Abstract
B-type cyclin-dependent kinases (CDKs) are unique to plants and are assumed to be involved in the control of the G2-to-M phase progression and mitotic events. However, little is known about their cyclin partners. In Arabidopsis, we isolated cDNA encoding the D-type cyclin CYCD4;1 by a yeast (Saccharomyces cerevisiae) two-hybrid screening using CDKB2;1 as bait. In vitro pull-down assay showed that CYCD4;1 bound to CDKB2;1 and CDKA;1. Protein complexes of CYCD4;1-CDKA;1 and CYCD4;1-CDKB2;1 in insect cells exhibited histone H1-kinase activity. Promoter analysis using the luciferase reporter gene showed that CDKB2;1 was expressed from early G2 to M phase, whereas CYCD4;1 was expressed throughout the cell cycle. In situ hybridization of plant tissues revealed that both CDKB2;1 and CYCD4;1 transcripts accumulated in the shoot apical meristem, leaf primordia, vasculature of leaves, and tapetal cells in anthers. Our results suggest that CDKB2;1 and CYCD4;1 may form an active kinase complex during G2/M phase and control the development of particular tissues.
Progression through the eukaryotic cell cycle is controlled by a family of cyclin-dependent kinases (CDKs). The kinase activity of CDKs is dependent on binding to cyclins. As in animals, plants have several types of CDKs and cyclins; thus, distinct CDK-cyclin complexes are involved in transition between different phases of the cell cycle (for review, see Mészáros et al., 2000; Stals and Inzé, 2001; Criqui and Genschik, 2002; Oakenfull et al., 2002).
Key checkpoints are assumed to operate at the G1/S and G2/M transitions. In animal cells, progression from G1 to S phase is mediated by complexes of CDK4 or CDK6 and D-type cyclins, which are induced by growth factors at the mRNA level. These complexes phosphorylate and inactivate the retinoblastoma protein (RB), and then active E2F transcription factors are released from binding with Rb to induce transcription of genes involved in S phase progression (for review, see Harbour and Dean, 2000). In Arabidopsis, CYCD2;1 and CYCD3;1 have been shown to interact with CDKA;1 in vivo (Healy et al., 2001), and RB-associated kinases include CDKA;1 and CYCD2;1 (Boniotti and Gutierrez, 2001). It also has been demonstrated that tobacco (Nicotiana tabacum) CDKA/CYCD3 complex purified from insect cells was able to phosphorylate RB in vitro (Nakagami et al., 1999). These results suggest that the regulatory mechanisms underlying the G1/S transition are well conserved among animals and plants.
Entry into mitosis is triggered by CDK1 (Cdc2) in animal cells, whereas two different types of CDKs, A-type CDK (CDKA) and B-type CDK (CDKB), are assumed to play a role in mitotic entry and progression in plants (for review, see Stals and Inzé, 2001; Potuschak and Doerner, 2001; Criqui and Genschik, 2002). CDKA has a conserved PSTAIRE motif, which is responsible for binding to cyclins. It is expressed throughout the cell cycle and closely related to the yeast Cdc2/Cdc28, thus in fact functionally complemented cdc2 mutants (Hirayama et al., 1991). Overexpression of a dominant negative type of CDKA in planta revealed that CDKA is involved in controlling both the G1/S and G2/M transitions (Hemerly et al., 1995). In contrast, CDKB is a plant-specific CDK in the sense that it has altered PSTAIRE motif, and its expression is under strict cell cycle control. CDKB is further classified into two groups: CDKB1 with the PPTALRE motif is expressed from S to M phase, and CDKB2 with the P(S/P) TTLRE motif is expressed in a more restricted period from G2 to M phase (Umeda et al., 1999b; Mészáros et al., 2000; Menges and Murray, 2002; Oakenfull et al., 2002). Recent studies showed that overexpression of a dominant negative type of Arabidopsis CDKB1;1 delayed the G2-to-M transition in tobacco cells (Porceddu et al., 2001), suggesting that at least CDKB1 is involved in mitotic entry. Mitotic cyclins, such as A-type cyclin (CYCA) and B-type cyclin (CYCB), are assumed to make complexes with CDKA or CDKB2 during G2/M phase (Mészáros et al., 2000; Roudier et al., 2000), but thus far, relatively little information is available regarding complexes between CDKA/B and mitotic cyclin.
Here, we show that Arabidopsis B2-type CDK CDKB2;1 can interact with CYCD1;1 and CYCD4;1 in vitro, and the CDKB2;1-CYCD4;1 complex purified from insect cells has a histone H1 kinase activity. Analysis of promoter activities of CDKB2;1 and CYCD4;1 demonstrated that CDKB2;1 is expressed from early G2 to M phase, whereas CYCD4;1 is expressed throughout the cell cycle. The results of in situ hybridization revealed that CDKB2;1 and CYCD4;1 are transcribed in tissues overlapping each other, suggesting that CDKB2;1 and CYCD4;1 may form an active kinase complex to control G2/M phase transition and mitotic events.
