Abstract
Mercury (Hg), especially in organic form, is a highly toxic pollutant affecting plants, animals, and man. In plants, the primary target of Hg damage is the chloroplast; Hg inhibits electron transport and photosynthesis. In the present study, chloroplast genetic engineering is used for the first time to our knowledge to enhance the capacity of plants for phytoremediation. This was achieved by integrating a native operon containing the merA and merB genes (without any codon modification), which code for mercuric ion reductase (merA) and organomercurial lyase (merB), respectively, into the chloroplast genome in a single transformation event. Stable integration of the merAB operon into the chloroplast genome resulted in high levels of tolerance to the organomercurial compound, phenylmercuric acetate (PMA) when grown in soil containing up to 400 μm PMA; plant dry weights of the chloroplast transformed lines were significantly higher than those of wild type at 100, 200, and 400 μm PMA. That the merAB operon was stably integrated into the chloroplast genome was confirmed by polymerase chain reaction and Southern-blot analyses. Northern-blot analyses revealed stable transcripts that were independent of the presence or absence of a 3′-untranslated region downstream of the coding sequence. The merAB dicistron was the more abundant transcript, but less abundant monocistrons were also observed, showing that specific processing occurs between transgenes. The use of chloroplast transformation to enhance Hg phytoremediation is particularly beneficial because it prevents the escape of transgenes via pollen to related weeds or crops and there is no need for codon optimization to improve transgene expression. Chloroplast transformation may also have application to other metals that affect chloroplast function.
Mercury (Hg) pollution of soil and water is a world-wide problem (Dean et al., 1972; Krämer and Chardonnens, 2001). The extent to which Hg is harmful depends on the form of mercury present in the ecosystem. Inorganic forms of Hg are less harmful than organic forms partly because they bind strongly to the organic components of soil. For this reason, Hg does not tend to contaminate the ground water except when it leaches from a municipal landfill (U.S. Environmental Protection Agency, 1984). Organomercurial compounds, on the other hand, may be 200 times more toxic than inorganic Hg (Patra and Sharma, 2000) and methyl-Hg is especially toxic (Meagher and Rugh, 1997).
The principal forms of organomercurial compounds are alkyl mercurials (methyl- and ethyl-Hg), aryl mercurials (phenyl-Hg), and alkoxy alkyl Hg diuretics. The excessive use of organomercurial compounds (e.g. in fertilizers and pesticides) is known to have severe effects on plants. The main site of action of Hg damage appears to be the chloroplast thylakoid membranes and photosynthesis. Organomercurial compounds have been shown to strongly inhibit electron transport, oxygen evolution (Bernier et al., 1993), Hill reaction, photophosphorylation, and to quench chlorophyll fluorescence in photosystem II (Kupper et al., 1996). Furthermore, Prasad and Prasad (1987) showed that Hg might replace Mg from the chlorophyll moiety, leading to a reduction in chlorophyll content. Sen and Mondal (1987) and Sinha et al. (1996) reported a 26% (w/v) reduction of chlorophyll content in Salvia natans and 35% (w/v) in Bacopa monnieri at 5 μg mL–1 HgCl2, even though these plants have a natural tolerance to Hg.
Current remediation methods to clean up heavy metal-contaminated soils include soil flushing, chemical reduction/oxidation and excavation, retrieval, and offsite disposal, all of which are expensive, environmentally invasive, and labor intensive (Kärenlampi et al., 2000). An alternative and more cost-effective approach is phytoremediation, i.e. the use of plants to clean up contaminated environments (Lin et al., 1995; Salt et al., 1995; Terry et al., 2000). With the aid of genetic engineering, plants can be genetically modified to substantially improve phytoremediation. Expression of several plant and bacterial genes in transgenic plants has significantly enhanced these plant remediation systems (Meagher, 2000; Doucleff and Terry, 2002). Several studies have successfully integrated bacterial genes into nuclear genomes to produce plants that were specifically engineered for phytoremediation of metal-polluted environments (Heaton et al., 1998; Rugh et al., 1998; Nies, 1999). With respect to Hg, plants have been engineered with modified bacterial mercuric ion reductase (merA) and organomercurial lyase (merB) genes; these enzymes are capable of converting highly toxic methyl-Hg into the much less toxic Hg(0), which may then be volatilized (Rugh et al., 1996; Bizily et al., 1999, 2000).
All of the attempts to genetically engineer plants with improved phytoremediation have previously been based on transformation of the nuclear genome. An alternative and novel approach is to engineer the chloroplast genomes of higher plants. This approach offers several advantages over nuclear transformation, i.e. very high levels of transgene expression (up to 46% (w/w) of total protein; De Cosa et al., 2001), uniparental plastid gene inheritance (in most crop plants) that prevents pollen transmission of foreign DNA (Daniell et al., 1998; Daniell, 2002; Daniell and Parkinson, 2003), the absence of gene silencing (Lee et al., 2003) and positioning effect (Daniell et al., 2001a), the ability to express multiple genes in a single transformation event (De Cosa et al., 2001; Daniell and Dhingra, 2002), the ability to express bacterial genes without codon optimization (McBride et al., 1995; Kota et al., 1999; De Cosa et al., 2001), integration via a homologous recombination process that facilitates targeted transgene integration (Daniell et al., 2002), and sequestration of foreign proteins in the organelle, which prevents adverse interactions with the cytoplasmic environment (Daniell et al., 2001a; Lee et al., 2003). Engineering the chloroplast genome has successfully conferred insect resistance (McBride et al., 1995; Kota et al., 1999; De Cosa et al., 2001), herbicide resistance (Daniell et al., 1998), disease resistance (De Gray et al., 2001), drought tolerance (Lee et al., 2003), and expression of edible vaccines (Daniell et al., 2001a), monoclonals (Daniell, 2003), and biopharmaceuticals (Guda et al., 2000; Staub et al., 2000; De Gray et al., 2001; Fernandez-San Millan et al., 2003).
