressing OsGA2ox1, its main GA inactivation enzyme (Sakamoto et al., 2001). Consistent with the expectation that the mutant phenotype was caused by perturbation of the GA metabolic pathway, we recovered a nearby genomic sequence and then isolated a corresponding cDNA copy of the gene that showed high homology to GA2ox genes from a number of plant species. The putative poplar PtaGA2ox1 is phylogenetically closest to pea PsGA2ox2 (Lester et al., 1999) and two as yet uncharacterized Arabidopsis GA2oxs (AtGA2ox4 and AtGA2ox5; Hedden and Phillips, 2000a). Three other GA2-oxidase genes from Arabidopsis have been identified and functionally characterized (Thomas et al., 1999), but the presence of up to six has been suggested (Hedden and Phillips, 2000a), and recently, two novel GA2ox genes have been identified (Schomburg et al., 2002). Several enzymes from the final stage of GA metabolism, including GA2ox, are encoded by small gene families in a number of plant species and show tissue and environment-specific expression (Yamaguchi and Kamiya, 2000). Thus, future characterization of the poplar GA2ox gene family, and its regulatory mechanisms and functional roles, should enable more specific manipulation of GA-related traits in trees.
Control of flux in biosynthetic pathways is usually distributed between several steps, and changes in the level of any one enzyme may not influence the overall level of the end-product (Hedden and Phillips, 2000a). For example, the overproduction of entcopalyl diphosphate synthase did not result in altered GA levels, and transgenic plants were indistinguishable from WT (Sun and Kamiya, 1994). In contrast, manipulation of the level of enzymes from the later cytoplasmic stages of the biosynthesis and the catabolic branch resulted in dramatic shifts in GA levels and evoked multiple phenotypic alterations in mutant and transgenic plants (Coles et al., 1999; Eriksson et al., 2000; Carrera et al., 2000; Niki et al., 2001). Thus, from a biotechnological perspective, these enzymes appear to provide powerful tools for manipulating GA levels. Our observations with the poplar mutant, which appears to result from hyperactivation of PtaGA2ox1, supports this view.
Analysis of the GA content in the mutant indicates severalfold decreases of the bioactive GAs (GA1 and GA4) and severalfold increases of their main C-hydroxylated inactive catabolites (GA8 and GA34). We also detected a nearly 2 (1.8)-fold increase of GA29, the catabolite of GA20 (the main GA1 precursor). Thus, our data are consistent with the expected function and biochemical activity of GA2ox (Ross et al., 1995). GA2ox may also be a natural regulator of poplar stature because GA8 is better correlated than other GAs with shoot elongation of saplings grown in different environments (Rood et al., 2002). Interestingly, in our present study, the level of GA20 was least affected and was approximately equal in the mutant and WT plants. Similarly, GA20 concentration was almost unaffected in transgenic rice plants overexpressing the rice OsGA2ox1 (Sakamoto et al., 2001). This may be the result of feedback regulation increasing transcriptional activity of biosynthetic gene(s) triggered by the low levels of GA1/4 in the mutant plant. Many genes encoding enzymes from the final steps of GA metabolism are subject to feedback regulation (Phillips et al., 1995; Chiang et al., 1995; Toyomasu et al., 1997; Cowling et al., 1998). Thus, we anticipate that the low bioactive GA1/4 levels may have caused up-regulation of a GA 20-oxidase gene, compensating for the increased catabolism of GA20 by the overexpressed PtaGA2ox1. Nevertheless, this putative compensatory mechanism obviously did not make up for the high turnover of the bioactive forms that resulted from the hyperactivated GA2ox gene in the mutant.
Manipulation of plant stature has long been a major goal in agriculture, horticulture, and silviculture. It previously has involved classical plant breeding and use of plant growth regulators produced by the chemical industry. These are exogenously applied to promote or retard elongation, often through chemical alteration of GA biosynthesis (Rademacher, 2002). However, stature control through “anti-GA” plant growth retardants requires repeated application of synthetic chemicals that is costly, variable in effectiveness, and can have undesired environmental consequences or public perceptions. Biotechnological manipulation of GA levels provides an alternative approach and can be achieved through various means, including up- or down-regulating genes encoding enzymes involved in GA biosynthesis and catabolism (Hedden and Phillips, 2000b).