RESULTS
Identification of CYCD4;1 as an Interacting Protein with CDKB2;1 in Yeast Cells
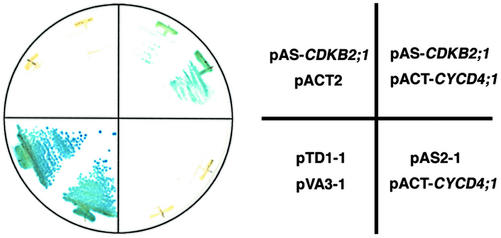
To identify proteins that interact with CDKB2;1 in Arabidopsis, we carried out a yeast two-hybrid screening. The full-length coding region of the CDKB2;1 cDNA was fused in-frame with the GAL4 DNA-binding domain and used as bait. Screening was performed with an Arabidopsis cDNA library derived from mRNA of suspension cultured cells. About 2.1 × 105 clones were screened on a medium lacking His, and, finally, 98 clones turned out to be His+ and LacZ+. Among them, 81 clones encoded a homolog of yeast p13Suc1, named Csk1At (De Veylder et al., 1997), and eight clones contained the full-length cDNA of CYCD4;1 (De Veylder et al., 1999). Cks1At is known to interact with CDKA;1, CDKB1;1, CDKB1;2, and CDKB2;1 (De Veylder et al., 1997; Boudolf et al., 2001), suggesting that our screening was working properly. As shown in Figure 1, expression of neither binding doman-CDKB2;1 nor CYCD4;1 fused to the GAL4 activation domain resulted in the LacZ+ phenotype, whereas co-expression of both proteins induced transcription of the marker gene, indicating that CDKB2;1 and CYCD4;1 interacted with each other in yeast cells.
Figure 1.
Interaction between CDKB2;1 and CYCD4;1 in yeast. Yeast strain Y190 was transformed with different combinations of pAS-CDKB2;1, pACT-CYCD4;1, or the empty vector pAS2–1 or pACT2. As a positive control, pTD1–1 carrying SV40 large T antigen and pVA3–1 carrying murine p53 were transformed. β-Galactosidase activity of yeast cells grown on a minimal medium was observed by the colony filter assay.
CYCD4;1 Interacts with CDKA;1 and CDKB2;1 in Vitro
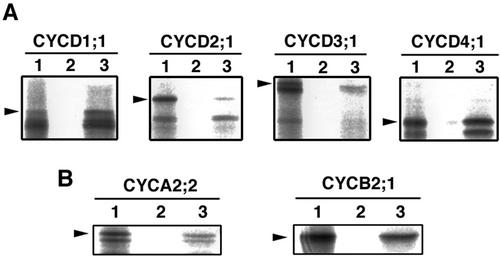
Arabidopsis encodes mainly four classes of D-type cyclins (CYCD1–4) on the genome (Vandepoele et al., 2002). To examine the specificity of interaction between CDKB2;1 and each class of CYCD, we performed in vitro pull-down assay. CDKB2;1 fused to glutathione S-transferase (GST) was expressed in Escherichia coli and immobilized on glutathione Sepharose beads. Four types of CYCD (CYCD1;1, CYCD2;1, CYCD3;1, and CYCD4;1) were transcribed and translated from cDNAs with the rabbit reticulocyte lysate in the presence of [35S]Met and incubated with the GST-CDKB2;1 beads. As shown in Figure 2A, CYCD1;1 and CYCD4;1 bound to GST-CDKB2;1 but not to GST only, whereas only a small amount of CYCD2;1 and CYCD3;1 was retained on the GST-CDKB2;1 resin. It is likely that the lower bands detected on autoradiography represent degradation products of cyclins, which contain the PEST sequence responsible for unstable nature of D-type cyclins (Rechsteiner and Rogers, 1996). Mitotic cyclins are also possible partners of CDKB2; thus, we applied the same assay as above to CYCA2;2 and CYCB2;1. The results showed that both mitotic cyclins bound to CDKB2;1, and the binding efficiency was the same or rather lower compared with the case of CYCD1;1 or CYCD4;1 (Fig. 2B). These findings suggest that CDKB2;1 exhibited significant level of binding activity to CYCD1;1 or CYCD4;1 in vitro.
Figure 2.
Arabidopsis cyclins interacting with CDKB2;1. In vitro pull-down assay was conducted with D-type and mitotic cyclins. A, [35S]Met-labeled CYCD1;1, CYCD2;1, CYCD3;1, and CYCD4;1 were incubated with GST (lane 2) or GST-CDKB2;1 (lane 3) immobilized on glutathione Sepharose beads. Proteins bound to the beads were separated by SDS-PAGE and autoradiographed. One-microliter input of the translation products is included as a control (lane 1). Arrowheads indicate the position of in vitro translated cyclins. B, CYCA2;2 and CYCB2;1 were incubated with the beads as described above.
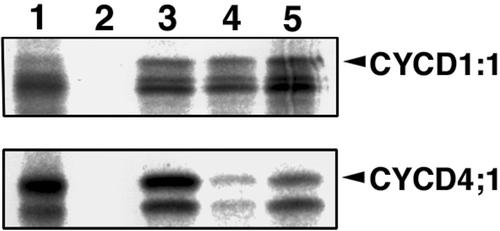
Next, we determined the type of CDK that interacts with CYCD1;1 or CYCD4;1 in the pull-down assay. For this purpose, CDKA;1, CDKB1;1, and CDKB2;1 were fused to GST and immobilized on the beads. As shown in Figure 3, CYCD1;1 was retained on any of the CDKs to almost the same extent, whereas CYCD4;1 tightly bound to CDKA;1 and CDKB2;1 but very weakly to CDKB1;1. These results indicate that CYCD4;1 can be a partner of CDKB2;1 and CDKA;1 in vitro.