This is the first report where the chloroplast genome was engineered to enhance the capacity of plants for phytoremediation and where a native bacterial operon was used for expression in plants without codon optimization. Phenylmercuric acetate (PMA) was chosen to test the chloroplast transformation method because of the importance of toxicity of organomercurial compounds as environmental contaminants and because the site of action of organomercurial damage is the chloroplast (see above). The approach we used was to integrate a native operon containing the merA and merB genes, coding for mercuric ion reductase and organomercurial lyase, respectively, into tobacco (Nicotiana tabacum) chloroplast genomes. The results show that the chloroplast transgenic plants were substantially more resistant than wild type to the highly toxic organomercurial compound, PMA.
RESULTS AND DISCUSSION
Chloroplast Vectors and Bacterial Resistance Assays
The bacterial native genes, merA (1.69 kb) and merB (638 bp) that encode the mercuric ion reductase and the organomercurial lyase, respectively, were amplified by PCR from Escherichia coli strains harboring plasmids NR1 (containing the full-length merA) and R831b (containing the full-length merB). The PCR gene products were successively cloned into the pLD-vector, which is a chloroplast-specific vector used in previous publications from this laboratory (De Cosa et al., 2001; Daniell et al., 2001b). This vector contains the homologous recombination sequences (flanking sequences) that allow site-specific integration of the operon containing the aadA, merB, and merA genes into the inverted repeat region of the chloroplast genome in between the trnI (tRNA Ile) and trnA (tRNA Ala) genes (Daniell et al., 1998; Guda et al., 2000). The chloroplast 16S ribosomal RNA gene constitutive promoter (Prrn) drives the transcription of all downstream genes that include the aadA (aminoglycoside 3′-adenylyltransferase) gene conferring resistance to spectinomycin, the merA, and merB genes. Two versions of the chloroplast vector were made with the presence or absence of the 3′-untranslated region (UTR) from the chloroplast psbA gene that was expected to confer stability to transcripts, and they were designated pLDR-MerAB-3′-UTR and pLDR-MerAB, respectively (Fig. 1A). The pLDR-MerAB-3′-UTR and the pLDR-MerAB chloroplast vectors also contain the E. coli origin of replication and the ampicillin selectable marker that facilitates E. coli expression studies.
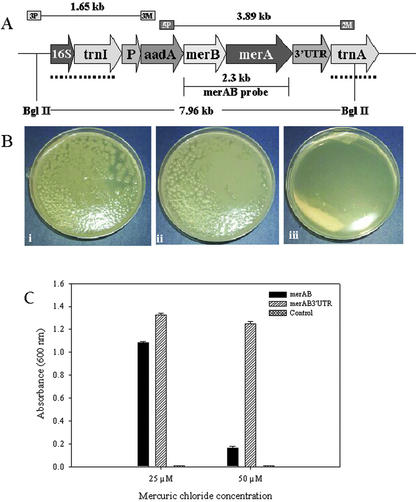
Figure 1.
Bacterial bioassay. A, Schematic representation of the transformed chloroplast genome: The map shows the transgenic chloroplast genome containing the pLDR-MerAB-3′-UTR construct. The site-specific integration between trnI and trnA chloroplast genes is shown by the dotted line, specifying the homologous recombination sequences in the pLDR-MerAB-3′-UTR and pLDR-MerAB. Landing sites for the 3P/3M and 5P/2M primer pairs used in PCR confirmation of integration, and expected sizes of products are shown. BglII restriction digestion sites and the merAB probe used in the Southern-blot analyses are shown. A fragment of 7.96 kb should be produced after restriction digestion of the transgenic chloroplast genome. B, Transformed E. coli grown in 100 μm HgCl2. i, Transformed E. coli cells containing the vectors pLDR-MerAB; ii, pLDR-MerAB-3′-UTR grown in Luria-Bertani at 100 μm HgCl2; iii, untransformed control (E. coli). C, Effect of mercuric chloride on E. coli cell proliferation. The transgenic clone pLDR-MerAB and pLDR-MerAB-3′-UTR and the control E. coli cells were grown on liquid Luria-Bertani medium with 25 and 50 μm of HgCl2 for 24 h at 37°C. The A600 was measured.
The transformed bacterial cells harboring pLDR-MerAB and pLDR-MerAB-3′-UTR, and the control untransformed cells (E. coli) were grown on Luria-Bertani medium in the presence of different concentrations of mercuric chloride. Bacterial cells containing the pLDR-MerAB and pLDR-MerAB-3′-UTR were able to grow in concentrations of HgCl2 of up to 100 μm on solid agar plates (Fig. 1B). Untransformed E. coli cells were unable to grow even at a concentration of 25 μm. Although transformed cells were able to grow in liquid broth at concentrations of 25 and 50 μm HgCl2, differences in the rate of growth between the clone transformed with the plasmid containing the 3′ terminator and the clone that lacked the terminator region were examined (Fig. 1C). It is known from previous studies that the 3′-UTRs in E. coli are engaged in the termination of transcription. The pLDR-MerAB-3′-UTR was expected to grow better in the presence of Hg because, by terminating effectively, more copies of a shorter transcript containing the merAB operon would be made, in contrast to fewer long transcripts in the case of the pLDR-MerAB clone. The Hg bioassay showed that indeed E. coli cells transformed with the pLDR-MerAB-3′-UTR vector resulted in higher bacterial growth when compared with the bacterial cells containing the vector lacking a 3′ psbA-UTR (Fig. 1C).
Transformation, Selection, and Characterization of Chloroplast Transgenic Plants
Chloroplast-transgenic plants were obtained as described (Daniell, 1997). More than 20 positive independent transgenic lines were obtained with each construct. In this report, we show the results of two transgenic lines that were transformed with the pLDR-MerAB vector and the pLDR-MerAB-3′-UTR, respectively. The variability in expression levels among independent chloroplast-transgenic lines were minimal, as reported previously (Daniell et al., 2001a), and the results shown here correlate well with the results of other transgenic lines with the same chloroplast vectors.