Exogenous application of GA3, which is resistant to catabolism by GA2ox, rapidly restored normal development to the mutant, strongly supporting the hypothesis that the mutant phenotype is a result of the deficiency of the bioactive GAs, GA1 and GA4. The rapid reversion to normal growth by exogenous application of GA that is resistant to the action of the enzyme also provides a potential method for control of transgenic plants overexpressing GA2ox genes during horticultural manipulation. For example, the rate of growth during commercial propagation could be greatly increased by GA application, allowing rapid nursery production. Once GA application ceases after transplanting, the slow growth and dwarf form would resume. Landscape managers might also choose to speed early growth via GA application, thereby allowing growth rate to attenuate only after plants reach a desired size.
Insertional mutagenesis using transposons or T-DNA has become an extremely valuable research tool for model plant systems (Hayashi et al., 1992; Martienssen, 1998; Krysan et al., 1999), but its utility for isolation of economically valuable genes in crop plants—for which transformation is often difficult— has been extremely limited (Brutnell, 2002; Zubko et al., 2002). For trees, poplars provide a marked exception. Due to its ease of transformation, small genome, rapid growth, amenability to clonal propagation, and extensive genomic resources (including the upcoming complete genome sequence), poplar is the consensus model organism for molecular tree biology (Wullschleger et al., 2002). Its amenability to transformation allowed us to employ activation tagging to produce a population of transgenic trees with dominantly inherited, tagged phenotypes. Because of the many large differences in development between annuals and perennial plants, including vegetative dormancy, delayed onset of flowering, extended periods of secondary (woody) growth, and gradual vegetative maturation, forward genetic approaches may uncover many types of regulatory genes that would be missed in reverse genetic experiments.
Employing the same activation tagging vector that has been extensively used in Arabidopsis (Kardailsky et al., 1999) and other species (Zubko et al., 2002), we observed a similar rate of morphological mutant recovery (approximately 1%). Because the poplar genome is approximately 4-fold larger than Arabidopsis (Bradshaw et al., 2001), this supports the observations that T-DNA insertion is strongly directed toward expressed sectors of the genome (Szabados et al., 2002) and suggests that forward genetic approaches may be useful in plant species with a wide variety of genome sizes—so long as at least one highly transformable genotype can be identified. We believe that isolation of the gene causing the stumpy phenotype is the first case, to our knowledge, of successful tagging of a developmental regulatory gene in any tree species.
MATERIALS AND METHODS
Plant Transformation, Tissue Culture, and Growth Conditions
We transformed activation tagging vectors pSKI015 and pSKI074 (Weigel et al., 2000) into hybrid aspen clone INRA 717-IB4 (Populus tremula × Populus alba) via Agrobacterium tumefaciens mediated transformation procedure (Han et al., 2000) to generate a population of activation-tagged lines. After regeneration on selection media, all lines were verified via PCR for presence of the 35S enhancer-tetramer and were transferred to greenhouse and 1 year later to a field plantation. Each line was represented by at least four ramets in all experiments.
To transform the bean (Phaseolus vulgaris) GA2ox gene (PcGA2ox1, accession no. AJ132438) into poplar, we used the binary vector pLARS124, constructed and kindly provided by Dr. Peter Hedden (Institute of Arable Crop Research Long Ashton Research Station, University of Bristol, Long Ashton, UK). pLARS124 was obtained by substituting the GUS gene in pGPTV-Kan (Becker et al., 1992) with three copies of 35S and inserting the PcGA2ox1 cDNA immediately downstream of the 35S promoters and upstream of the nos terminator. pLARS124 was transformed into the same poplar genotype using the transformation procedure cited above.
DNA and RNA Extraction
DNA was extracted from approximately 0.5 g of expanding shoots including the subtending leaves using the DNeasy Plant Maxi Kit (Qiagen, Valencia, CA). RNA was extracted from approximately 0.2 g of the same tissues using a modified Qiagen RNeasy Mini kit protocol. Tissue was ground with a mortar and pestle to a fine powder in liquid nitrogen, lysis buffer was added, and the slurry was homogenized using a polytron. A 0.4 volume of 5 m K-acetate was added to the homogenate and incubated on ice for 15 min. The extracts were spun for 15 min at 4°C at top speed in a tabletop centrifuge. A one-half volume of 100% (w/v) ethanol was added to the supernatant, and the mix was applied to the RNeasy mini column. We followed the remaining kit procedures precisely. RNA and DNA concentrations were measured using a DU 640 spectrophotometer (Beckman Coulter, Fullerton, CA), and Mr was checked on 1% (w/v) agarose Trisacetate EDTA ethidium bromide gels.