Figure 3.
Arabidopsis CDKs interacting with CYCD1;1 or CYCD4;1. In vitro pull-down assay was conducted with CDKAs and CDKBs. [35S]Met-labeled CYCD1;1 or CYCD4;1 was incubated with GST (lane 2), GST-CDKA;1 (lane 3), GST-CDKB1;1 (lane 4), or GST-CDKB2:1 (lane 5) immobilized on glutathione Sepharose beads. Proteins bound to the beads were separated by SDS-PAGE and autoradiographed. One microliter of in vitro-translated products is included as a control (lane 1).
CYCD4;1 Binds to and Activates CDKA;1 and CDKB2;1 in Insect Cells
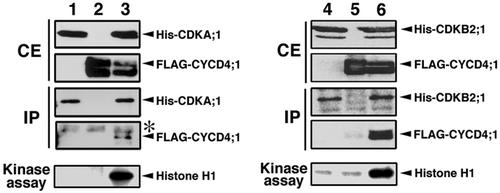
In the next step, we determined whether activation of CDKA;1 or CDKB2;1 was dependent on interaction with CYCD4;1. CDKA;1 or CDKB2;1 fused to 6× His tag and/or CYCD4;1 fused to FLAG tag were expressed in insect cells via a baculovirus-mediated system. Immunoblotting showed that each protein was properly produced with expected molecular size (Fig. 4). To test whether CDKA;1 or CDKB2;1 makes protein complexes with CYCD4;1, protein extract was immunoprecipitated with anti-CDKA;1 or anti-CDKB2;1 antibody and assayed by western blotting. As shown in Figure 4, His-CDKA;1 or His-CDKB2;1 was equally immunoprecipitated with the antibody, and FLAG-CYCD4;1 was included in the immunoprecipitates in the case of co-expression. These results indicate that His-CDKA;1 or His-CDKB2;1 formed a complex with FLAG-CYCD4;1 in insect cells.
Figure 4.
Activation of CDKA;1 and CDKB2;1 by interaction with CYCD4;1. Insect Sf-9 cells were transfected with baculovirus to express His-CDKA;1 (lane 1), His-CDKB2;1 (lane 4), FLAG-CYCD4;1 (lanes 2 and 5), or to co-express His-CDKA;1 and FLAG-CYCD4;1 (lane 3) or His-CDKB2;1 and FLAG-CYCD4;1 (lane 6). Crude extracts (CE) from insect cells were immunoblotted with anti-CDKA;1 antibody (lanes 1–3), anti-CDKB2;1 antibody (lanes 4–6), or anti-FLAG M2 antibody (lanes 1–6). Insect proteins were immunoprecipitated with anti-CDKA;1 antibody (lanes 1–3) or with anti-CDKB2;1 antibody (lanes 4–6), and the immunoprecipitates (IP) were detected by immunoblotting with anti-CDKA;1 antibody (lanes 1–3), anti-CDKB2;1 antibody (lanes 4–6), or anti-FLAG M2 antibody (lanes 1–6). The faint bands indicated with asterisks represent nonspecific cross-reaction with the rabbit IgG. The immunoprecipitates prepared as described above were used as enzyme in histone H1 kinase assay.
The same immunoprecipitates were subjected to kinase assay using histone H1 as a substrate. Almost no phosphorylation was detected with extracts from either CDK- or cyclin-expressing cells, whereas an intense band of phosphorylation was noted with immunoprecipitates containing His-CDKA;1 or His-CDKB2;1 and FLAG-CYCD4:1 (Fig. 4). These results indicate that His-CDKA;1 or His-CDKB2;1 was activated by making a complex with FLAG-CYCD4;1 in insect cells.
Expression of CDKB2;1 and CYCD4;1 during the Cell Cycle
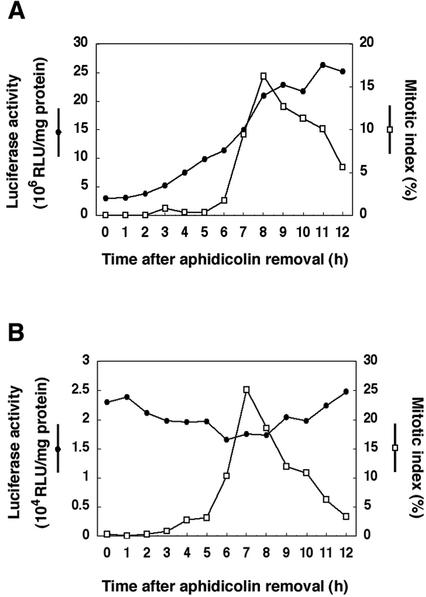
It has been reported that CDKA;1 is expressed throughout the cell cycle (Hemerly et al., 1993; Menges and Murray, 2002), indicating that CYCD4;1 can be a partner of CDKA;1 whenever it is expressed. In contrast, the expression of CDKB2;1 is assumed to be dependent on the cell cycle (Joubès et al., 2000; Mészáros et al., 2000). Therefore, we investigated the promoter activities of CDKB2;1 and CYCD4;1 during the cell cycle in tobacco Bright Yellow-2 (BY-2) cells.