The primer pair 3P and 3M was used to test integration of the transgene cassette into the chloroplast genome at very early stages during the selection process. The 3P primer lands in the native chloroplast genome and the 3M primer lands in the aadA gene that is present within the gene cassette (Fig. 1A). If integration has occurred, a 1.65 kb PCR product should be obtained (Fig. 2A). The untransformed control and the mutants (caused by the spontaneous mutation of the 16S rRNA gene that confers resistance to spectinomycin) did not show any product, confirming that these plants are negative for integration of transgenes (Fig. 2A). The integration of transgenes (aadA, merA, and merB) was further tested by using the 5P/2M primers and PCR analysis. The 5P and 2M primers annealed to the internal region of the aadA and trnA genes, respectively (Fig. 1A). The product size of positive transgenic clones was 3.89 kb, whereas the mutants and untransformed control did not show any PCR product (Fig. 2B). The DNA from full-grown T0 and T1 generation plants was extracted and used for the Southern-blot analysis (Fig. 3). The 0.81 kb flanking sequence probe that hybridizes with the trnI and trnA genes (Fig. 3A) allowed detection of the site-specific integration of the gene cassette into the chloroplast genome. The transformed chloroplast genome digested with BglII restriction enzyme produced a fragment of 7.96 kb (Figs. 1A, and 3, B and C). The untransformed chloroplast genome digested with BglII yielded a 4.47-kb fragment (Fig. 3, A–C).
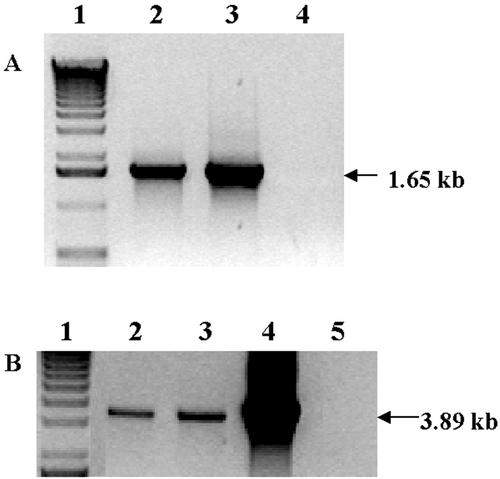
Figure 2.
PCR analysis of control and putative transformants. A, PCR products (1.65 kb) using 3P/3M primers show integration into the chloroplast genome. Lane 1, Marker; lane 2, pLDR-MerAB transgenic line; lane 3, pLDR-MerAB-3′-UTR transgenic line; lane 4, untransformed wild type. B, PCR products (3.8 kb) using 5P/2M primers confirm merAB integration. Lane 1, Marker; lane 2, pLDR-MerAB transgenic line; lane 3, pLDR-MerAB-3′-UTR transgenic line; lane 4, positive control (pLDR-MerAB plasmid DNA); lane 5, untransformed wild-type tobacco.
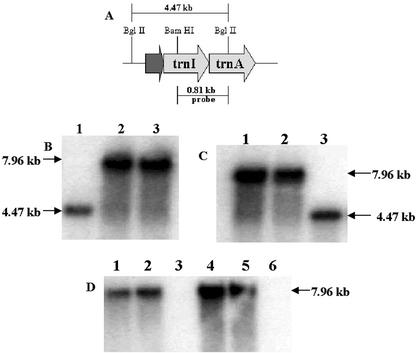
Figure 3.
Southern-blot analysis using the flanking sequence probe and the merAB probe. A, The map shows the wild-type chloroplast genome, restriction digestion sites used for Southern-blot analysis, and the 0.81-kb flanking sequence probe. B, Transgenic lines (T0 generation) for the pLDR-MerAB (lane 2) and the pLDR-MerAB-3′-UTR (lane 3) show the expected size fragment of 7.96 kb; the untransformed control (lane 1) shows the 4.47-kb fragment. C, Lanes 1 and 2, T1 generation transgenic lines; lane 3, the untransformed control. B and C, The flanking sequence probe was used. D, T0 transgenic lines, pLDR-MerAB (lane 1), pLDR-MerAB-3′-UTR (lane 2), and their respective T1 generation transgenic lines (lanes 4 and 5) show the 7.96-kb fragment. Lanes 3 and 6, Untransformed wild type. The merAB probe was used in D.
The flanking sequence probe also showed that homoplasmy of the chloroplast genomes was achieved through the selection process. Southern blots confirmed stable integration of foreign genes into all of the chloroplast genomes confirming homoplasmy. T0 and T1 generation transgenic plants only showed a single fragment of 7.96 kb. The absence of any detectable native untransformed chloroplast genomes not only confirmed homoplasmy, but also facilitated detection of transgene copy numbers in each cell. It is known that mature leaf cells in tobacco contain about 10,000 copies of chloroplast genomes per cell (Bendich, 1987). By virtue of achieving homoplasmy, it is inferred that there are 10,000 copies of transgenes per cell. Southern blots detected with the merAB probe (2.3 kb in size) showed integration of specific genes, merA and merB, as a single fragment of 7.96 kb (Figs. 1A and 3D). The control untransformed tobacco plants and mutants did not show this fragment (Fig. 3D). If the merAB probe would have detected any unexpected size fragments, it might be a nonspecific integration into other plant genomes (nuclear or mitochondria) as discussed elsewhere (Daniell and Parkinson, 2003), but this was not observed. The transgenic plants were fully characterized via PCR and Southern-blot analysis, which showed site-specific integration of the genes into the chloroplast genome and achievement of homoplasmy, even at very early stages of selection (T0). No difference in homoplasmy was detected among plants transformed with the pLDR-MerAB or pLDR-MerAB-3′-UTR vector.