Plasmid Rescue
Plasmid rescue was conducted essentially as previously described (Weigel et al., 2000). Ligation of EcoRI- and SpeI-digested and purified genomic DNA was performed in a 250-μL volume overnight at 16°C, using 25 units of T4 DNA ligase (Invitrogen, Carlsbad, CA). The purified ligation reactions (5 μL) were transformed into SURE electrocompetent cells (Stratagene, La Jolla, CA) by electroporation using TransPorator Plus (BTX, San Diego) set at 1.7kV. Ampicillin-resistant colonies were used to inoculate overnight bacterial cultures, and plasmid DNA from approximately 30 minipreps from each ligation was digested using KpnI and SpeI (Invitrogen).
Sequence Analysis
Sequencing was performed in the Central Services Laboratory (Oregon State University Center for Gene Research and Biotechnology, Corvallis) using capillary 3100 Genetic Analyzers (Applied Biosystems, Foster City, CA) and ABI Prism BigDye Terminator Cycle Sequencing v2.0 Ready Reaction with AmpliTaq DNA Polymerase (Applied Biosystems). The EcoRI-rescued plasmid was initially sequenced using primer pSK0015E1 (5′ATGGATAAATAGCCTTGCTTCC-3′). The poplar PtaGA2ox1 cDNA was sequenced in both directions. Sequence homology searches and sequence analyses were performed using the National Center for Biotechnology Information BLAST server and the University of Wisconsin Genetics Computer Group (Madison, WI) software package (Devereux et al., 1984). Sequence alignments were carried out by the ClustalW method (Thompson et al., 1994) and using the EMBL server (http://www.ebi.ac.uk/clustalw/).
RT-PCR and cDNA Isolation
RT-PCR was performed on 3.0 μg of total RNA using a Gene Racer Kit (Invitrogen), the GeneRacer 3′ primer, and the GA2oxF1 primer (5′-ATGGTAGTGGCATCCCCAACTC-3′). The amplified fragment was gel purified using the QIAquick Spin kit (Qiagen) and cloned into pCR-TOPO using the TOPO TA Cloning kit (Invitrogen). Ubiquitin cDNA fragment was PCR amplified using the same RT reaction and the following primers: forward, 5′-CTCAAAGTGAAAGGCCAGGATG-3′; and reverse, 5′-ACTGTCAAAGCTCTTGGTGAG-3′.
GA3 Application
Ten microliters of 3 mm aqueous solution of GA3 (Sigma, St. Louis) was applied to the shoot apex at 4-d intervals for 2 weeks.
Quantification of GAs
Approximately 2.0 to 3.0 g of fresh leaf and stem tissue from field- and greenhouse-grown poplar plants was collected, immediately frozen in liquid nitrogen, and subsequently lyophilized. Extraction, purification, and analysis of GA content were performed as described by Pearce et al. (2002) with [2H2]GA internal standards and quantification by gas chromatography-mass spectrometry with selected ion monitoring. All measurements were repeated three times in independent extractions from three different ramets of the control and mutant genotypes.
Accession Numbers
The accession numbers of the sequences used in this study are as follows: AtGA2ox1 (AJ132435), AtGA2ox2 (AJ132436), AtGA2ox3 (AJ132437), AtGA2ox4 (AC051631), AtGA2ox5 (AC064879), AtGA2ox7 (AC079284), AtGA2ox8 (AL021960), lettuce (Lactuca sativa; AB031206), wild cucumber (Marah macrocarpus; Y09113), OsGA2ox1 (AB059416), PcGA2ox1 (AJ132438), PsGA2ox1 (AF056935), and PsGA2ox2 (AF100954).
Acknowledgments
We thank Dr. Peter Hedden for providing the binary vector pLARS124, Dr. Detlef Weigel for providing the activation tagging vectors pSKI015 and pSKI074, and Jace Carson for the technical assistance in sample collection and preparation of the manuscript.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.020354.