First, we determined the transcription start site of CDKB2;1 and CYCD4;1 genes by 5′-RACE method. Nucleotide sequences of amplified fragments showed that the transcripts of CDKB2;1 and CYCD4;1 start from the adenine nucleotides 89 and 129 bp upstream of the start codon, respectively. Therefore, an Arabidopsis genomic DNA containing the promoter region of CDKB2;1 (0.9 kb) or CYCD4;1 (2.5 kb) was fused to the luciferase reporter gene (LUC), and the promoter-LUC constructs were introduced into BY-2 cells by Agrobacterium tumefaciens-mediated transformation. Stably transformed cell lines were treated with aphidicolin to arrest cells at the early S phase, and the LUC activity was measured after removal of aphidicolin. The mitotic index showed a peak 7 to 8 h after aphidicolin removal in transgenic BY-2 cells (Fig. 5). The LUC activity driven by the CDKB2;1 promoter increased from 2 to 3 h, and a marked increase was observed at 7 to 8 h (Fig. 5A), suggesting that the transcripts of CDKB2;1 accumulated from early G2 to M phase. In the case of CYCD4;1 promoter, a low but significant level of LUC activity was observed throughout the cell cycle, and it showed a slight peak from G1 to S phase, namely 0 to 1 h and 9 to 12 h after aphidicolin removal (Fig. 5B). These results indicate that the dynamics of expression of CDKB2;1 and CYCD4;1 overlap each other, suggesting that they form an active kinase complex and function during G2 to M phase.
Figure 5.
Expression of CDKB2;1 and CYCD4;1 during the cell cycle. Tobacco BY-2 cells were transformed with the CDKB2;1 (A) or CYCD4;1 (B) promoter fused to the luciferase (LUC) reporter gene and synchronized with aphidicolin. After removal of aphidicolin, cell cycle progression was monitored by counting the mitotic index (white square). Cells were harvested at 1-h intervals and examined for luciferase activity (black circles). RLU, Relative light unit.
Spatial Expression Patterns of CDKB2;1 and CYCD4;1 in Planta
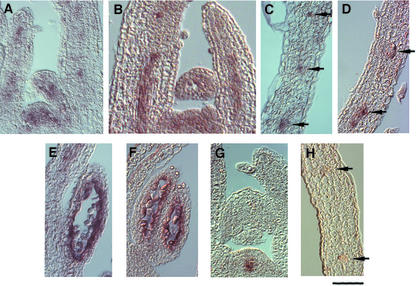
To study the spatial expression patterns of CDKB2;1 and CYCD4;1, we performed in situ hybridizations using probes specific for transcripts of CDKB2;1 and CYCD4;1. RNA probes were prepared from cDNAs and labeled with digoxygenin. By using an antisense probe of CDKB2;1, a patchy pattern of hybridization signals was observed in the vegetative shoot apical meristem and young leaf primordia (Fig. 6A). It is likely that the patchy pattern reflects the G2/M phase-specific expression of CDKB2;1 as described above. The signal was also seen in the middle of growing leaves and tended to correlate with the provascular strands (Fig. 6A). Hybridization of transverse leaf sections with the same probe confirmed CDKB2;1 expression in vascular tissues (Fig. 6C, arrows). Similar results were obtained with the probe of CYCD4;1. The transcripts of CYCD4;1 were detected in the shoot apical meristem, leaf primordia (Fig. 6B), and vascular tissues (Fig. 6D). The control WUSCHEL probe produced hybridization signals underneath the outer layer of the shoot apex as described previously (Mayer et al., 1998; Fig. 6G) but not in vascular tissues of leaves (Fig. 6H, arrows). These findings suggest that the signals of CDKB2;1 and CYCD4;1 in vascular tissues were not derived from nonspecific hybridization of probes. During flower development, the transcripts of CDKB2;1 and CYCD4;1 were detected in tapetum of anthers (Fig. 6, E and F) but not in gynoecium (data not shown). No signal was detected with the control WUSCHEL probe in tapetum (data not shown).
Figure 6.
Spatial expression patterns of CDKB2;1 and CYCD4;1 in planta. In situ hybridization was performed with specific probes for transcripts of CDKB2;1 (A, C, and E), CYCD4;1 (B, D, and F), and WUSCHEL (G and H). Antisense riboprobes were labeled with digoxygenin. Hybridization signals are visible as brownish-purple staining. A, B, and G, Longitudinal sections through shoot apices of 7-d-old seedlings. C, D, and H, Transverse sections through young rosette leaves of 20-d-old seedlings. E and F, Longitudinal sections through anthers. Arrows indicate the vascular tissue. Bar = 50 μm.
DISCUSSION
In yeast two-hybrid screening with CDKB2;1 as bait, we identified a D-type cyclin, CYCD4;1, and an Arabidopsis homologue of yeast p13Suc1, Cks1At. However, we could not isolate cDNA clones encoding mitotic cyclins, although at least two mitotic cyclins, CYCA2;2 and CYCB2;1, bound to CDKB2;1 in in vitro pull-down assay (Fig. 2B). This might be due to the toxic effect of some of the plant mitotic cyclins on yeast growth (Umeda et al., 1999a); hence, cells overexpressing these cyclins might not be able to survive on selection media.
The result of in vitro pull-down assay showed that CDKB2;1 efficiently bound to CYCD1;1 and CYCD4;1, whereas CYCD4;1 interacted with both CDKA;1 and CDKB2;1 but not with CAKB1:1. Mészáros et al. (2000) showed that alfalfa (Medicago sativa) CycD4 interacted with a B2-type CDK, Cdc2MsF, in yeast cells. This D-type cyclin also bound to Cdc2MsA (CDKA) and Cdc2MsD (B1-type CDK) in the two-hybrid system, but neither CycD3 nor a new D-type cyclin, CycD5, interacted with Cdc2MsF. These results indicate that some but not all D-type cyclins can be a partner of B2-type CDKs.