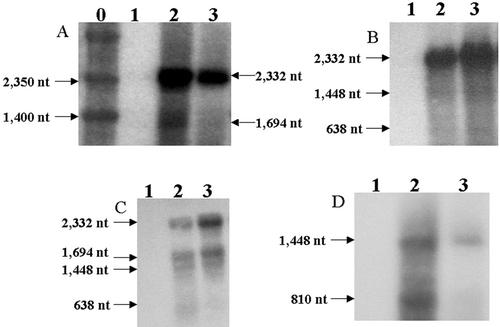
Total RNA from T0 and T1 plants transformed with the pLDR-MerAB-3′-UTR and the pLDR-MerAB was extracted and used to perform the northern-blot analysis with four different probes (the merA, merB, merAB, and aadA probes). The merA probe clearly showed the dicistron containing the merB and merA genes with sizes of 2,332 nucleotides and also a minor transcript for the merA monocistron of 1,694 nucleotides (Fig. 4A). The merB probe showed the merAB dicistron (2,332 nucleotides) plus a less abundant transcript (1,448 nucleotides) containing the aadA and merB genes, and the monocistron corresponding to the merB (638 nucleotides) transcript (Fig. 4B). The merAB probe helped to visualize different transcripts in a single blot, the merB and merA dicistronic transcript (2,332 nucleotides), the merA monocistron (1,694 nucleotides), the aadA and merB dicistron (1,448 nucleotides), and the merB monocistron (638 nucleotides; Fig. 4C). The aadA probe showed transcripts for the dicistron containing the aadA and merB genes and also the aadA monocistron of 810 nucleotides (Fig. 4D). The northern-blot analyses showed that the most abundant transcript is the dicistron (2,332 nucleotides) containing the merA and merB genes. Less abundant transcripts corresponding to the aadA/merB dicistron (1,448 nucleotides), the merA monocistron (1,694 nucleotides), the merB monocistron (638 nucleotides), and to the aadA monocistron (810 nucleotides) were also detected. The high abundance of the merAB dicistron in the pLDR-MerAB or the pLDR-MerAB-3′-UTR plants is an interesting observation. Contrary to the current dogma in the literature, these transcripts were stable even in the absence of a 3′-UTR believed to be required for transcript stability. In addition, there is an indication that processing occurs in between transgenes in transgenic chloroplasts even though no such processing sequences were engineered. Even though all three transgenes are transcribed from a single promoter, no tricistrons containing the aadA, merB, and merA genes were detected. Observed processing between transgenes might be due to recognition of bacterial intergenic sequences by the chloroplast protein synthesis machinery.
Figure 4.
Northern-blot analysis. A, The merA probe: transcripts of merAB dicistron (2,332 nucleotides) and the merA monocistron (1,694 nucleotides) are shown by arrows. B, The merB probe: transcripts for the merAB dicistron (2,332 nucleotides), the aadA/merB dicistron (1,448 nucleotides), and the merB monocistron (638 nucleotides) are shown. C, The merAB probe: transcripts of the merAB dicistron (2,332 nucleotides), the merA monocistron (1,694 nucleotides), the aadA/merB dicistron (1,448 nucleotides), and the merB monocistron (638 bp) are shown. D, The aadA probe: transcripts of the aadA/merB dicistron (1,448 nucleotides) and the aadA monocistron (810 nucleotides) are shown. 0, Marker; 1, wild-type, untransformed; 2, pLDR-MerAB transgenic line; 3, pLDR-MerAB-3′-UTR transgenic line.
Bioassays
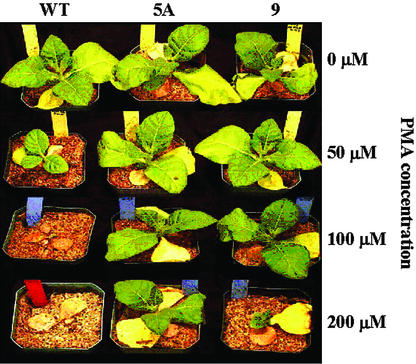
When 16-d-old tobacco plants were grown for 14 d in soil containing PMA concentrations of 0, 50, 100, and 200 μm, the merAB seedlings (pLDR-MerAB and pLDR-MerAB-3′-UTR clones) grew well at PMA concentrations up to 100 μm PMA, and survived the highest PMA concentration of 200 μm (Fig. 5). On the other hand, PMA concentrations of 100 and 200 μm PMA were lethal to wild-type plants, which barely survived 50 μm PMA (Fig. 5). There were no significant differences between transgenic lines with or without the 3′-UTR terminator.
Figure 5.
Effect of PMA concentration on the growth of wild-type and transgenic lines of tobacco plants. Seeds were germinated in vitro on Murashige and Skoog medium (without Suc and 0.5 g mL–1 spectinomycin). Seedling plants (10 d from germination) were transferred to a greenhouse and were grown in soil for 6 d. Plants were then treated by adding 200 mL of 0, 50, 100, and 200 μm PMA supplied in Hoagland nutrient solution. Photographs were taken 14 d after treatment. WT, Negative control cv Petit Havana; 5A, pLDR-MerAB transgenic line; 9, pLDR-MerAB-3′-UTR transgenic line.
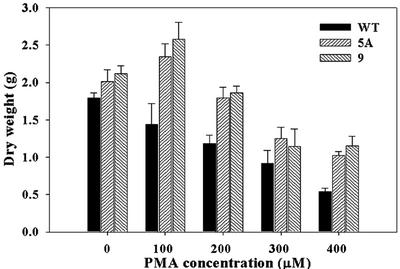
The effect of PMA on plant growth was determined by treating 24-d-old tobacco plants with PMA concentrations of 0, 100, 200, 300, and 400 μm in soil and measuring total plant dry weight at each concentration (Fig. 6). The total dry weight of wild-type plants decreased progressively with each increase in PMA from 0 to 400 μm. On the other hand, in the transgenic plants, there was no decrease in total dry weight with increase in PMA concentration until PMA reached 400 μm. Statistical analysis (unpaired t test) showed that the transgenic lines were substantially more resistant than wild type to concentrations of PMA of 100, 200, and 400 μm (Table I). These results indicate clearly that, compared with the wild type, the insertion of merA and merB into the chloroplast genome substantially increased the resistance of the transgenic plants to the toxic effects of PMA. There was no significant difference between the dry weights of the two clones, pLDR-MerAB and pLDR-MerAB-3′-UTR, at each concentration of PMA tested (Fig. 6).