This work was sponsored in part by the Tree Genetic Engineering Research Cooperative, by the National Science Foundation Industry/University Cooperative Research Centers Program (grant no. 9980423–EEC), by the Consortium for Plant Biotechnology Research, Inc. (Department of Energy Prime Agreement no. DEFG36–02GO12026), by the U.S. Department of Energy's Biomass Program (contract no. 4000014546 with Oak Ridge National Laboratory; Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. Department of Energy under contract no. DE–AC05–00OR22725), and by the Natural Sciences and Engineering Research Council of Canada (Discovery Grant to S.B.R).
References
- Atkinson C, Else M (2001) Understanding how rootstocks dwarf fruit trees. Compact Fruit Tree 34: 46–49 [Google Scholar]
- Becker D, Kemper E, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20: 1195–1197 [DOI] [PubMed] [Google Scholar]
- Bradshaw HD, Ceulemans R, Davis J, Stettler R (2001) Emerging model systems in plant biology: poplar (Populus) as a model forest tree. J Plant Growth Regul 19: 306–313 [Google Scholar]
- Bradshaw HD, Strauss SH (2001) Breeding strategies for the 21st century: domestication of poplar. In DI Dickman, JG Isebrands, JE Eckenwalder, J Richardson, eds, Poplar Culture in North America, Part B. NRC Research Press, Ottawa, pp 383–394
- Brutnell TP (2002) Transposon tagging in maize. Funct Integr Genomics 2: 4–12 [DOI] [PubMed] [Google Scholar]
- Carrera E, Bou J, Garcia-Martinez JL, Prat S (2000) Changes in GA 20-oxidase gene expression strongly affect stem length, tuber induction and tuber yield of potato plants. Plant J 22: 247–256 [DOI] [PubMed] [Google Scholar]
- Chiang HH, Hwang I, Goodman HM (1995) Isolation of the Arabidopsis GA4 locus. Plant Cell 7: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles JP, Phillips AL, Croker SJ, Garcia-Lepe R, Lewis MJ, Hedden P (1999) Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J 17: 547–556 [DOI] [PubMed] [Google Scholar]
- Cowling RJ, Kamiya Y, Seto H, Harberd NP (1998) Gibberellin dose-response regulation of GA4 gene transcript levels in Arabidopsis. Plant Physiol 117: 1195–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CC, Otsuka K (1994) Modern rice technology and income distribution in Asia. Lynne Reinner, Boulder, CO, pp 3–17
- Davies PJ (1995) Plant hormones: physiology, biochemistry, and molecular biology. Kluwer Academic Publishers, London, pp 6–7
- Devereux J, Haeberli P, Smithies O (1984) A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res 12: 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Ross JJ, Smith JL, Lester DR, Reid JB (2001) Feed-forward regulation of gibberellin deactivation in pea. J Plant Growth Regul 20: 87–94 [Google Scholar]
- Eriksson ME, Israelsson M, Olsson O, Moritz T (2000) Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat Biotechnol 18: 784–788 [DOI] [PubMed] [Google Scholar]
- Gressel J (1999) Tandem constructs: preventing the rise of superweeds. Trends Biotechnol 17: 361–366 [DOI] [PubMed] [Google Scholar]
- Han KH, Meilan R, Ma C, Strauss SH (2000) An Agrobacterium tumefaciens transformation protocol effective on a variety of cottonwood hybrids (genus Populus). Plant Cell Rep 19: 315–320 [DOI] [PubMed] [Google Scholar]
- Hayashi H, Czaja I, Schell J, Walden R (1992) Activation of a plant gene implicated in auxin signal transduction by T-DNA tagging. Science 258: 1350–1353 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL (2000a) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL (2000b) Manipulation of hormone biosynthetic genes in transgenic plants. Curr Opin Biotechnol 11: 130–137 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL, Rojas MC, Carrera E, Tudzynski B (2001) Gibberellin biosynthesis in plants and fungi: a case of convergent evolution? J Plant Growth Regul 20: 319–331 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Sheldon CC, Olive MR, Walker AR, Zeevaart JAD, Peacock WJ, Dennis ES (1998) Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc Natl Acad Sci USA 95: 9019–9024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janick J, Cummins JV, Brown SK, Hemmat M (1996) Apples. In J Janick, JN Moore, eds, Fruit Breeding. John Wiley & Sons, Inc., New York, pp 1–78
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Smith JJ, Elliott RC, Reid JB (1999) Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J 19: 65–73 [DOI] [PubMed] [Google Scholar]