At present, we do not have suitable antibodies against CYCD4;1; thus, the protein complex of CDKB2;1 and CYCD4;1 could not be identified by immunoprecipitation. Moreover, any effort to overproduce CYCD4:1 protein in plant cells failed, probably due to its unstable nature or toxic effect of overexpression. Instead, we showed that CYCD4;1 formed protein complexes with CDKA;1 and CDKB2;1 in insect cells, and they were active in terms of histone H1-kinase activity. These results indicate that CYCD4;1 functions as a cyclin subunit by controlling kinase activities of CDKA;1 and CDKB2;1 in living cells. To our knowledge, Arabidopsis CYCD4;1 is the first D-type cyclin that is shown to make an active kinase complex with B2-type CDK. Because histone H1 may not be an adequate substrate for the CDKB2;1/CYCD4;1 complex, it is important to identify another physiological substrates. Considering that the involvement of D-type cyclins during the G2/M phase has not been demonstrated in other organisms, the substrates for CDKB2;1/CYCD4;1 may be associated with some plant-specific events.
We analyzed the promoter activities of CDKB2;1 and CYCD4;1 in transgenic BY-2 cells. Our results showed that the promoter activity of CDKB2;1 was restricted from early G2 to M phase, whereas that of CYCD4;1 was observed throughout the cell cycle. Thus, there is an overlap of the dynamics of CDKB2;1 and CYCD4;1 expression, suggesting that CYCD4;1 could bind to and activate CDKB2;1 during G2/M phase. Menges et al. (2002) analyzed the cell cycle-regulated gene expression in Arabidopsis suspension-cultured cells by using the microarray technique and found that CYCD4;1 expression reaches a peak level at the G1 phase. Here, we observed a little higher LUC activity during the G1/S phase, suggesting that CYCD4;1 is also involved in the control of G1/S phase possibly by making a complex with CDKA;1. Menges and Murray (2002) also demonstrated that the transcripts of CDKB2;2, which shares 86% identity with CDKB2;1 at the amino acid level, accumulated mainly during M phase in Arabidopsis suspension-cultured cells. In contrast, our data showed that the expression of CDKB2;1 is induced early in the G2 phase and is clearly enhanced in the M phase. In support of our results, Himanen et al. (2002) reported that the mRNA level of CDKB2;1 started to increase from S/G2 phase when the cell cycle was synchronized during lateral root initiation. These results suggest that CDKB2;1 may be expressed earlier than CDKB2;2 regardless of their high similarity in primary sequence.
In planta, transcripts of both CDKB2;1 and CYCD4;1 were detected in shoot apical meristem, young leaf primordia, vascular tissues, and anthers. De Veylder et al. (1999) described accumulation of CYCD4;1 transcripts in vascular tissue of roots, lateral root primordial, and fertilized ovules, but not in stamen. The reason for this discrepancy in expression patterns, especially in reproductive organs, remains unknown. Among the other D-type cyclins, CYCD3;2 expression pattern was studied in detail (Swaminathan et al., 2000). CYCD3;2 is expressed in vegetative shoot apical meristem, young leaf or floral primordia, developing floral organs, microspores, megaspores, vasculature in the main stem, pedical, and floral organs. The transcript level of CYCD3;2 decreased to a background level with differentiation of the organs. Comparison with CYCD3;2, CYCD4;1 expression seems to be more restrictive in particular tissues.
Recently, the patterns of cell cycling during leaf vein development have been reported in Arabidopsis by using a CYCB1;1::GUS reporter construct (Kang and Dengler 2002). It has been demonstrated that at early stage of the leaf development, GUS-expressing cells were uniformly distributed throughout the leaf blade, but after the development of leaves, GUS-expressing cells were confined to the developing vascular tissue in the basal part of the leaf. Finally, CYCB1;1-expressing cells became localized to the region of meristematic cells adjacent to differentiated phloem. These results indicate that cells in the vascular tissue are actively dividing throughout leaf development, and cell divisions in the vasculature last longer than those in other areas of leaves. The localized expression of CYCD4;1 in the leaf vein suggests a role in cell division associated with vascular tissue development by making a complex with CDKA;1 or CDKB2;1.
Our results from in situ hybridization showed that both CYCD4;1 and CDKB2;1 were expressed in tapetum of anthers. The tapetal cells are initially uninucleate but become binucleate before meiosis of the pollen mother cells, (Misra, 1962). This indicates that during their cell differentiation, tapetal cells may stop cell cycling and remain in the M phase before cytokinesis. Uniform expression of CDKB2;1 in tapetum (Fig. 6E) may support this conclusion. The complex of CYCD4;1 and CDKB2;1 may be associated with cell cycle control during tapetal cell differentiation.