Figure 6.
Effect of PMA on the total dry weight per plant of 24-d-old wild-type and transgenic tobacco plant lines grown on soil containing 0, 100, 200, 300, and 400 μm PMA for 14 d. WT, Negative control cv Petit Havana; 5A, pLDR-MerAB transgenic line; 9, pLDR-MerAB-3′-UTR transgenic line. se shown, n = 5.
Table I.
Unpaired t test values comparing the differences in dry weight between each transgenic line of tobacco versus wild type
5A: pLDR-MerAB transgenic line; 9: pLDR-MerAB-3′UTR transgenic line. An asterisk indicates significance at P < 0.05; a double asterisk indicates significance at P < 0.001.
| PMA Concentration
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control
|
100
|
200
|
300
|
400
|
||||||
| 5A | 9 | 5A | 9 | 5A | 9 | 5A | 9 | 5A | 9 | |
| μM | ||||||||||
| Dry weight | 1.31 | 2.60* | 2.77* | 3.19** | 3.38** | 4.62** | 1.41 | 0.79 | 6.67** | 4.72** |
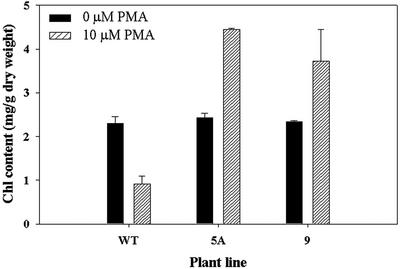
As discussed in the Introduction, previous research has shown that the main site of damage of organomercurial compounds is the chloroplast, and that chlorophyll synthesis, electron transport, and photosynthesis are all seriously affected. Therefore, the overexpression of merA and merB in the chloroplast should reduce the toxic effects of PMA directly on chloroplast function. To test this idea, we treated 15-mm diameter leaf discs excised from wild-type and transgenic plants with 10 μm PMA for 10 d and measured chlorophyll contents (Fig. 7). The results show that without PMA present, chlorophyll concentration did not differ between wild-type and the two transgenic lines. Surprisingly, when PMA was supplied to the leaf discs, the chlorophyll content was markedly increased in the transgenic lines, whereas in the wild type, chlorophyll content was reduced. These results are consistent with the view that PMA exerts a damaging effect on the chloroplasts of wild-type plants as expected, reducing chlorophyll content substantially, and that overexpression of merA and merB in the chloroplast genome appears to increase chloroplast resistance to PMA toxicity. However, because the overexpression of these genes results in an increase in chlorophyll content of the transgenic chloroplasts, it would appear that PMA could in fact stimulate chlorophyll synthesis in some way in these transgenic plants. In this regard, it is of interest that the leaf discs taken from the transgenic plants increased in size over the 10-d experimental period, whereas discs from the wild type decreased in size. Thus, it is possible that the increase in chlorophyll concentration with PMA in the transgenic plants was associated with an increase in chloroplast number and/or size.
Figure 7.
Effect of PMA on total chlorophyll content (milligrams per gram of dry weight) of 15-mm diameter leaf discs excised from wild-type and transgenic lines of tobacco and treated with 0 and 10 μm PMA for 6 d. WT, Negative control cv Petit Havana; 5A, pLDR-MerAB transgenic line; 9, pLDR-MerAB-3′-UTR transgenic line. se shown, n = 5.
Levels of transgene expression in chloroplasts could be further enhanced by introducing appropriate UTRs instead of the ribosome-binding site (RBS) used in the present study. For example, we have recently shown that use of the psbA 5′-UTR instead of RBS resulted in a 500-fold increase in the expression of human serum albumin in transgenic chloroplasts (Fernandez-San Millan et al., 2003). The most significant advantage is the ability to introduce the mer operon in a single transformation event in contrast to nuclear transgenic plants that required introduction of single genes followed by time consuming back-crosses to reconstitute the entire pathway. In addition, prokaryotic genes do not require codon optimization when expressed in transgenic chloroplasts (Kota et al., 1999; De Cosa et al., 2001).
This is the first report on the use of chloroplast transformation using multigene engineering for the phytoremediation of toxic compounds. Because of the containment of transgenes and high levels of expression via chloroplast genomes, the chloroplast transformation approach is highly suitable for phytoremediation, especially for toxic agents that affect chloroplast function. Although 3′-UTR is believed to stabilize chloroplast transcripts and to be essential for transgene expression, it may not be necessary for transcript stability in the context of a polycistron. Because there are more than 60 such polycistrons within the chloroplast genome (Sugita and Sugiura, 1996), this is a significant observation.
MATERIALS AND METHODS
Bacterial Plasmids That Contain Organomercurial and Hg Resistance Genes
Host Escherichia coli cells containing plasmids NR1 and R831b were kindly provided by Dr. Ann Summers (University of Georgia, Athens). These plasmids contain the mer operon with the complete and functional merA and merB genes, respectively (Jackson and Summers, 1982; Rinderle et al., 1983; Ogawa et al., 1984; Begley et al., 1986). Each of these plasmids confers resistance to at least one antibiotic that can be used as a selectable marker. Host bacterial containing plasmid NR1 was grown on solid Luria-Bertani media containing 100 μg mL–1 tetracycline; E. coli cells containing the plasmid R831b was cultured on solid Luria-Bertani media containing 12.5 μg mL–1 kanamycin and were grown overnight at 37°C.