- Martienssen R (1998) Functional genomics: probing plant gene function and expression with transposons. Proc Natl Acad Sci USA 95: 2021–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Hedden P (1999) The SLENDER gene of pea encodes a gibberellin 2-oxidase. Plant Physiol 121: 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki T, Nishijima T, Nakayama M, Hisamatsu T, Oyama-Okubo N, Yamazaki H, Hedden P, Lange T, Mander LN, Koshioka M (2001) Production of dwarf lettuce by overexpressing a pumpkin gibberellin 20-oxidase gene. Plant Physiol 126: 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14: S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce DW, Hutt OE, Rood SB, Mander LN (2002) Gibberellins in shoots and developing capsules of Populus species. Phytochemistry 59: 679–687 [DOI] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F et al. (1999) “Green revolution” genes encode mutant gibberellin response modulators. Nature 400: 256–261 [DOI] [PubMed] [Google Scholar]
- Perkins JH (1997) Geopolitics and the green revolution: wheat, genes, and the cold war. Oxford University Press, New York, pp 210–256
- Phillips AL, Ward DA, Uknes S, Appleford NE, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P (1995) Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol 108: 1049–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher W (2002) Growth retardants: effects on gibberellin biosynthesis and other metabolic pathways. Annu Rev Plant Physiol Plant Mol Biol 51: 501–531 [DOI] [PubMed] [Google Scholar]
- Reid JB, Ross JJ, Swain SM (1992) Internode length in Pisum: a new, slender mutant with elevated levels of C19 gibberellins. Planta 188: 462–467 [DOI] [PubMed] [Google Scholar]
- Richardson DM (1998) Forestry trees as invasive aliens. Cons Biol 12: 18–26 [Google Scholar]
- Rood SB, Zanewich K, Stefura C, Mahoney JM (2002) Influence of water table decline on growth allocation and endogenous gibberellins in black cottonwood. Tree Physiol 20: 831–836 [DOI] [PubMed] [Google Scholar]
- Ross JJ, Reid JB, Swain SM, Hasan O, Poole AT, Hedden P, Willis CL (1995) Genetic regulation of gibberellin deactivation in Pisum. Plant J 17: 241–250 [Google Scholar]
- Sakamoto T, Kobayashi M, Itoh H, Tagiri A, Kayano T, Tanaka H, Iwahori S, Matsuoka M (2001) Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol 125: 1508–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JAD, Amasino RM (2002) Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmeyer W, Ellis MH, Chandler PM (2002) Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci USA 99: 9043–9048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Kamiya Y (1994) The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6: 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados L, Kovacs I, Oberschall A, Abraham E, Kerekes I, Zsigmond L, Nagy R, Alvarado M, Krasovskaja I, Gal M et al. (2002) Distribution of 1000 sequenced T-DNA tags in the Arabidopsis genome. Plant J 32: 233–242 [DOI] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P (1999) Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA 96: 4698–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu T, Kawaide H, Sekimoto H, vonNumers C, Phillips AL, Hedden P, Kamiya Y (1997) Cloning and characterization of a cDNA encoding gibberellin 20-oxidase from rice (Oryza sativa) seedlings. Physiol Plant 99: 111–118 [Google Scholar]
- Webster T (2002) Dwarfing rootstocks: past, present and future. Compact Fruit Tree 35: 67–72 [Google Scholar]
- Weigel D, Ahn JH, Blazquez J, Borevitz JO, Christensen SK, Frankhauser C, Ferrandiz C, Kardailsky I, Neff MM, Nguyen JT et al. (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger SD, Jansson S, Taylor G (2002) Genomics and forest biology: Populus emerges as the perennial favorite. Plant Cell 14: 2651–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Kamiya Y (2000) Gibberellin biosynthesis: its regulation by endogenous and environmental signals. Plant Cell Physiol 41: 251–257 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Sun T, Kawaide H, Kamiya Y (1998) The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol 116: 1271–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko E, Adams CJ, Machaekova I, Malbeck J, Scollan C, Meyer P (2002) Activation tagging identifies a gene from Petunia hybrida responsible for the production of active cytokinins in plants. Plant J 29: 797–808 [DOI] [PubMed] [Google Scholar]