In conclusion, Arabidopsis CYCD4;1 probably forms an active kinase complex with CDKB2;1 during G2/M phase in specific tissues. The cell cycle is integrated into complex pathways of morphogenesis and histogenesis in plants (for review, see Meijer and Murray, 2001). To determine the relationship between the cell cycle and the regulatory mechanism underlying plant development, it is important to study the spatial expression patterns of core cell cycle regulators. Even in Arabidopsis, however, the information is quite limited. For example, neither CYCD2;1 nor CYCD3;1 have been subjected to detailed expression analysis in planta, although their expression patterns during the cell cycle have been well described (Riou-Khamlichi et al., 1999, 2000; Cockcroft et al., 2000; Healy et al., 2001). Isolation of loss-of-function mutants and detailed expression analysis should reveal the cross talk between signals regulating cell division and differentiation during plant development.
MATERIALS AND METHODS
Yeast (Saccharomyces cerevisiae) Two-Hybrid Screening
The entire open reading frame (ORF) of CDKB2;1 was amplified from an Arabidopsis cDNA mixture by PCR with primers that included the recognition sequence for EcoRI at both the N- and C-terminal ends. The amplified fragment was cloned into the pBluescript II SK– (Stratagene, La Jolla, CA) to produce pBSII-CDKB2;1, and its nucleotide sequence was confirmed. After digestion with EcoRI, the fragment was subcloned into the EcoRI site of pAS2–1 (CLONTECH Laboratories, Palo Alto, CA). The resultant plasmid pAS-CDKB2;1 was introduced into the yeast strain Y190, which was then transformed with the Arabidopsis cDNA library that was prepared in the pACT2 vector (CLONTECH) derived from mRNA of suspension-cultured cells (Németh et al., 1998). The transformants were screened on minimal medium containing 20 mm 3-aminotriazole and lacking Leu, Trp, and His. Yeast colonies were grown on nylon membrane filters on minimal medium with 20 mm 3-aminotriazole to perform LacZ filter lift assays as described in the protocol of the Matchmaker Gal4 two-hybrid system (CLONTECH).
In Vitro Pull-Down Assay
In vitro translation of Arabidopsis cyclins was conducted with [35S]Met by using the TNT Coupled Reticulocyte Lysate Systems (Promega, Madison, WI). cDNAs of D-type cyclins CYCA2;2 and CYCB2;1 were subcloned into the pBluescript II SK– vector and used as template for in vitro transcription. The ORFs of CDKA;1 and CDKB1;1 were amplified by PCR with primers that included the recognition sequence for EcoRI at both the N- and C-terminal ends. The fragments were ligated to pBluescript II SK– to produce pBSII-CDKA;1 and pBSII-CDKB1;1, and their nucleotide sequences were confirmed. After digestion with EcoRI, the fragments and the ORF of CDKB2;1 were subcloned into the EcoRI site of pGEX-1λT (Pharmacia, Piscataway, NJ). GST fusion proteins were expressed in Escherichia coli and purified with glutathione Sepharose (Amersham Pharmacia Biotech, Piscataway, NJ) as described previously (Yamaguchi et al., 1998). To prepare the affinity resin, 1 mg of GST or GST-CDK proteins were incubated with 100 μL of glutathione Sepharose in 1 mL of phosphate-buffered saline (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, and 1.8 mm KH2PO4 [pH 7.3]). Ten microliters of affinity resin was incubated with 20 μL of translated product in 200 μL of binding buffer (50 mm Tris-HCl [pH 7.5], 5 mm MgCl2, 100 mm NaCl, 10% [v/v] glycerol, 5 mm β-mercaptoethanol, and 0.5 mg mL–1 bovine serum albumin) for 120 min at 4°C. After washing the resin four times with 500 μL of binding buffer, proteins retained on the resin were eluted with 20 μL of elution buffer (50 mm Tris-HCl [pH 7.5], 5 mm MgCl2, 100 mm NaCl, 10% [v/v] glycerol, 5 mm β-mercaptoethanol, and 20 mm glutathione) and separated by SDS-PAGE.
In Vitro Kinase Assay
The plasmids for expression in baculovirus-infected insect cells were constructed as follows. pBSII-CDKA;1 and pBSII-CDKB2;1 were digested with EcoRI, and the insert fragments were cloned into the EcoRI site of pFASTBAC HTa (Gibco BRL, Gaithersburg, MD) to be in frame with the 6× His. The ORF of CYCD4;1 was amplified by PCR with primers that included the recognition sequence for EcoRI at both the N- and C-terminal ends. The fragment was cloned into pBluescript II SK– to produce pBSII-CYCD4;1, and the nucleotide sequence was confirmed. After digested with EcoRI, the fragment was subcloned into the EcoRI site of pFAST-BAC-FLAG1 (Yamaguchi et al., 2000) to be in frame with the FLAG tag.
Transfection of insect Sf9 cells was performed as described by Yamaguchi et al. (2000). Thirty micrograms of total protein extracted from insect cells was used for immunoprecipitation with anti-CDKA;1 or anti-CDKB2;1 antibody. Anti-CDKA;1 antibody was described previously (Umeda et al., 2000). Anti-CDKB2;1 antibody was raised against the carboxyl-terminal CFDDLPEKSSL peptide of Arabidopsis CDKB2;1. The immunoprecipitates were eluted with 0.1 m sodium citrate (pH 3.0), neutralized with 1 m Tris-HCl (pH 8.8), and separated by SDS-PAGE. Immunoblotting was conducted with anti-CDKA;1, anti-CDKB2;1, or anti-FLAG M2 antibody (Sigma Chemical Co., St. Louis) by using an ECL Western Blotting Detection Kit (Amersham). For the kinase assay, the immunoprecipitates were subjected to phosphorylation reaction with 3 μg of histone H1 as substrate as described previously (Umeda et al., 1999b).