Chloroplast Vector Constructions
To amplify the merB gene from the native plasmid, a primer pair was designed to have a PstI restriction site followed by a chloroplast and bacterial functional RBS of sequence GGAGG in the 5′ primer, followed by a four-nucleotide spacer region upstream of the start codon. This primer had 20-nucleotide homology with the 5′ end of the gene and a total of 35 nucleotides. The 3′ primer was designed to have 20-nucleotide homology with the 3′ end of the gene and a ClaI restriction site. To amplify the merA gene from the native plasmid, a 5′ primer was designed to have a ClaI restriction site followed by the RBS sequence and a four-nucleotide spacer region before the start codon and the 20-nucleotide homology with the merA gene. All primer pairs were designed using the QUICKPRI program of the DNASTAR software. Two PCR reactions were done to amplify the merA and the merB genes individually from the plasmid NR1 that contained the complete and functional merA gene and the plasmid R831b that contained the full-length merB gene. The PCR products were cloned into suitable plasmid vectors.
pLDR-MerAB-3′-UTR Vector Construction
The functional merAB operon was amplified via PCR from the vector pCR2.1-MerAB and a new set of primers was made. The 5′ primer was designed to have an EcoRV site, an RBS, a spacer region of four nucleotides (attt) and 20 bases of homology to the merAB operon starting at the start codon (atg). The 3′ primer is a simple primer with 20 bases of homology to the 3′ end of the operon. After cloning, correct orientation was verified by restriction analyses.
Hg Resistance Bioassay in Bacteria
The bacterial clones pLDR-MerAB, pLDR-MerAB-3′-UTR, and the control E. coli XL1-blue cells were grown for 24 h at 37°C in 50 mL of Luria-Bertani broth with concentrations of HgCl2 of 0, 25, and 50 μm. The growth medium was autoclaved and cooled to 40°C before adding HgCl2, and was mixed thoroughly to provide an even concentration throughout the plate or growth medium. The bacterial clones pLDR-MerAB, pLDR-MerAB-3′-UTR, and the untransformed control E. coli cells were plated in solid Luria-Bertani medium containing HgCl2 concentrations of 0, 50, 100, and 500 μm. Plates were incubated for 24 h at 37°C.
Bombardment and Selection of Transgenic Plants
The steps involved in the gene delivery by particle bombardment and the selection process of the transgenic tobacco (Nicotiana tabacum var Petit Havana) clones were performed essentially as describe by Daniell (1997). Tobacco leaves were bombarded using a biolostic device (PDS-1000/He; Bio-Rad, Hercules, CA). After bombardment, leaves were placed on Regeneration Medium of Plants medium containing 500 μg mL–1 spectinomycin for two rounds of selection on plates and subsequently moved to jars on Murashige Skoog medium containing 500 μg mL–1 spectinomycin.
Confirmation of Chloroplast Integration by PCR
Plant DNA was isolated using the DNeasy Plant Mini kit (Qiagen, Valencia, CA). The PCR primer pairs 3P-3M and 5P-2M were used to confirm the integration of the gene cassette into the chloroplast and the presence of the genes of interest, respectively, essentially as described elsewhere (Guda et al., 2000). PCR analysis was performed using the Gene Amp PCR System 2400 (Perkin Elmer, Chicago).
Southern-Blot Analysis
The total plant DNA was obtained from transgenic T0 and T1 plants as well as from untransformed tobacco plants following the protocol previously explained (Daniell et al., 2001a,b). The plant DNA was digested with BglII and was separated on a 0.8% (w/v) agarose gel at 50 V for 2 h. The gel was soaked in 0.25 n HCl for 15 min and was then rinsed two times with water. The gel was then soaked in transfer buffer (0.4 n NaOH and 1 m NaCl) for 20 min and transferred overnight to a nitrocellulose membrane. The membrane was rinsed twice in 2× SSC (0.3 m NaCl and 0.03 m sodium citrate), dried on filter paper, and then cross-linked in the GS GeneLinker (Bio-Rad). The flanking sequence probe was obtained by BglII/BamHI digestion of the plasmid pUC-ct that contains the chloroplast-flanking sequences (trnI and trnA genes). The merAB probe was obtained by EcoRI digestion of plasmid pCR2.1-MerAB. Probes were labeled with 32P using Ready Mix and were purified by using Quant G-50 microcolumns (Amersham, Arlington Heights, IL), followed by radioisotope incorporation. The probe was quantified by using a scintillation counter (LS 5000TD; Beckman Instruments, Fullerton, CA). Prehybridization and hybridization were done using the Quick-Hyb solution (Stratagene, La Jolla, CA). The membrane was washed twice in 2× SSC with 0.1% (w/v) SDS for 15 min at room temperature, followed by two additional washes in 0.1× SSC with 0.1% (w/v) SDS for 15 min at 60°C (to increase the stringency). Blots were exposed to x-ray films and were developed in a SRX-101A (Konica, Tokyo).
Northern-Blot Analysis
The RNeasy Mini kit and protocol was used to isolate total RNA from plant tissues (Qiagen). The merA, merB, aadA, and merAB probes were used to probe different RNA blots. The merA probe was made by cutting out the merA gene from the pCR2.1-MerA vector with EcoRI. The merB probe was made by cutting out the merB gene from the pCR2.1-MerB vector with EcoRI. The aadA probe was amplified by PCR from the pLD-ctv vector with a specific primer pair (5′-ccatggcagaagcggtaatcg/3′-aagatttatttgccgactacctt). The merAB probe was made digesting the pCR2.1-MerAB vector with EcoRI. Restriction fragments were cut out and eluted from the gels. The probe-labeling reaction, prehybridization/hybridization steps, membrane washing step, and autoradiography were performed as explained in the Southern-blot section in “Materials and Methods.”
PMA Treatments
Seeds of wild-type tobacco and two transgenic lines (pLDR-MerAB and pLDR-MerAB-3′-UTR) were surface-sterilized in 7% (w/v) sodium hypochlorite containing 0.1% (v/v) Tween 20. Seeds were kept on a rocking platform for 20 min and were rinsed in sterile distilled water at least three times. Sterilized seeds were transferred to plates containing one-half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) with 0.5 mg mL–1 spectinomycin and 0.3% (w/v) phytoagar, pH 5.7. Plates were incubated in the dark at 4°C for 3 d, and were then maintained in a controlled growth chamber at a temperature of 22°C to 24°C, relative humidity of 75% to 90%, and a photon flux density of 750 μE m–2 supplied over a 16-h daylength. After germination (approximately 10 d), seedlings were transferred to soil (sand:Davis Mix, 50:50) in the greenhouse at 22°C using a 16-h photoperiod. Five replicate pots each contained a single seedling, wild-type or transgenic plant. All pots were watered twice a week with one-half-strength Hoagland solution.