Promoter Analysis in Transgenic BY-2 Cells
The plasmid pDO432 (Ow et al., 1986) was digested with BamHI and SacI to obtain the luciferase (LUC) cording region. To isolate the fragment of nopaline synthase terminator, pBI221 was digested with SacI and EcoRI. These fragments were cloned into the BamHI/EcoRI sites of the binary cloning vector pPZP211 (Hajdukiewicz et al., 1994) to produce pPZP211-LUC.
Transcription start sites of CDKB2;1 and CYCD4;1 were determined by 5′-RACE using the RML-RACE kit (Ambion, Austin, TX), according to the instructions provided by the manufacturer. The promoter region of CDKB2;1 (–835 to +50 bp) was amplified with a primer that included the recognition sequence for BamHI at the 3′ end. After confirming the nucleotide sequence, the fragment was digested with BamHI and HindIII, whose recognition sequence resides 835 bp upstream of the transcription start site, and the HindIII-BamHI fragment was cloned into the HindIII/BamHI sites of pPZP211-LUC. The promoter region of CYCD4;1 (–2,362 to +129 bp) was amplified by PCR with primers that included the recognition sequence for SalI at both the 5′ and 3′ ends. The fragment was cloned into the pCR-XL-TOPO vector (Invitrogen, San Diego), and the nucleotide sequence was confirmed. After digestion with SalI, the fragment was subcloned into the SalI site of pPZP211-LUC. The resultant plasmids were used to transform tobacco (Nicotiana tabacum) BY-2 cells. For synchronization of BY2 cells, 7-d-old culture was diluted to 1:10 (v/v), mixed with 5 mg L–1 aphidicolin, and cultured for 24 h. To restart the cell cycle, aphidicolin was removed by washing the cells with 1,000 mL of fresh medium. LUC assay was performed as described by Ito et al. (1998).
In Situ RNA Hybridization
Arabidopsis tissues were fixed in FAA (50% [v/v] ethanol, 5% [v/v] acetic acid, and 3.7% [v/v] formaldehyde), and 8-μm paraffin sections were hybridized with digoxygenin-labeled probes as described previously (Braissant and Wahli, 1998). The CDKB2;1 and CYCD4;1 probes were antisense strands corresponding to the region 460 to 804 of the CDKB2;1 ORF and to 816 to 927 of the CYCD4;1 ORF, respectively. The WUSCHEL probe corresponds to 101 to 1,202 of the cDNA (DDBJ/EMBL/GenBank accession no. AJ012310).
Acknowledgments
We thank Dr. Csaba Koncz for the Arabidopsis cDNA library for yeast two-hybrid screening. We are also grateful to Prof. Dirk Inzé and Dr. Christiane Genetello for cyclin cDNAs and to Dr. Takashi Araki for the WUSCHEL cDNA fragment.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.020644.
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (grant no. 14036212) and by Research for the Future from the Japan Society for the Promotion of Science.
References
- Boniotti MB, Gutierrez C (2001) A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J 28: 341–350 [DOI] [PubMed] [Google Scholar]
- Boudolf V, Rombauts S, Naudts M, Inzé D, De Veylder L (2001) Identification of novel cyclin-dependent kinases interacting with the CKS1 protein of Arabidopsis. J Exp Bot 52: 1381–1382 [PubMed] [Google Scholar]
- Braissant O, Wahli W (1998) Differential expression of peroxisome proliferator-activated receptor-alpha, -beta, and -gamma during rat embryonic development. Endocrinology 139: 2748–2754 [DOI] [PubMed] [Google Scholar]
- Cockcroft CE, den Boer BG, Healy JM, Murray JAH (2000) Cyclin D control of growth rate in plants. Nature 405: 575–579 [DOI] [PubMed] [Google Scholar]
- Criqui MC, Genschik P (2002) Mitosis in plants: how far we have come at the molecular level? Curr Opin Plant Biol 5: 487–493 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Segers G, Glab N, Casteels P, Van Montagu M, Inzé D (1997) The Arabidopsis Cks1At protein binds the cyclin-dependent kinases Cdc2aAt and Cdc2bAt. FEBS Lett 412: 446–452 [DOI] [PubMed] [Google Scholar]
- De Veylder L, De Almeida Engler J, Burssens S, Manevski A, Lescure B, Van Montagu M, Engler G, Inzé D (1999) A new D-type cyclin of Arabidopsis thaliana expressed during lateral root primordia formation. Planta 208: 453–462 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Harbour JW, Dean DC (2000) The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev 14: 2393–2409 [DOI] [PubMed] [Google Scholar]
- Healy JMS, Menges M, Doonan JH, Murray JAH (2001) The Arabidopsis D-type cyclins CycD2 and CycD3 both interact in vivo with the PSTAIRE cyclin-dependent kinase Cdc2a but are differentially controlled. J Biol Chem 276: 7041–7047 [DOI] [PubMed] [Google Scholar]
- Hemerly AS, de Almeida Engler J, Bergounioux C, Van Montagu M, Engler G, Inzé D, Ferreira P (1995) Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J 14: 3925–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly AS, Ferreira P, de Almeida Engler J, Van Montagu M, Engler G, Inzé D (1993) cdc2a expression in Arabidopsis is linked with competence for cell division. Plant Cell 5: 1711–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Imajuku Y, Anai T, Matsui M, Oka A (1991) Identification of two cell-cycle-controlling cdc2 gene homologs in Arabidopsis thaliana. Gene 105: 159–165 [DOI] [PubMed] [Google Scholar]