Effect of PMA on Seedling Germination
To determine the inhibitory concentration of PMA on seedling germination, three different concentrations of PMA were applied to pots containing 16-d-old plants from wild-type and two transgenic lines in three replicates. PMA stock solutions were prepared as 10 mm in dimethyl sulfoxide. Different PMA concentrations (50–200 μm) were added to each pot in 100 mL of one-half-strength Hoagland solution. Control pots received the same volume of Hoagland solution without PMA. All plants were grown in the greenhouse under the same conditions as described above.
Effect of PMA on Potted Plants
Pots of five replicates representing the wild-type and the two transgenic lines (of approximately the same size) were transferred to Poly Vinyl Chloride plastic trays 3 inches high. Different concentrations of PMA (in micromoles) were prepared (100, 200, 300, and 400) using a stock solution of one-half-strength Hoagland solution. For each treatment, a single tray maintained approximately 200 mL (to about one-half of the pot's height) of the PMA-Hoagland's solution. All plants in the same treatment were exposed to exactly the same concentration of PMA. The control tray was filled with one-half-strength Hoagland solution without metal. After about 14 d, plants were harvested, washed thoroughly with distilled water, and the length of the longest root and shoot of the plants were measured. Shoots and roots were separated and dry weights were determined.
Determination of Chlorophyll Content in Leaf Discs Treated with PMA
Leaf discs were cut out with a cork-borer (15-mm diameter) from the youngest and fully expanded leaves on 3-week-old plants grown in the soil with no PMA. Discs of wild-type and different transgenic plants were placed in petri dishes containing solidified Murashige and Skoog medium (pH 5.7 with no Suc) supplemented with different concentrations of PMA ranging from 0.1 to 1 μm, 10 to 100 μm, and 200 to 500 μm. Plates with no PMA were used as controls. The effect of Hg stress was assessed by the loss of chlorophyll in leaf discs. Leaf discs were collected after 6 d of exposure to PMA. They were immediately extracted in 80% (v/v) chilled acetone for determination of total chlorophyll content following the protocol from Current Protocols in Food Analytical Chemistry Online (http://www.mrw2.interscience.wiley.com).
Acknowledgments
We thank Dr. Ann Summers (University of Georgia) for providing bacterial strains used in this study.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.020958.
This work was supported in part by funding from Chlorogen Inc. (St. Louis).
References
- Begley TP, Walts AE, Walsh CT (1986) Bacterial organomercurial lyase: overproduction, isolation, and characterization. Biochemistry 25: 7186–7192 [DOI] [PubMed] [Google Scholar]
- Bendich AJ (1987) Why do chloroplasts and mitochondria contain so many copies of their genome? Bioassays 6: 279–282 [DOI] [PubMed] [Google Scholar]
- Bernier M, Popovic R, Carpentier R (1993) Mercury inhibition of photosystem II. FEBS Lett 32: 19–23 [DOI] [PubMed] [Google Scholar]
- Bizily S, Rugh CC, Meagher RB (2000) Phytoremediation of hazardous organomercurials by genetically engineered plants. Nat Biotechnol 18: 213–217 [DOI] [PubMed] [Google Scholar]
- Bizily S, Rugh CC, Summers AO, Meagher RB (1999) Phytoremediation of methylmercury pollution: merB expression in Arabidopsis thaliana plants confer resistance to organomercurial Proc Natl Acad Sci USA 96: 6808–6813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H (1997) Transformation and foreign gene expression in plants mediated by microprojectile bombardment. Methods Mol Biol 62: 453–488 [DOI] [PubMed] [Google Scholar]
- Daniell H (2002) Molecular strategies for gene containment in transgenic crops. Nat Biotechnol 20: 581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H (2003) Medical molecular pharming: expression of antibodies biopharmaceuticals and edible vaccines via the chloroplast genome. In K Vasil, ed, Plant Biotechnology 2002 and Beyond. Kluwer Academic Publisher, Dordrecht, The Netherlands, pp 371–376
- Daniell H, Datta R, Varma S, Gray S, Lee SB (1998) Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat Biotechnol 16: 345–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Dhingra A (2002) Multigene engineering: dawn of an exciting new era in biotechnology. Curr Opin Biotechnol 13: 136–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Khan MS, Allison L (2002) Milestones in chloroplast genetic engineering: an environmental friendly era in biotechnology. Trends Plant Sci 7: 84–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Panchal T, Weibe PO (2001a) Expression of the native cholera toxin B subunit gene and assembly as functional oligomers in transgenic chloroplasts. J Mol Biol 311: 1001–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Muthukumar B, Lee SB (2001b) Marker free transgenic plants: engineering the chloroplast genome without the use of antibiotic selection. Curr Genet 39: 109–116 [DOI] [PubMed] [Google Scholar]
- Daniell H, Parkinson L (2003) Jumping genes and containment. Nat Biotechnol 21: 374–375 [DOI] [PubMed] [Google Scholar]
- Dean JG, Bosqui FL, Lanouette VH (1972) Removing heavy metals from waste water. Environ Sci Technol 6: 518–522 [Google Scholar]
- De Cosa B, Moar W, Lee SB, Miller M, Daniell H (2001) Hyper-expression of the Bt Cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat Biotechnol 19: 71–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gray G, Rajasekaran K, Smith F, Sanford J, Daniell H (2001) Expression of an antimicrobial peptide via chloroplast genome control phytopathogenic bacteria and fungi. Plant Physiol 127: 852–862 [PMC free article] [PubMed] [Google Scholar]
- Doucleff M, Terry N (2002) Pumping out the arsenic. Nat Biotechnol 20: 1094–1095 [DOI] [PubMed] [Google Scholar]
- Fernandez-San Millan A, Mingo-Castel A, Miller M, Daniell H (2003) A chloroplast transgenic approach to hyper-express and purify human serum albumin, a protein highly susceptible to proteolytic degradation. Plant Biotechnol J 1: 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guda C, Lee SB, Daniell H (2000) Stable expression of biodegradable protein base polymer in tobacco chloroplasts. Plant Cell Rep 19: 257–262 [DOI] [PubMed] [Google Scholar]
- Heaton ACP, Rugh CL, Wang NJ, Meagher RB (1998) Phytoremediation of mercury and methylmercury polluted soils using genetically engineered plants. J Soil Contam 7: 497–509 [Google Scholar]
- Jackson WJ, Summers AO (1982) Biochemical characterization of HgCl2-inducible polypeptides encoded by the mer operon of plasmid R100. J Bacteriol 151: 962–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärenlampi S, Schat H, Vangronsveld J, Verkleij JAC, Lelie D, Mergeay M, Tervahauta AI (2000) Genetic engineering in the improvement of plants for phytoremediation of metal polluted soils. Environ Pollut 107: 225–231 [DOI] [PubMed] [Google Scholar]
- Kota M, Daniell H, Varma S, Garczynski F, Gould F, Moar WJ (1999) Overexpression of the Bacillus thuringiensis Cry2A protein in chloroplast confers resistance to plants against susceptible and Bt-resistant insects. Proc Natl Acad Sci USA 96: 1840–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer U, Chardonnens AN (2001) The use of transgenic plants in the bioremediation of soil contaminated with trace elements. Appl Microbiol Biotechnol 55: 661–672 [DOI] [PubMed] [Google Scholar]
- Kupper H, Kupper F, Spiller M (1996) Environmental relevance of heavy metal substituted chlorophylls using the example of water plants. J Exp Bot 47: 259–266 [Google Scholar]
- Lee SB, Kwon S, Park S, Jeong M, Han S, Byun M, Daniell H (2003) Accumulation of trehalose within transgenic chloroplasts confers drought tolerance. Mol Breed 11: 1–13 [Google Scholar]
- Lin ZQ, Hansen D, Zayed AM, Terry N (1995) Biological selenium volatilization: method of measurement under field conditions. J Environ Qualt 28(1): 309–315 [Google Scholar]
- McBride KE, Svab Z, Schaaf DJ, Hogen PS, Stalker DM, Maliga P (1995) Amplification of a chimeric Bacillus genes in chloroplasts leads to an extraordinary level of an insecticidal protein in tobacco. BioTechnology 13: 362–365 [DOI] [PubMed] [Google Scholar]
- Meagher RB (2000) Phytoremediation of toxic elemental and organic pollutants. Curr Opin Plant Biol 3: 153–162 [DOI] [PubMed] [Google Scholar]
- Meagher RB, Rugh CL (1997) Phytoremediation of heavy metal pollution: ionic and methyl mercury. In Organization for Economic Cooperation and Development Document: Biotechnology for Water Use and Conservation. The Mexico'96 Workshop, Organization for Economic Cooperation and Development, Paris, pp 305–321
- Murashige T, Skoog F (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Planta 15: 473–497 [Google Scholar]
- Nies DH (1999) Microbial heavy metal resistance. Appl Microbiol Biotechnol 51: 730–750 [DOI] [PubMed] [Google Scholar]
- Ogawa HI, Tolle CL, Summers AO (1984) Physical and genetic map of the organomercury resistance (Omr) and inorganic mercury resistance (Hgr) loci of the IncM plasmid R831b. Gene 32: 311–320 [DOI] [PubMed] [Google Scholar]
- Patra M, Sharma A (2000) Mercury toxicity in plants. Bot Rev 66: 379–422 [Google Scholar]
- Prasad DDK, Prasad ARK (1987) Altered o-aminolevulinic acid metabolism by lead and mercury in germination seedlings of bajra (Pennisetum typoideum). J Plant Physiol 127: 241–249 [Google Scholar]
- Rinderle SJ, Booth JE, Williams JW (1983) Mercuric reductase from R-plasmid NR1: characterization and mechanistic study. Biochemistry 22: 869–876 [DOI] [PubMed] [Google Scholar]
- Rugh CL, Senecoff JF, Meagher RB, Merkle SA (1998) Development of transgenic yellow poplar for mercury phytoremediation. Nat Biotechnol 16: 925–928 [DOI] [PubMed] [Google Scholar]
- Rugh CL, Wilde HD, Stack NM, Thompson DM, Summers AO, Meagher RB (1996) Mercuric ion reduction and resistance in transgenic Arabidopsis thaliana plants expressing a modified bacterial merA gene. Proc Natl Acad Sci USA 93: 3182–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt DE, Smith RD, Raskin I (1995) Phytoremediation: a novel strategy for the removal of toxic metals from the environment using plants. BioTechnology 13: 468–474 [DOI] [PubMed] [Google Scholar]
- Sen AK, Mondal NG (1987) Salvia natans as the scavenger of Hg (II). Water Air Soil Pollut 34: 439–446 [Google Scholar]
- Sinha S, Gupta M, Chandra P (1996) Bioaccumulation and biochemical effects of mercury in the plant Bacopa monnieri (L). Environ. Toxicol Water Qualt 11: 105–112 [Google Scholar]
- Staub JM, Garcia B, Graves J, Hajdukiewicz PTJ, Hunter P, Nehra N, Paradkar V, Schlittler M, Carol JA, Spatola L et al. (2000) High-yield production of human therapeutic protein in tobacco chloroplast. Nat Biotechnol 18: 333–338 [DOI] [PubMed] [Google Scholar]
- Sugita M, Sugiura M (1996) Regulation of genes expression in chloroplast of higher plants. Plant Mol Biol 32: 315–326 [DOI] [PubMed] [Google Scholar]
- Terry N, Zayed AM, De Souza MP, Tarun AS (2000) Selenium in higher plants. Annu Rev Plant Physiol Plant Mol Biol 51: 401–432 [DOI] [PubMed] [Google Scholar]
- U.S. Environmental Protection Agency (1984) Health effects assessment of mercury. Environmental Criteria and Assessment Office, Cincinnati, OH