- Ito M, Iwase M, Kodama H, Lavisse P, Komamine A, Nishihama R, Machida Y, Watanabe A (1998) A novel cis-acting element in promoters of plant B-type cyclin genes activates M phase-specific transcription. Plant Cell 10: 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J, Chevalier C, Dudits D, Heberle-Bors E, Inzé D, Umeda M, Renaudin JP (2000) CDK-related protein kinases in plants. Plant Mol Biol 43: 607–620 [DOI] [PubMed] [Google Scholar]
- Kang J, Dengler N (2002) Cell cycling frequency and expression of the homeobox gene ATHB-8 during leaf vein development in Arabidopsis. Planta 216: 212–219 [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Meijer M, Murray JAH (2001) Cell cycle controls and the development of plant form. Curr Opin Plant Biol 4: 44–49 [DOI] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JAH (2002) Cell cycle-regulated gene expression in Arabidopsis. J Biol Chem 277: 41987–42002 [DOI] [PubMed] [Google Scholar]
- Menges M, Murray JAH (2002) Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J 30: 203–212 [DOI] [PubMed] [Google Scholar]
- Mészáros T, Miskolczi P, Ayaydin F, Pettkó-Szandtner A, Peres A, Magyar Z, Horváth GV, Bakó L, Fehér A, Dudits D (2000) Multiple cyclin-dependent kinase complexes and phosphatases control G2/M progression in alfalfa cells. Plant Mol Biol 43: 595–605 [DOI] [PubMed] [Google Scholar]
- Misra RC (1962) Contribution to the embryology of Arabidopsis thalianum (Gay and Monn.) Agra Univ J Res 11: 191–199 [Google Scholar]
- Nakagami H, Sekine M, Murakami H, Shinmyo A (1999) Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D in vitro. Plant J 18: 243–252 [DOI] [PubMed] [Google Scholar]
- Németh K, Salchert K, Putnoky P, Bhalerao R, Koncz-Kálmán Z, Stankovic-Stangeland B, Bakó L, Mathur J, Ökrész L, Stabel S et al. (1998) Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev 12: 3059–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakenfull EA, Riou-Khamlichi C, Murray JAH (2002) Plant D-type cyclins and the control of G1 progression. Philos Trans R Soc Lond B Biol Sci 357: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow DW, Wood KV, DeLuca M, de Wet JR, Helinski DR, Howell SH (1986) Transient and stable expression of the firefly luciferase gene in plant cells and transgenic plants. Science 234: 856–859 [DOI] [PubMed] [Google Scholar]
- Porceddu A, Stals H, Reichheld JP, Segers G, De Veylder L, De Pinho Barrôco R, Casteels P, Van Montagu M, Inzé D, Mironov V (2001) A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J Biol Chem 276: 36354–36360 [DOI] [PubMed] [Google Scholar]
- Potuschak T, Doerner P (2001) Cell cycle controls: genome-wide analysis in Arabidopsis. Curr Opin Plant Biol 4: 501–506 [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW (1996) PEST sequences and regulation by proteolysis. Trends Biochem Sci 21: 267–271 [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JAH (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Menges M, Healy JM, Murray JAH (2000) Sugar control of the plant cell cycle: differential regulation of Arabidopsis D-type cyclin gene expression. Mol Cell Biol 20: 4513–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F, Fedorova E, Gyorgyey J, Feher A, Brown S, Kondorosi A, Kondorosi E (2000) Cell cycle function of a Medicago sativa A2-type cyclin interacting with a PSTAIRE-type cyclin-dependent kinase and a retinoblastoma protein. Plant J 23: 73–83 [DOI] [PubMed] [Google Scholar]
- Stals H, Inzé D (2001) When plant cells decide to divide. Trends Plant Sci 6: 359–364 [DOI] [PubMed] [Google Scholar]
- Swaminathan K, Yang Y, Grotz N, Campisi L, Jack T (2000) An enhancer trap line associated with a D-class cyclin gene in Arabidopsis. Plant Physiol 124: 1658–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda M, Iwamoto N, Umeda-Hara C, Yamaguchi M, Hashimoto J, Uchimiya H (1999a) Molecular characterization of mitotic cyclins in rice plants. Mol Gen Genet 262: 230–238 [DOI] [PubMed] [Google Scholar]
- Umeda M, Umeda-Hara C, Uchimiya H (2000) A cyclin-dependent kinase-activating kinase regulates differentiation of root initial cells in Arabidopsis. Proc Natl Acad Sci USA 97: 13396–13400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda M, Umeda-Hara C, Yamaguchi M, Hashimoto J, Uchimiya H (1999b) Differential expression of genes for cyclin-dependent protein kinases in rice plants. Plant Physiol 119: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Fabian T, Sauter M, Bhalerao RP, Schrader J, Sandberg G, Umeda M, Uchimiya H (2000) Activation of CDK-activating kinase is dependent on interaction with H-type cyclins in plants. Plant J 24: 11–20 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Umeda M, Uchimiya H (1998) A rice homolog of Cdk7/MO15 phosphorylates both cyclin-dependent protein kinases and the carboxy-terminal domain of RNA polymerase II. Plant J 16: 613–619 [DOI] [PubMed] [Google Scholar]